Abstract
Members of the AP2 family of transcription factors, such as BABY BOOM (BBM), play important roles in cell proliferation and embryogenesis in Arabidopsis thaliana (AtBBM) and Brassica napus (BnBBM) but how this occurs is not understood. We have isolated three AP2 genes (GmBBM1, GmAIL5, GmPLT2) from somatic embryo cultures of soybean, Glycine max (L.) Merr, and discovered GmBBM1 to be homologous to AtBBM and BnBBM. GmAIL5 and GmPLT2 were homologous to Arabidopsis AINTEGUMENTA-like5 (AIL5) and PLETHORA2 (PLT2), respectively. Constitutive expression of GmBBM1 in Arabidopsis induced somatic embryos on vegetative organs and other pleiotropic effects on post-germinative vegetative organ development. Sequence comparisons of BBM orthologues revealed the presence of ten sequence motifs outside of the AP2 DNA-binding domains. One of the motifs, bbm-1, was specific to the BBM-like genes. Deletion and domain swap analyses revealed that bbm-1 was important for somatic embryogenesis and acted cooperatively with at least one other motif, euANT2, in the regulation of somatic embryogenesis and embryo development in transgenic Arabidopsis. The results provide new insights into the mechanisms by which BBM governs embryogenesis.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-010-9674-8) contains supplementary material, which is available to authorized users.
Keywords: AP2/ANT, Arabidopsis, BABYBOOM, Embryogenesis, Soybean, Transcription factor
Introduction
BABY BOOM (BBM) is a member of the AP2 family of transcription factors which have diverse functions in plant development (Nole-Wilson et al. 2005; Floyd and Bowman 2007; Feng et al. 2005). The AP2 family belongs to the AP2/ERF superfamily. This is one of the largest groups of plant transcription factors and has undergone extensive duplication and domain shuffling during its evolution (Riechmann et al. 2000; Kim et al. 2006; Nakano et al. 2006). Members have double AP2/ERF domains in the AP2 family, single AP2/ERF DNA-binding domains in the ERF family and single AP2/ERF domains together with a B3 DNA-binding domain in the RAV family (Riechmann and Meyerowitz 1998). The superfamily consists of 147 members in Arabidopsis, 157 members in rice (Nakano et al. 2006) and 148 members in soybean (Zhang et al. 2008). Members function in diverse processes fundamental to plant growth, reproduction and environmental interactions (Riechmann and Meyerowitz 1998; Feng et al. 2005; Nole-Wilson et al. 2005). In Arabidopsis, rice and soybean (Sakuma et al. 2002; Gong et al. 2004; Nakano et al. 2006; Zhang et al. 2008) the members can be grouped by sequence similarity into the same family and subfamily groupings (Floyd and Bowman 2007). BBM clusters within one of the sublineages, euANT, which appears to have specialized in meristem differentiation and maintenance (Floyd and Bowman 2007).
Functional studies of the AP2 family members, such as APETALA2 (AP2), AINTEGUMENTA (ANT), BABY BOOM (BBM), PLETHORA1 (PLT1), PLETHORA2 (PLT2) and the AINTEGUMENTA-like (AIL) genes, have revealed diverse transcriptional networks and developmental processes that the family controls as well as redundancies that exists among and within the different groups. APETALA 2 (AP2), the first member of the family to be reported, functions independently in the specification of floral organ identity (Jofuku et al. 1994; Okamuro et al. 1997, Maes 1999) and in the maintenance of the stem cell niche of the shoot meristem (Würschum et al. 2006). AINTEGUMENTA (ANT) is required for ovule development and floral organ growth (Elliott et al. 1996, Klucher et al. 1996). ANT can act redundantly with AP2 in floral development (Krizek et al. 2000). BABY BOOM (BBM) has been implicated in the differentiation of embryonal stem cells from somatic cells (Boutilier et al. 2002) and clusters within the same clade as PLETHORA (PLT1 and PLT2) which controls root stem cell identity and maintenance (Aida et al. 2004). PLT1, PLT2, BBM and PLT3/AIL6 function redundantly in root meristem and embryo differentiation (Galinha et al. 2007). They are also closely related to a number of other AINTEGUMENTA-like (AIL) genes (Nole-Wilson et al. 2005; Tsuwamoto et al. 2010) which are generally involved in the specification of meristems or division-competent states (Nole-Wilson et al. 2005).
Members of the AP2 family share two highly-conserved AP2 DNA-binding domain repeats separated by a linker region; however, the N-terminal and C-terminal sequences are very distinct. The domains within these regions have not been studied but they are likely to be important for the specific transcriptional activities, protein interactions and nuclear localizations that confer the unique functions associated with each member (Nakano et al. 2006). Sequence comparisons of the AP2/ERF superfamily members from soybean, Arabidopsis and rice have revealed the presence of many conserved motifs outside of the AP2/ERF DNA binding domain raising the possibility that shared conserved motifs may form the basis for functional similarities among different groups (Zhang et al. 2008).
BABY BOOM (BnBBM) was cloned from Brassica napus microspore embryo cultures and was shown to induce somatic embryos when ectopically overexpressed in Arabidopsis or B. napus (Boutilier et al. 2002). The acquisition of totipotency through this process was accompanied by a number of pleiotropic effects on plant development (Boutilier et al. 2002). In transgenic tobacco, BnBBM expression induced pleiotropic effects on vegetative growth and development but did not induce embryogenesis (Srinivasan et al. 2007) indicating that embryogenic pathways differ among species or that the domains within BBM that govern embryogenesis have diverged in sequence among plants and were not recognized. It is currently believed that BnBBM enhances cell proliferations that can result in different developmental outcomes including organogenesis or embryogenesis (Srinivasan et al. 2007). The variety of different pleiotropic effects on plant development that were observed with ectopically-expressed BnBBM may indicate broad redundancies among AP2 family members beyond those already demonstrated with the PLT-like and BBM genes (Galinha et al. 2007) or the AP2 and ANT genes (Krizek et al. 2000).
In this study we examined BBM orthologues from a non-cruciferous species, soybean, to determine if the same developmental pathways were induced in transgenic plants; to identify conserved motifs in BBM-like genes; and to locate the determinants of embryogenesis. GmBBM1 was identified as the functional orthologue of AtBBM and BnBBM through both structural and functional studies. The modular domain structure of BBM-like genes was also analyzed and revealed motifs that were important for their specificity in the induction of somatic embryogenesis.
Materials and methods
Plant material
Embryos were isolated from soybean, Glycine max (L.) Merril genotype X5, formerly called X2650-7-2-3 (Simmonds and Donaldson 2000). Donor plants were grown under a 16 h photoperiod in the greenhouse as previously reported (Simmonds and Donaldson 2000). Embryogenic cultures were established as previously described (Finer 1988; Finer and Nagasawa 1988). Immature cotyledons, 4 and 5.9 mm in size, were placed abaxial side-down on MSD40 medium (Finer and Nagasawa 1988) containing MS salts, B5 vitamins, 6%(w:v) sucrose, 40 mg/l 2,4-D and 0.2% Gelrite, pH 5.8 and cultured at 27°C under a photoperiod of 16 h with a light intensity of 60–70 µmol m−2 s−1 provided by cool-white fluorescent lamps. Secondary globular embryos developing on the cultured cotyledons were transferred to 125 ml Erlenmeyer flasks with 30 ml of 10A40N medium (Finer and Nagasawa 1988) containing modified MS salts (with MS nitrogen replaced by 10 mM NH4NO3 and 30 mM KNO3), B5 vitamins, 6% sucrose, 5 mg/l 2,4-D, and 5 mM asparagine, pH 5.8 and cultured as above at 30 µmol m−2 s−1 on a rotary shaker at 125 rpm. Embryogenic tissue (30–75 mg) was subcultured to fresh medium every 2–4 weeks to maintain proliferation. Proliferation was stopped and embryo maturation was initiated by culturing embryogenic tissue for 4 weeks on solid OMSM6 medium without charcoal under a light intensity of 70 µmol m−2 s−1. Embryos were desiccated in 85% relative humidity and germinated on B5 medium with 3% sucrose and 0.6% Phytagar or 0.2% Gelrite, pH 5.8. Plants with two trifoliate leaves were transplanted to soil and grown under a 16-h photoperiod prior to transfer to a 12-h photoperiod for seed formation.
Root tissues were harvested from 1-week old seedlings. Leaf and stem tissues were from 3-weeks old seedlings. Flowers were harvested 2 days after full anthesis. Developing seeds and pods were harvested from different stages between 6 and 24 days post anthesis (dpa) at 3 days intervals. To study the expression of GmBBM1 genes in developing seeds tissues from seed coat, embryo and pods were collected separately and RNA was extracted using the RNeasy Kit (Qiagen, Canada). Tissues from soybean roots, leaves, stems and flowers were also collected, homogenized in liquid nitrogen and total RNA was extracted using Trizol (Invitrogen, Canada).
RT-PCR
Probes generated by RT-PCR were used for library screening and expression profiling. Primers specific to each of the three soybean genes, GmBBM1, GmPLT2 and GmAIL5, are shown in Supplementary Table 1. The SoyTub2 gene was used as positive control. PCR conditions for GmPLT2 and GmAIL5 were as follows: 3 min at 95°C; 30 cycles [1 min 95°C, 1 min specific annealing temperature (Supplementary Table 1), 1 min 72°C] and 5 min extension at 72°C. PCR conditions for GmBBM1 and SoyTub2 were as follows: 31 min at 50°C, 15 min at 95°C; 30 cycles (30 s 94°C, 30 s specific annealing temperature (Supplementary Table 1), 30 s 72°C) and 2 min extension at 72°C.
Library construction
Total RNA (2 mg/g fresh tissue) was extracted from cultured embryogenic soybean suspension cultures and mRNA (0.2% of total RNA) was purified using the Quik mRNA Isolation Kit (Stratagene/VWR, Canada). A cDNA library was constructed using Lambda-ZAPII cDNA synthesis kit (Stratagene/VWR, Canada). Phages (1.6 × 106pfu) were plated according to the manufacturer’s instructions. The amplified cDNA library titre was 2.5 × 109pfu. The cDNA library phage plaques were hybridized using a probe containing the AP2 domain and linker using primers designed from the alignment of EST AW200688 coding for a soybean AP2 protein and BnBBM1 from Brassica napus as described in Results. cDNA were excised from the lambda-ZAPII vectors following the conversion of the lambda-ZAP clone plasmids pBluescript SK DNA. Sequences were determined with the ABI PRISM TM dye terminator cycle sequencing kit (PE Applied Biosystems). Data was analyzed using DNAsis (Hitachi Corporation 2003) Genejumper primer insertion kit for sequencing version B (Invitrogen, Canada) was used in order to be able to sequence the full-length clones.
Arabidopsis transformation and assessment
Plant transformations were carried out according to Clough and Bent (1998). GmBBM1 was cloned into the pBINPLUS vector and expression was driven by the double 35S promoter and alfalfa mosaic virus (AMV) translational enhancer as described in Boutilier et al. (2002). Selection was performed on 50 mg/l kanamycin. Thousands of seeds were screened until a collection of approximately 35 lines with somatic embryos were recovered. Of these, six representative lines were studied in greater detail over successive generations.
BnBBM1 mutants were generated by inverse PCR reactions on the BnBBM1 PUC19 plasmid, using Expand Long Template PCR System (Roche) and synthetic primers with 10-bp sequence including a BamHI or XhoI sites in their 5′-ends that replaces the original sequences. The following primers were used for euANT2 (N1BamHIF 5′-CGGGATCCCTGAGAAATCAACCCGTGGATAATG-3′ and N1BamHIR5′-CTGGATCCGCCACCACCACCGTCTCCTCCTC-3′) and for bbm-1 (N2XhoIF5′-CGGCTCGAGCCTTATGAACAAAATCACCATCG-3′ and N2XhoIR5′-CCGCTCGAGGGTGGAAGTATTTGAAAGAAAT-3′). These constructs were obtained by KpnI/SalI digestion and cloned into the binary vector pCAMBIA1300, along with the wild-type BnBBM1 gene as the control. They were introduced into Agrobacterium tumefaciens GV3101 for plant transformation and selection was performed with 30 mg/L hygromycin. Approximately, 3,000–4,000 seeds in total were screened in triplicate experiments to assess the phenotypes of the transgenic lines.
For the domain swap analysis, the PLT2, AIL5, and AIL7 genes from Arabidopsis were synthesized with the BnBBM1 bbm-1 motif (SLGLSMIKTWLRNQP) as shown in Fig. 2A by GenScript Corporation (Piscataway, NJ). They were cloned into pCAMBIA1300 under control of the double 35S promoter. As above the constructs were introduced into Agrobacterium tumefaciens GV3103 for plant transformation and screened.
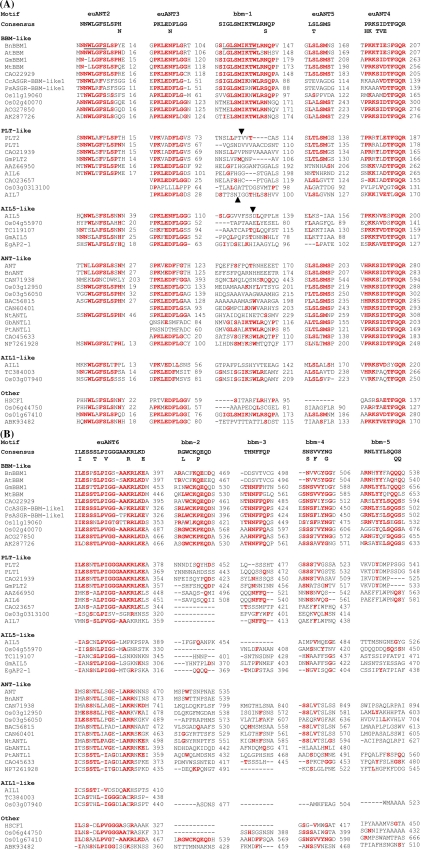
Fig. 2.
Motifs in the N-terminal (A) or C-terminal (B) sequences of the BBM-like genes and their conservation in the euANT subgroup of the AP2 family. Residues that match the consensus sequence are in red. Sequences are grouped into clades as indicated in the phylogenetic tree. The sequences of the euANT2 and bbm-1 motifs in the BnBBM1 sequence that were deleted to test their function are underlined. For the domain swap analysis, the site of insertion of the bbm-1 motif from BnBBM1 (SLGLSMIKTWLRNQP) in PLT2, AIL5, and AIL7 are indicated with arrowheads
Seeds from each transgenic seed line were sterilized in a solution of 70% ethanol for 30 s followed by a solution of 25% javex plus 0.05% Triton for 20–30 min. The seeds were shaken for the duration. They were then rinsed 4 times with sterile distilled water and placed in a 0.1% solution of agarose at 4°C overnight. The next day, the seeds were plated on ½ MSB5 media (Sigma Chemical Co) with 3% sucrose and 0.6% agarose along with selectable agents: 50 mg/l kanamycin for pBINPLUS or 30 mg/l hygromycin for pCAMBIA1300. Seedlings were then surveyed for phenotype over a 2–3 week period. At 3–4 weeks growth they were transferred to soil and brought to flowering stage.
Phylogenetic analysis
Sequences of double AP2 domain proteins similar to AtBBM were mined from GenBank® (National Institutes of Health genetic sequence database at National Center for Biotechnology Information) using the Entrez browser (available at: http://www.ncbi.nlm.nih.gov/) and from the TIGR Gene Indices. Proteins that were not full-length were not included as we intended to eventually assess the N-terminal and the C-terminal sequences also. To identify those sequences belonging to the euANT subgroup, sequences were screened for the 10 amino acid insertion in the first AP2 domain characteristic of this group as well as a predomain region longer than 127 amino acids, as described by Kim et al. (2006). WRI1 from the basalANT subgroup, a separate ANT lineage, was included as an outgroup. The mined amino acid sequences were aligned using CLUSTALW (Thompson et al. 1994) and further improved by visual examination and editing using GeneDoc© Version 2.6.002 (Nicholas and Nicholas 1997). The AP2 domain was used for alignment as the full length sequences were too divergent to successfully align all the sequences retrieved from GeneBank® and TIGR Gene Indices database. We limited the boundaries of the double AP2 domain to the sequences starting 5 amino acids upstream and approximately 10 amino acids downstream of the conserved double AP2 domains. In total, 53 sequences (Supplementary Table 2) were analyzed.
The aligned sequences were first subjected to a tool for the selection of the best fit model from among 112 models of protein evolution, using ProtTest (Abascal et al. 2005) version 2.2. After the likelihood statistics were completed three statistical frameworks were selected consecutively one at a time to determine which candidate model fits the data best. Although a tree using the model-averaged estimate of the parameters was obtained in the analysis and examined, we carried a separate Maximum Likelihood (ML) phylogenetic analysis using the PROML program (PROtein Maximum Likelihood program) in the PHYLIP package (Felsenstein 2008) with the selected evolutionary model along with its specific parameters, previously obtained from ProtTest. In the analysis 100 bootstrap trees were produced from which a majority rule tree was computed to obtain bootstrap support for the branches on the ML tree and a strict consensus tree was also computed for examination. Another similar bootstrap analysis was carried with MEGA4 (Nei and Kumar 2000) with 1,000 repeats.
Conserved motifs were identified through a combination of ClustalW alignment, MEME version 4.0.0 (Bailey and Elkan 1994), and Block Maker (Henikoff et al. 1995). The Eukaryotic Linear Motif resource for Functional Sites in Protein (http://elm.eu.org/links.html) was used to predict motif function.
Results
AP2 family genes cloned from soybean somatic embryos
Soybean cDNAs with sequences similar to BnBBM1, were isolated from immature somatic embryos of soybean, G. max cv Merril genotype X5 (Simmonds and Donaldson 2000). The probe used to screen the cDNA library was strategically designed using PCR primers (Supplementary Table 1) targeted to sequences identified from alignments of BnBBM1 and a partial soybean putative AP2 protein EST AW200688 from a cDNA library constructed from cotyledons of 3- and 7-day-old seedlings of cultivar Williams (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucest&val=6481417) (Shoemaker R, The Public Soybean EST Project) (Biogenetics Services, SD, USA) sequences.
Two clones, GmBBM1 (2.5 kb, 707 aa, GenBank Accession HM775856) and GmAIL5 (2.3 kb, 558 aa, GenBank Accession HM775857), were identified and isolated. Another cDNA clone (2 Kb, 560 aa), GmPLT2, was first identified as two 5′-truncted cDNA clones that later appeared as a full length clone (EU677381). All possessed high levels of sequence similarity in the double AP2 domain region (97 %) which decreased in the N- and C-terminal regions (Supplementary Fig. 1A). The soybean GmBBM1 sequence was similar to those of the cruciferous genes, BnBBM1 and AtBBM, with more than 91% similarity in the double AP2 domains (Supplementary Fig. 1B). This decreased in the N- and C-terminal sequences to 48 and 30% respectively (Supplementary Fig. 1B).
Phylogenetic analysis of GmBBM1, GmAIL5 and GmPLT1
To examine the relationships of the three soybean genes with other AP2 family members all available genes in The Arabidopsis Information Resource (http://www.arabidopsis.org/), GenBank, DFCI (Dana Farber Cancer Institute) Gene Index (http://compbio.dfci.harvard.edu/tgi/; formerly TIGR gene index), and the TIGR Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu) at the time of the study with sequence similarity to BBM genes were retrieved using the double AP2 domain region of the AtBBM sequence. Only full-length sequences and those with at least 60% similarity in the double AP2 domain were retained. Sequences belonging to the euANT subgroup are characterized by the presence of a 10 amino acid insert in the first AP2 domain region and a relatively long predomain region (Kim et al. 2006). These two characteristics were used to select 49 sequences belonging to the euANT subgroup. WRI1, belonging to the basalANT subgroup, was included as an outgroup. A total of 53 sequences (Supplementary Table 2) were included in the phylogenetic analysis. The alignment was carried out using the region spanning the two AP2 domains, including the linker region, starting 5 residues upstream of the first conserved AP2 domain and finishing approximately 10 amino acids downstream of the second AP2 domain. The evolutionary model that best fit this data was the JTT + G model (i.e. the Jones-Taylor-Thornton model) under the three statistical frameworks analyzed, i.e. AIC, AICc and BIC, and its parameter G = 0.558, i.e. gamma shape with 4 rate categories.
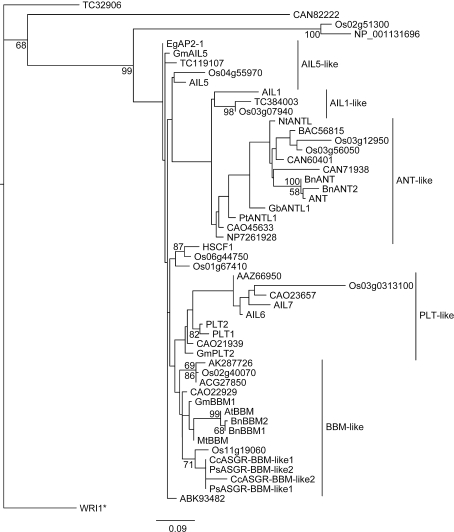
Figure 1 shows the relationship of the three soybean genes to the euANT subgroup based on the AP2-linker-AP2 sequences. GmBBM1 is closest to the other legume BBM gene, MtBBM, from Medicago truncatula and also clusters closely with the BBM genes from the Brassicacea and Poaceae families. GmAIL5 is grouped with the oil palm EgAp2-1 gene and is close to AIL5 from Arabidopsis. GmPLT2 is grouped with the Arabidopsis PLT1 and PLT2 genes along with a gene from Vitis vinifera, all of which form the larger PLT-like grouping with Arabidopsis AIL6 and AIL7 and sequences from rice, Brassica rapa, and another sequence from Vitis vinifera.
Fig. 1.
Maximum likelihood tree of the euANT subgroup of the double AP2 family
Motif composition and identification of a BBM-specific motif
To further distinguish the genes in the BBM-like grouping from other euANT genes, the more variable N-terminal and C-terminal protein sequences were aligned to search for conserved motifs (see Supplementary Fig. 1C). In the N-terminal sequences, five motifs were identified in most members of the BBM-like grouping. Three of these motifs were previously identified in the euANT lineage (euANT2, 3, and 4; Kim et al. 2006) and were found throughout most of the sequences analyzed (Fig. 2A). The fourth motif (bbm-1) was specific to the BBM-like genes and absent from the closely-related PLT-like genes (Fig. 2A). This motif was weakly conserved in some of the ANT-like genes, particularly GbANTL1, PtANTL1, and CAO45633. It was also found in Os01g67410, which was not associated with any of the identified groupings. The fifth motif (euANT5) was also identified in the majority of the members of the BBM-like grouping but was absent in the BBM-like genes from Cenchrus ciliaris and Pennisetum squamulatum (Fig. 2A). This motif is also conserved in the ANT-like genes as well as the AIL1-like genes, and representatives containing this motif can also be found amongst the PLT-like genes, although it is completely absent from the AIL5-like genes. Interestingly the bbm-1 and euANT5 motifs bore some similarity, both including the sequence LSM.
In the C-terminal sequences five other motifs were identified (Fig. 2B). A motif (euANT6) was identified in all of the BBM-like genes immediately following the second AP2 domain. It was identified in representatives from all of the major euANT lineages, although the degree of conservation varied considerably (Fig. 2B). This motif contains a number of Lys and Arg residues, suggesting that it may at least partially function as a nuclear localization signal, as has been suggested for the PLT genes (Aida et al. 2004). The four additional motifs in the C-terminal sequences of the BBM-like genes were conserved with a low degree of specificity. The bbm-2 motif was only weakly conserved in the Arabidopsis and B. napus BBM genes and bbm-3 was completely absent. Some of these motifs were weakly conserved in sequences outside of the BBM-like grouping particularly bbm-3 and bbm-4 in some of the PLT-like sequences. As with the bbm-1 motifs, Os01g67410 also contained the bbm-2 and bbm-4 motifs, although the bbm-3 motif was only weakly conserved and the bbm-5 motif was absent.
Several of the motifs, including euANT2, bbm-1, euANT5, and euANT6, contain consensus sequences for phosphorylation. euANT5 also contains a consensus sequence for sumoylation. The euANT2 motif also conforms to a TRFH domain docking motif and a WW ligand motif.
Expression profile of GmBBM1, GmAIL5 and GmPLT1
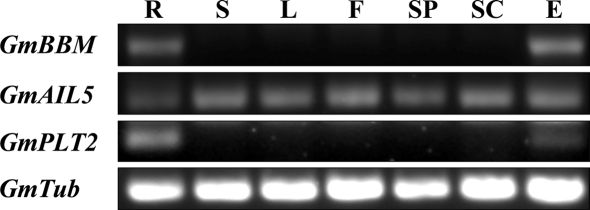
In soybean plants, the expression of GmBBM1 paralleled that of AtBBM in Arabidopsis (Boutilier et al. 2002). It was selectively expressed in soybean embryos and young roots (Fig. 3). GmAIL5 also possessed the same expression pattern as AIL5 in Arabidopsis (Nole-Wilson et al. 2005). It was expressed in all of the soybean organs undergoing growth and development that were examined (Fig. 3). GmPLT2 had the same expression pattern as PLT1 and PLT2 in Arabidopsis (Aida et al. 2004) and M. truncatula (Imin et al. 2007; Holmes et al. 2008). It was expressed predominantly in soybean roots and to a lesser extent in soybean embryos (Fig. 3). The data supported the assignment of GmBBM1 as a BBM homologue, GmAIL5 as an AIL5 homologue and GmPLT2 as a PLT homologue based on the gene phylogeny, conservation of the N-terminal and C-terminal sequences and the expression patterns.
Fig. 3.
Accumulation of GmBBM1, GmAIL5, GmPLT1 and tubulin (accession number M21297) mRNA in soybean cv Merril genotype X5 detected by RT-PCR. Roots at 1 week (R), stem at 3 weeks (S), leaf at 3 weeks (L), flowers at 2 dpa (F), seed pod at 21 dpa (SP), seed coats at 21 dpa (SC), embryos at 21 dpa (E)
Phenotypes generated by BBM-like genes in transgenic Arabidopsis
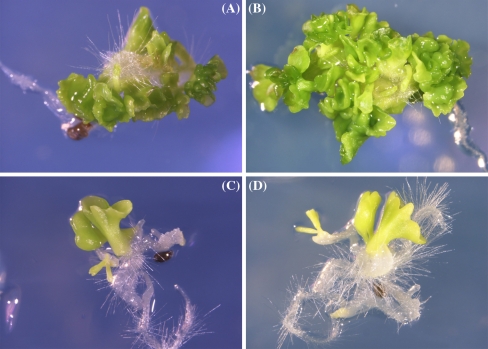
A comparison of the embryogenic phenotype and pleiotropic effects conferred by GmBBM1 and BnBBM1 was carried out in transgenic Arabidopsis using the pBINPLUS vector system used previously for ectopic overexpression of BnBBM1 (Boutilier et al. 2002) (Fig. 4). We confirmed previous descriptions (Boutilier et al. 2002) that showed BnBBM1 expression in Arabidopsis induced somatic embryos and cotyledon-like structures on post-germination organs of transgenic seedlings as well as a unique set of pleiotropic effects on both the vegetative and reproductive organs (Table 1, BnBBM1 lines 1–3). Approximately 35 independent transgenic Arabidopsis lines expressing GmBBM1 embryos on the cotyledons were examined and six representative lines were followed into the T2 generations (Fig. 4A–C). Table 1 show the data for 3 of the GmBBM1 lines. Somatic embryogenesis generally occurred at a lower frequency than with BnBBM1 (Table 1). The highest frequency was 28% in the GmBBM1 line 1, followed by 17% in the GmBBM1 line 2 and 5% in the GmBBM1 line 3 (n > 100 for each line). Embryos also emerged from the shoot apical meristem (Fig. 4D) and hypocotyls (Fig. 4E). Pleiotropic effects on development that were typical of BnBBM1 overexpression were again evident with GmBBM1 overexpression. These included short roots (80% penetrance, GmBBM1 line3; n = 100), short hypocotyls (greater than 80% penetrance in all GmBBM1 lines tested), elongated cotyledons (low penetrance phenotype <25% in all GmBBM1 lines) on seedlings (Fig. 4E, F). Later in development a range of pleiotropic effects were noted including altered leaf morphologies and thickened floral stems. Increased numbers of inflorescences were also noted (data not shown). During the reproductive phase of growth, GmBBM1 and BnBBM1 both induced delayed-flowering time; increased floral organ size; thickened and increased numbers of inflorescences; decreased silique size; and decreased seed production (Table 1).
Fig. 4.
Phenotypes induced by expression of GmBBM1 in transgenic Arabidopsis seedlings. Somatic embryos differentiating from the tips of cotyledons (A, B, C). Ectopic roots differentiating from the embryogenic tissues (A, C). Somatic embryos differentiating from the shoot apical meristem (D) and hypocotyls (E). Alterations in seedling development characterized by short roots, swollen hypocotyls and elongated cotyledons (E, F). Un-transformed control seedlings for comparison (G)
Table 1.
Phenotypes of individual transgenic lines expressing BnBBM1 and GmBBM1 constitutively in transgenic Arabidopsis
| Description of phenotype | Transgenic BnBBM1 lines | Transgenic GmBBM1 lines | ||||
|---|---|---|---|---|---|---|
| Line 1 | Line 2 | Line 3 | Line 1 | Line 2 | Line 3 | |
| Somatic embryo formation on cotyledons | ++++ | ++++ | +++ | ++ | + | + |
| Short roots, fibrous roots | +++ | ++++ | + | + | + | ++++ |
| Elongated roots | – | – | – | ++ | + | – |
| Thick, short hypocotyls | + | + | + | ++++ | ++++ | ++++ |
| Elongated cotyledons | + | – | ++ | + | – | + |
| Delayed flowering time | ++++ | ++++ | +++ | ++ | + | + |
| Reduced seed set | + | + | + | +++ | +++ | +++ |
| Increased floral organ size | + | + | + | + | ++ | ++ |
The phenotypes in bold at the seedling stage of development and used for the initial screening of lines. The frequency of occurrence of the phenotype was recorded for each line using at least 100 plantlets
+ (>0<25%), ++ (25–50%), +++ (50–75%), ++++ (75–100%)
Although many of the phenotypes were similar for BnBBM1 and GmBBM1 some interesting differences were observed. For example, in GmBBM1 transgenic lines ectopic roots occassionally developed on cotyledons along with embryos (Fig. 4A, C), a feature that was not reported with BnBBM1 (Boutilier et al. 2002). Furthermore, elongated roots were seen on plantlets of the GmBBM1 line in contrast to the short-root phenotype observed in BnBBM1 plants (Table 1).
Function of the bbm-1 motif in embryogenesis
The significance of the bbm-1 sequence motif to BnBBM1 function was examined by creating a 9-amino acid deletion mutation in the BnBBM1 gene (Fig. 2A) and assessing functionality in transgenic Arabidopsis (Fig. 5). For comparison, a deletion of motif euANT2 (Fig. 2A) was also created singly and together with a deletion of bbm-1. The induction of somatic embryos on cotyledons by wild-type BnBBM1 was first confirmed in 22 transgenic lines as controls using the pCAMBIA1300 vector (Fig. 5C). Shoots subsequently developed in culture (Fig. 5D) and plantlets regenerated as described previously by Boutilier et al. 2002 (data not shown). Somatic embryogenesis was not eliminated by deletion of euANT2 (Fig. 5G, H) or euANT2 and bbm-1 together (Fig. 5I, J). However, the mutant phenotypes differed in that the embryo-like structures tended to proliferate and shoots did not emerge over time (Fig. 5H, J) as with wild-type BnBBM1 (Fig. 5D). This phenotype was confirmed in 14 and 25 transgenic lines, respectively and was in striking contrast to the phenotype generated by deletion of the bbm-1 motif alone which was characterized by the complete loss of somatic embryogenesis on cotyledons as well as the accompanying pleiotropic effects on seedlings (Fig. 5E, F). Pleiotropic effects associated with this mutation emerged at later stages of plant development. Some were similar to the class II pleiotropic effects described for BnBBM1 by Boutilier et al. (2002). For example, flowers with short sepals and petals relative to carpels were frequently observed (Fig. 6A) compared with wild type flowers (Fig. 6B). The changes in leaf morphology described for BnBBM1 (Boutilier et al. 2002) were infrequently observed; however, serrated margins were occasionally observed (Fig. 6C) in plants expressing the deletion mutant but not wild type plants (Fig. 6D).
Fig. 5.
Phenotype of un-transformed Arabidopsis seedlings at 14 days (A) and 21–28 days (B) post germination compared with the phenotype of 14 day and 21–28 day transgenic Arabidopsis ectopically expressing wild-type BnBBM1 (C, D respectively), the BnBBM1 deletion mutant without bbm-1 (E, F respectively), the BnBBM1 deletion mutant without euANT2 (G, H respectively), the BnBBM1 deletion mutant without euANT2 and bbm-1 (I, J respectively)
Fig. 6.
Pleiotropic effects on transgenic Arabidopsis flower (A) and leaf (C) morphologies generated by the BnBBM1 deletion mutant that lacks the bbm-1 motif compared with wild type flowers (B) and leaf (D)
The bbm-1 motif was inserted into Arabidopsis PLT2, AIL5 and AIL7 (Fig. 2A) to determine if other euANT members possessed the complementary elements needed for somatic embryogenesis in transgenic Arabidopsis. AIL7 + bbm-1 and AIL5 + bbm-1 failed to generate ectopic embryos in experiments parallel to those above (data not shown). The ectopic PLT2 phenotype was previously characterized by the proliferation of ectopic roots and root hairs on germinating embryos or seedlings (Aida et al. 2004). PLT2 + bbm-1 generated a range of phenotypes in five different transgenic lines. Rapid and strong tissue proliferation followed by copious embryo differentiation was observed in 4 of the 5 lines (Fig. 7A, B). This phenotype resembled the BBM phenotype (Ben Scheres and colleagues, personal communication; Fig. 7). Smaller undefined green tissues had also been described as part of the PLT2 ectopic phenotype (Aida et al. 2004) In 2 of the lines, both BBM and PLT2 phenotypes were expressed simultaneously. In these lines embryo differentiation was less prolific and root differentiation was very abundant (Fig. 7C, D).
Fig. 7.
Phenotypes of transgenic Arabidopsis seedlings, in which the bbm-1 motif from BnBBM1was inserted into the corresponding position of the PLT2 gene, at 11 days (A, B), 13 days (C) and 17 days (D) post germination
Discussion
The phenotypes associated with the AP2/ANT family frequently involve undifferentiated cell proliferation and differentiation of stem cell niches within meristems (Nole-Wilson et al. 2005). Recently, a role in embryogenesis has also emerged. Most of the information has been generated by the induction of somatic embryogenesis by ectopic overexpression of BnBBM1 in Arabidopsis and B napus (Boutilier et al. 2002; Passarinho et al. 2008). GmBBM1 was shown here to induce similar developmental events in Arabidopsis. Compared with meristematic cell types, the differentiation of embryonal stem cells from somatic cells is poorly understood. Studies have shown that somatic embryogenesis may be activated through a number of different pathways and by a number of different genes (Passarinho et al. 2008; reviewed by Verdeil et al. 2007). In this study, we have identified specific features of the BBM-like homologues that are responsible for the induction of embryogenesis and that may separate the BBM-like genes from closely related members of the AP2/ANT family that function in vegetative development yet act redundantly in embryo development.
To understand how BBM-like genes function in the conversion of somatic cells to embryonal cells and the development of the resulting embryo it may be important to understand the relationships among the AP2/ANT family members. The members of the euANT lineage, such as the BBM-like, PLT-like and AIL5-like genes, are very closely related phylogenetically and structurally. Our analysis has revealed domain conservation among them but also domain specificity. For example, the bbm-1 motif was specific to all of the BBM-like genes. The different members are known to have specific roles in development but evidence for redundancy is also emerging. For example, the mutant phenotypes of bbm and plt2 have shown that AtBBM and PLT2 function as redundant partners in early Arabidopsis embryogenesis (Galinha et al. 2007). AtBBM, PLT1, PLT2 and PLT3/AIL6 are expressed in root primordia where they function as redundant partners in root stem cell differentiation in embryos and root differentiation (Aida et al. 2004; Galinha et al. 2007). The demonstration that the addition of the bbm-1 motif to PLT2 enhanced the capacity to induce somatic embryogenesis confirmed the close relationship between these genes and may have revealed a key structural feature of BBM important for its ability to function in embryogenesis. The PLT2 ectopic phenotype was not lost when the BnBBM1 ectopic phenotype was expressed but the strength of the PLT2 and BnBBM1 ectopic phenotypes were inversely related indicating that the embryogenic and root meristematic pathways could be competing in these transgenic lines.
How AIL5 functions, particularly in embryogenesis, is unclear. AIL5 appears to be involved in cell proliferation activities in many organs and may generate enlarged organs, for example large flowers, when ectopically expressed (Nole-Wilson et al. 2005). This phenotype is similar to that of ANT, which generates large flowers (Nole-Wilson et al. 2005; Mizukami and Fischer 2000) and AP2 which generates large embryos (Ohto et al. 2005) when ectopically expressed. Since AIL5 mutants have no apparent phenotype it is believed that it acts redundantly with other relatives (Nole-Wilson et al. 2005). AIL5 and BBM double mutants appear to lack defects in embryo or root meristem development therefore additional unidentified partners may exist if they act in the same pathways (Tsuwamoto et al. 2010). The finding that ectopic over-expression of AIL5 yielded phenotypes similar to BBM over-expression, including somatic embryogenesis (Tsuwamoto et al. 2010), indicates that it could be involved in the proliferation of the totipotent somatic cells. The embryonic structures that emerged exhibit elevated expression of AGL15, LEC1, LEC2, FUS3 and BBM (Tsuwamoto et al. 2010) all of which have the capacity to induce somatic embryogenesis when ectopically expressed. Interesting, the oil palm AIL5-like gene, EgAp2-1, is also expressed in proliferating tissues, especially embryonic tissues but induces callus growth and shoot organogenesis rather than embryos in Arabidopsis (Morcillo et al. 2007). BnBBM1 also induces shoot organogenesis in tobacco rather than embryogenesis revealing that BBM-like genes have the capacity to induce both shoot meristem activity as well as embryogenesis depending on the genetic and cellular environment of the proliferating cells (Srinivasan et al. 2007). Unlike PLT2, the addition of the bbm-1 motif to AIL5 did not generate the capacity to induce embryogenesis in transgenic Arabidopsis in this study. Whether this alteration disrupted the capacity of AIL5 to generate somatic embryos as described by Tsuwamoto et al. (2010) is unknown and points to complex roles for AIL5 in plant development.
Conserved sequence motifs have been recognized among soybean, rice and Arabidopsis AP2/ANT genes and soybean-specific motifs are among them (Zhang et al. 2008). Previous studies have suggested that within subfamilies conserved sequence motifs are abundant and likely reflect shared functions (Nakano et al. 2006). Unlike some of the well-characterized motifs of the ERF family, sequence motifs within the AP2/ANT family have not yet been assigned specific roles in the transcriptional regulation of developmental processes (Nakano et al. 2006; Zhang et al. 2008). The characterization of the GmBBM1 gene here allowed us to align the BBM-like genes from divergent species which are known to generate ectopic embryos in Arabidopsis and thus provide a functional data set that was used to identify 10 conserved sequence motifs outside of the AP2-linker-AP2 region. Among the BBM-like, PLT-like and AIL5-like genes the motif composition was very similar, however, the bbm-1 motif was the most specific to the BBM-like genes in the N-terminal region. In the C-terminal region sequences with a lower degree of conservation among the BBM-like genes exist. Of these bbm-2, bbm-3 and bbm-4 are poorly conserved in the cruciferous genes and thus unlikely to be essential for embryogenesis. The motif, bbm-5, is also poorly conserved relative to bbm-1.
Both deletion and domain swap analyses revealed that the bbm-1 motif was needed for BnBBM1 to induce somatic embryogenesis in Arabidopsis. Further examination revealed that bbm-1 activity is intimately linked to the activities of other motifs within these transcription factors, especially euANT2, which is found in almost all other genes in the euANT lineage including PLT2 and AIL5. Deletion of euANT2 singly or euANT2 and bbm-1 simultaneously prevented the somatic embryos from generating shoots even after prolonged times in culture. Among the many possible explanations for bbm-1 function is the possibility that the euANT2 motif functions in vegetative pathways of development and that bbm-1 acts to restrain the vegetative activity of BnBBM1. If correct, then a mechanism must exist to relieve the restraints on euANT2 in BnBBM1 during the later stages of embryo development when the shoot and root meristems differentiate and develop. Furthermore, if a single domain, bbm-1, can separate the ectopic phenotypes of redundant partners such as BBM and PLT2 then it would seem possible that the shared motifs could provide the basis for functional redundancy. A hierarchical process must exist in the cellular environment to recruit and coordinate the different AP2/ANT members to the different developmental pathways in an orderly manner. As both the bbm-1 and euANT2 motifs have the consensus sequence for phosporylation post-translational processes may be involved in the regulation of such interactions. For PLT-like genes, expression gradients appear to play an important in the transition from root meristem initiation to root development (Galinha et al. 2007).
Any general model for the role of AP2/ANT transcription factors in somatic embryogenesis should consider the observations that BBM-like genes may play distinct roles at different stages of development; interact with redundant partners; and induce different pathways of development depending on the genetic and cellular environment. The bbm-1 motif of BBM-like genes appears to play an early role in the conversion of somatic cells to neoplastic embryonal cells. This may occur by suppressing the activity of other motifs, such as euANT2, which function in vegetative cell differentiation. The shared motifs among the AP2/ANT members may provide a mechanism for BBM to recruit the activities of members with meristematic activities essential for the development of the embryo once embryogenesis is established. As new members are recruited to the developing embryo dilution of the bbm-1 motif may release the cells from the neoplastic state and allow the meristems to develop. Sustained ectopic expression of BBM genes during plant maturation results in a range of pleiotropic effects. This may occur through similar unintended interactions with AP2/ANT proteins through shared motifs that disrupt their normal patterns of activity. The different pleiotropic effects observed for GmBBM1 and BnBBM1 may reveal differences in the efficiencies of interactions resulting from sequence divergence. If BBM-like genes function by utilizing redundancies with other AP2/ANT transcription factors to recruit developmental pathways needed for embryogenesis and embryo development in seeds then strict spatial control of BBM-like gene expression would be essential for normal plant development. As shown here and in all other studies of BBM–like gene expression (Boutilier et al. 2002; Nole-Wilson et al. 2005; Imin et al. 2007) expression in seeds is under strict spatial and developmental regulation as are the other AP2/ANT family members (Feng et al. 2005).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 3 A. Alignment of the predicted amino acid sequences of the three BBM-like genes isolated from soybean. B. Alignment of the predicted amino acid sequence of GmBBM1 with BBM sequences from Brassica napus and Arabidopsis thaliana. Dotted lines above the sequences indicate motifs that were identified in the BBM-like genes. Motifs bbm-2, bbm-3 and bbm-4 were only weakly conserved in the cruciferous genes and therefore are only indicated for GmBBM1. In A. and B., residues identical in all three sequences are in black boxes and residues similar in all three sequences are in gray boxes. The two AP2 domains are indicated with a bar above the sequences. C. Schematic diagram illustrating the structure of the three BBM-like genes isolated from soybean. Black boxes represent the AP2 domains, coloured boxes represent conserved motifs identified in BBM sequences. Light grey boxes represent the untranslated regions of the isolated cDNAs. The black bar indicates the location that would hybridize to the AP2 domain probe. Arrows indicate the position of primers described in Supplementary Table 1.(TIFF 7505 kb)
Acknowledgments
The authors are very grateful to Li Wang for Arabidopsis transformation, Michelle Lopez for the transgenic studies, Wayne Stangle for the construct, Daina Simmonds, Sheryl Hubbard and Suquin Zheng for the soybean tissues, Réjean Desagagné and David Gagné for their support and cooperation and Ben Scheres and his lab for examination of our data prior to publication. The research was funded by the Canadian Biotechnology Strategy.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Souad El Ouakfaoui, Jaimie Schnell, Ashraf Abdeen and Adam Colville have contributed equally.
Contributor Information
Souad El Ouakfaoui, Email: Souad.ElOuakfaoui@ec.gc.ca.
Jaimie Schnell, Email: Jaimie.Schnell@inspection.gc.ca.
Ashraf Abdeen, Email: Ashraf.Abdeen@mail.mcgill.ca.
Adam Colville, Email: Adam.Colville@IOGEN.ca.
Hélène Labbé, Email: labbehe@agr.gc.ca.
Shuyou Han, Email: hans@agr.gc.ca.
Bernard Baum, Email: bernard.baum@agr.gc.ca.
Serge Laberge, Email: laberges@agr.gc.ca.
Brian Miki, Phone: +1-613-7591685, FAX: +1-613-7591701, Email: mikib@agr.gc.ca.
References
- Abascal F, Zardoya R, Posada, D (2005) ProtTest selection of best fit models of protein evolution. Bioinformatics 21:2104–2105. http://darwin.uvigo.es/software/prottest.html Version 2.2 accessed 28 Sept. 2008 [DOI] [PubMed]
- Aida M, Beis D, Heidstra R, Willemsen V, Bliloum I, Galinham C, Nussaumem L, Nohm Y-S, Amasinom R, Scheres B. The PLETHORA genes mediate patterning of the arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman R, Brutlag D, Karp P, Lathrop R, Searls D, editors. Proceedings of the second international conference on intelligent systems for molecular biology. Menlo Park, CA: American Association for Artificial Intelligence Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, Custers JB, van Lookeren Campagne MM. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium- mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQJ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (2008) PHYLIP, Phylogeny Inference Package, version 3.68. Available at http://evolution.gs.washington.edu/phylip.html. Accessed November 2008
- Feng J-X, Liu D, Pan Y, Gong W, Ma L-G, Luo J-C, Deng XW, Zhu Y-X. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol Biol. 2005;59:853–868. doi: 10.1007/s11103-005-1511-0. [DOI] [PubMed] [Google Scholar]
- Finer JJ. Apical proliferation of embryogenic tissue of soybean [Glycine max (L.) Merrill] Plant Cell Rep. 1988;7:238–241. doi: 10.1007/BF00272532. [DOI] [PubMed] [Google Scholar]
- Finer JJ, Nagasawa A. Development of an embryogenic suspension culture of soybean [Glycine max (L.) Merrill] Plant Cell Tissue Org Cult. 1988;15:125–136. doi: 10.1007/BF00035754. [DOI] [Google Scholar]
- Floyd SK, Bowman JL. The ancestral developmental tool kit of land plants. Int J Plant Sci. 2007;168:1–35. doi: 10.1086/509079. [DOI] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1056. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gong W, Shen Y-P, Ma L-G, Pan Y, Du Y-L, Wang D-H, Yang J-Y, Hu L-D, Liu X-F, Dong C-X, Ma L, Chen Y-H, Yang X-Y, Gao Y, Zhu D, Tan X, Mu J-Y, Zhang D-B, Liu Y-L, Dinesh-Kumar SP, Li Y, Wang X-P, Gu H-Y, Qu L-J, Bai S-N, Lu Y-T, Li J-Y, Zhao J-D, Zuo J, Huang H, Deng XW, Zhu Y-X. Genome-wide ORFeome cloning and analysis of arabidopsis transcription factor genes. Plant Physiol. 2004;135:773–782. doi: 10.1104/pp.104.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-P. [DOI] [PubMed] [Google Scholar]
- Holmes P, Goffard N, Weiller GF, Rolfe BG, Imin N. Transcriptional profiling of Medicago truncatula meristematic root cells. BMC Plant Biol. 2008;8:21. doi: 10.1186/1471-2229-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Nizamidin M, Wu T, Rolfe BG. Factors involved in root formation in Medicago truncatula. J Exp Bot. 2007;58:439–451. doi: 10.1093/jxb/erl224. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, Boer B, Montagu MV, Okamuro JK. Control of arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Soltis PS, Wall K, Soltis DE. Phylogeny and domain evolution in the APETALA2-like gene family. Mol Biol Evol. 2006;23:107–120. doi: 10.1093/molbev/msj014. [DOI] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. The AINTEGUMENTA gene of arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A. AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell. 2000;12:1357–1366. doi: 10.1105/tpc.12.8.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes T. The inflorescence architecture of Petunia hybrida is modified by the Arabidopsis thaliana Ap2 gene. Dev Genet. 1999;25:199–208. doi: 10.1002/(SICI)1520-6408(1999)25:3<199::AID-DVG3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo F, Gallard A, Pillot M, Jouannic S, Aberlene-Bertossi F, Collin M, Verdeil JL, Tregear JW. EgAP2–1, and AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm. Planta. 2007;226:1353–1362. doi: 10.1007/s00425-007-0574-3. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Nicholas KB, Nicholas HB Jr (1997) GeneDoc©: a tool for editing and annotating multiple sequence alignments. Distributed by the authors. www.psc.edu/biomed/genedoc/ Accessed Oct-Dec 2008
- Nole-Wilson S, Tranby TL, Krizek BA. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol. 2005;57:613–628. doi: 10.1007/s11103-005-0955-6. [DOI] [PubMed] [Google Scholar]
- Ohto M, Fischer RL, Goldberg RB, Nakamura K, Harada JJ. Control of seed mass by APETALA2. Proc Natl Acad Sci USA. 2005;102:3123–3128. doi: 10.1073/pnas.0409858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarinho P, Ketelaar T, Xing M, van Arkel J, Maliepaard C, Hendriks MW, Joosen R, Lammers M, Herdies L, den Boer B, van der Geest L, Boutilier K. BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol. 2008;68:225–237. doi: 10.1007/s11103-008-9364-y. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G–L. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Simmonds DH, Donaldson PA. Genotype screening for proliferative embryogenesis and biolistic transformation of short-season soybean genotypes. Plant Cell Rep. 2000;19:485–490. doi: 10.1007/s002990050760. [DOI] [PubMed] [Google Scholar]
- Srinivasan C, Liu Z, Heidmann I, Supena ENJ, Fukuoka H, Joosen R, Lambalk J, Angenent G, Scorza R, Custers JBM, Boutilier K. Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L) Planta. 2007;225:341–351. doi: 10.1007/s00425-006-0358-1. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto R, Yokoi S, Takahata Y (2010) Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Mol Biol. doi: 10.1007/s11103-010-9634-3 [DOI] [PubMed]
- Verdeil J-L, Alemanno L, Niemenak N, Tranbarger TJ. Pluripotent versus totipotent plant stem cells: dependence versus autonomy? Trends Plant Sci. 2007;12:1360–1385. doi: 10.1016/j.tplants.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Würschum T, Groβ-Hardt R, Laux T. APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell. 2006;18:295–307. doi: 10.1105/tpc.105.038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chen M, Chen X, Xu Z, Guan S, Li L-C, Li A, Guo J, Mao L, Ma Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) J Exp Bot. 2008;59:4095–4107. doi: 10.1093/jxb/ern248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 3 A. Alignment of the predicted amino acid sequences of the three BBM-like genes isolated from soybean. B. Alignment of the predicted amino acid sequence of GmBBM1 with BBM sequences from Brassica napus and Arabidopsis thaliana. Dotted lines above the sequences indicate motifs that were identified in the BBM-like genes. Motifs bbm-2, bbm-3 and bbm-4 were only weakly conserved in the cruciferous genes and therefore are only indicated for GmBBM1. In A. and B., residues identical in all three sequences are in black boxes and residues similar in all three sequences are in gray boxes. The two AP2 domains are indicated with a bar above the sequences. C. Schematic diagram illustrating the structure of the three BBM-like genes isolated from soybean. Black boxes represent the AP2 domains, coloured boxes represent conserved motifs identified in BBM sequences. Light grey boxes represent the untranslated regions of the isolated cDNAs. The black bar indicates the location that would hybridize to the AP2 domain probe. Arrows indicate the position of primers described in Supplementary Table 1.(TIFF 7505 kb)