Abstract
Ethylene, a regulator of plant growth and development, is perceived by specific receptors that act as negative regulators of the ethylene response. Five ethylene receptors, i.e., ETR1, ERS1, EIN4, ETR2, and ERS2, are present in Arabidopsis and dominant negative mutants of each that confer ethylene insensitivity have been reported. In contrast, maize contains just two types of ethylene receptors: ZmERS1, encoded by ZmERS1a and ZmERS1b, and ZmETR2, encoded by ZmETR2a and ZmETR2b. In this study, we introduced a Cys to Tyr mutation in the transmembrane domain of ZmERS1b and ZmETR2b that is present in the etr1-1 dominant negative mutant and expressed each protein in Arabidopsis. Mutant Zmers1b and Zmetr2b receptors conferred ethylene insensitivity and Arabidopsis expressing Zmers1b or Zmetr2b were larger and exhibited a delay in leaf senescence characteristic of ethylene insensitive Arabidopsis mutants. Zmers1b and Zmetr2b were dominant and functioned equally well in a hemizygous or homozygous state. Expression of the Zmers1b N-terminal transmembrane domain was sufficient to exert dominance over endogenous Arabidopsis ethylene receptors whereas the Zmetr2b N-terminal domain failed to do so. Neither Zmers1b nor Zmetr2b functioned in the absence of subfamily 1 ethylene receptors, i.e., ETR1 and ERS1. These results suggest that Cys65 in maize ZmERS1b and ZmETR2b plays the same role that it does in Arabidopsis receptors. Moreover, the results demonstrate that the mutant maize ethylene receptors are functionally dependent on subfamily 1 ethylene receptors in Arabidopsis, indicating substantial functional conservation between maize and Arabidopsis ethylene receptors despite their sequence divergence.
Keywords: Ethylene, Ethylene receptors, etr1-1, Maize, Arabidopsis, Signal transduction
Introduction
The plant hormone ethylene regulates diverse aspects of plant growth and development, including germination; leaf, stem, and root growth; sex determination; fruit ripening; organ abscission; leaf and flower senescence; and cell death of the cereal endosperm (Mattoo and Suttle 1991; Abeles et al. 1992; Dolan 1998; Bleecker and Kende 2000; Schiefelbein 2000; Young and Gallie 2000; Klee 2004; Lin et al. 2009). Ethylene also regulates responses to adverse growth conditions, such as hypoxia, mechanical impedance, and pathogen attack (Feldman 1984; Ecker and Davis 1987; Mattoo and Suttle 1991; Abeles et al. 1992; Morgan and Drew 1997).
Ethylene is produced from methionine in which the latter is converted initially to S-adenosylmethionine (AdoMet) by S-adenosylmethionine synthase, which is then converted to 1-aminocyclopropane-1-carboxylate (ACC) by ACC synthase (ACS). The generation of ethylene results from the oxidation of ACC by ACC oxidase (ACO) in a reaction that also produces CO2 and HCN (Yang and Hoffman 1984). Both ACS and ACO are encoded by multigene families: e.g., the Arabidopsis genome contains nine ACS genes that exhibit cell specific and overlapping expression (Liang et al. 1992; Zarembinski and Theologis 1994; Tsuchisaka and Theologis 2004). The ACO gene family may be composed of up to 17 members although not all may function as ACC oxidases (Tsuchisaka et al. 2009). The ACS and ACO gene families are considerably smaller in maize, with just three members comprising the ZmACS family and four members comprising the ZmACO family (Gallie and Young 2004).
Following its production, ethylene is perceived by binding to endoplasmic reticulum-localized receptors (Chen et al. 2002), of which five different types (i.e., ETR1, ERS1, EIN4, ETR2, and ERS2) are present in Arabidopsis (Bleecker et al. 1998; Chang and Shockey 1999; Chang and Stadler 2001; Wang et al. 2002; Chang and Bleecker 2004; Stepanova and Alonso 2005; Lin et al. 2009). As negative regulators, the receptors, in conjunction with the CTR1 Raf-like kinase, repress the activity of the downstream components of ethylene signaling in the absence of ethylene (Kieber et al. 1993; Hua and Meyerowitz 1998; Clark et al. 1998). Binding of ethylene to the N-terminal membrane domain of the receptors relieves the repression of the downstream components of the signaling pathway resulting in the activation of EIN2 and the downstream transcriptional factors including EIN3/EIL and ERF (Chao et al. 1997; Solano et al. 1998; Alonso et al. 1999).
Ethylene receptors share structural similarity with two-component regulators present in bacteria and yeast which are characterized by domains for signal input and output and have His-kinase activity (Schaller 1997; Chang and Stewart 1998; Chang and Stadler 2001; Lohrmann and Harter 2002). In Arabidopsis, ETR1, ETR2, and EIN4 contain a C-terminal receiver domain that follows the His-kinase domain whereas ERS1 and ERS2 do not. ETR1 and ERS1 possess the amino acid sequences and motifs within the His-kinase domain that are necessary for His-kinase activity and exhibit such activity whereas EIN4, ETR2, and ERS2 lack some or most of these required sequences and instead exhibit Ser-Thr kinase activity, which ERS1 also exhibits (Chang et al. 1993; Hua et al. 1995, 1998; Gamble et al. 1998; Sakai et al. 1998; Moussatche and Klee 2004). Because of their demonstrated His-kinase activity and lack of an obvious N-terminal signal peptide, ETR1 and ERS1 have been classified as subfamily I receptors whereas EIN4, ETR2, and ERS2 represent subfamily II receptors (Wang et al. 2003). Despite the distinction between these two subfamilies based on His-kinase activity, mutants of ETR1 lacking His-kinase activity remain competent to rescue the etr1-7;ers1-2 mutant phenotype in which ETR1 expression is lacking and ERS1 expression is substantially reduced (Wang et al. 2003). Subfamily I receptors appear to be functionally distinct from subfamily II receptors in that loss of their expression results in a severe constitutive ethylene response (Hall and Bleecker 2003; Wang et al. 2003) and ectopic expression of any subfamily II receptor fails to rescue the etr1-7;ers1-2 mutant (Wang et al. 2003). ETR1 has been shown to form covalently linked dimers through a disulfide bond formed between Cys-4 and Cys-6 and the oligomerization of ethylene receptors may play a role in their function (Schaller et al. 1995; O’Malley et al. 2005; Chen et al. 2010).
Mutations of ethylene receptors resulting in constitutive signaling have been described (Bleecker et al. 1988; Chang et al. 1993; Hua et al. 1995, 1998). One such mutant, etr1-1, has a Cys to Tyr mutation at residue 65 in the N-terminal transmembrane domain that results in a dominant negative effect and confers a strong ethylene insensitive phenotype (Bleecker et al. 1988; Guzmán and Ecker 1990; Chang et al. 1993; Chen and Bleecker 1995). The N-terminal 349 amino acid residues are sufficient to confer ethylene insensitivity (Gamble et al. 2002). Loss of EIN2 expression also disrupts ethylene signaling and results in ethylene insensitivity (Alonso et al. 1999). In contrast, loss of CTR1 disrupts the ability of the receptors to repress the activity of the downstream components of the ethylene signaling pathway, resulting in constitutive ethylene signaling.
As with the ZmACS and ZmACO gene families, the ethylene receptor gene family is smaller in maize with fewer types of ethylene receptors than in Arabidopsis. Maize lacks homologs for ETR1, ERS2, or EIN4 and expresses just two types of ethylene receptors based on sequence conservation and domain structure: the first type with homology to Arabidopsis ERS1 is referred to as ZmERS1 and the second type that is the likely homolog of Arabidopsis ETR2 is referred to as ZmETR2 (Gallie and Young 2004). The presence of two genes encoding ZmERS1 (i.e., ZmERS1a and ZmERS1b) and two genes encoding ZmETR2 (i.e., ZmETR2a and ZmETR2b) in maize is consistent with the allotetraploid nature of its genome (Anderson 1945; Rhoades 1951). Rice also has two types of ethylene receptors. The subfamily I receptors, OsERS1 and OsERS2, are ERS1-like whereas the subfamily II receptors, OsETR2, OsETR3, and OsETR4, are ETR2-like (Yau et al. 2004), suggesting that the family structure of ethylene receptors in maize may be a general feature of monocots.
In this report, we have investigated the conservation of maize ethylene receptor function with those in Arabidopsis. A Cys to Tyr mutation was introduced at amino acid 65 in the transmembrane domain of ZmERS1b and ZmETR2b to generate mutant maize receptors that copy the mutation present in the etr1-1 dominant negative mutant. The effect of the mutation in the maize receptors was determined following their expression in Arabidopsis. The expression of each mutant maize receptor conferred a state of ethylene insensitivity in Arabidopsis and resulted in many of the phenotypes characteristic of ethylene insensitive Arabidopsis mutants, including increased leaf size and delayed leaf senescence. The mutant maize receptors were dominant and therefore functioned when present in a hemizygous state. Dominance over endogenous Arabidopsis ethylene receptors was observed when just the Zmers1b N-terminal transmembrane domain was expressed. Interestingly, expression of the mutant Zmetr2b N-terminal domain did not confer a state of ethylene insensitivity in Arabidopsis. The mutant maize receptors were dependent on subfamily 1 receptors to function in Arabidopsis as neither mutant maize receptor functioned to confer a state of ethylene insensitivity in the absence of subfamily 1 ethylene receptor expression. These results suggest that Cys65 in maize ZmERS1 and ZmETR2 plays the same role that it does for Arabidopsis receptors and that the dominance of both mutant maize receptors is dependent on subfamily 1 ethylene receptors in Arabidopsis, indicating substantial functional conservation between maize and Arabidopsis ethylene receptors.
Materials and methods
Plasmid constructs and mutagenesis
The cDNAs of ZmETR2b and ZmERS1b were obtained by RT–PCR from B73 maize RNA and cloned into pGEM-T Easy (Promega, Madison WI, USA). To generate the Zmetr2b and Zmers1b mutants, mutagenesis was performed using the GeneEditor™ in vitro Site-Directed Mutagenesis System (Promega, Madison WI, USA). The DNA template was denatured by alkaline treatment, the mutagenic and selection oligonucleotides were annealed, and the mutant strand was synthesized with T4 DNA polymerase and T4 DNA ligase. The DNA was then transformed into BMH 71-18 mutS cells which were grown overnight with the GeneEditor™ Antibiotic Selection Mix. Plasmid DNA was isolated and transformed into JM109 cells. The mutagenic oligonucleotides used were ZmETR2b: 5′-CATCGTGCTCTACGGCCTCACGC-3′ and ZmERS1b: 5′-GTTTATAGTTCTCTATGGGGCAAC-3′, in which the mutation (i.e., G to A) is underlined. The region encoding Zmetr2b(1-386) or Zmers1b(1-350) were obtained by PCR from the full length Zmetr2b and Zmers1b constructs, respectively. The primer sets used to generate each were ZmETR2-F1/ZmETR2-R1 and ZmERS1-F1/ZmERS1-R1, respectively (see below). Both sets of primers were also used for genotyping Arabidopsis containing the Zmetr2b or Zmers1b transgenes.
Plant material and transformation
Col-0 Arabidopsis was used throughout this study. After surface-sterilization and cold treatment at 4°C for 4 days in the dark, seeds were planted on 0.25× MS agar plates with or without ACC or AgNO3 at the concentrations indicated. For the triple response assay, seeds were germinated on medium in the dark for 4 days and the length of the seedling hypocotyl and root measured. For adult plants, seeds were germinated on medium for 1 week and transferred to soil and grown under a 24 h light cycle at 20°C in a plant growth room supplemented with Sylvania Gro-Lite fluorescent bulbs (Sylvania, Danvers MA, USA) at a photon flux density (PFD) of 100 μmol m−2 s−1. Arabidopsis was transformed with each transgene in the binary vector, pBI121, at bolting using Agrobacterium. The primary inflorescence was removed and secondary inflorescences were allowed to initiate before infiltration. Inverted plants were dipped into the infiltration medium containing the Aglo1 strain of Agrobacterium containing the transgene. Infiltrated plants were kept on their side for 1 day and allowed to continue to flower in an upright position in the same growth room. Seeds of infiltrated plants were collected and screened on 0.25× MS plates containing 50 μg/ml kanamycin and 500 μg/ml vancomycin.
Northern analysis
RNA was extracted by quick-freezing plant material in liquid nitrogen, grounding it to a fine powder, and resuspending 100 mg of the material in 1 ml TRIZOL® Reagent (Invitrogen, Carlsbad CA, USA). Following centrifugation, the supernatant was extracted with 200 μl chloroform and centrifuged to separate the phases. The RNA was precipitated from the aqueous phase by isopropyl alcohol, washed with 75% ethanol, resuspended in RNase-free H2O, and resolved on a 1.2% agarose-formaldehyde gel. PCR-generated Zmetr2b and Zmers1b fragments were radiolabeled with dCTP using Prime-a-Gene labeling system (Promega, Madison WI, USA) and used for hybridization with the membrane overnight at 38°C in 5× SSPE (150 mM NaCl, 10 mM NaH2PO4 · H2O, 1 mM EDTA), 5× Denhardt’s solution, 50% formamide, and 1.5% SDS. Blots were washed for 30 min at 45°C in 1× SSPE/0.1% SDS, 30 min at 50°C in 0.5× SSPE/0.1% SDS, and 30 min at 55°C in 0.2× SSPE/0.1% SDS. The membrane was then exposed to film at −80°C with an intensifier screen. Each Northern was repeated at least twice. The same membrane was stripped in 50% formamide, 2× SSPE at 65°C for 30–60 min until no signal could be detected. Where indicated, the membrane was reprobed for eEF1A mRNA using similar conditions.
PCR analysis
DNA was isolated by quick-freezing plant material in liquid nitrogen, grounding to a fine powder, and resuspending in 400 μl extraction buffer (100 mM Tris–Cl pH 9.0, 20 mM EDTA, 200 mM NaCl, 1% Sarcosyl, and 1% β-ME). Following centrifugation, the supernatant was extracted with 400 μl phenol: chloroform (1:1) and centrifuged to separate the phases. The DNA was precipitated from the aqueous phase by sodium acetate and isopropyl alcohol, washed with 75% ethanol and resuspended in H2O. PCR amplification was performed in 20 μl reactions containing 1× PCR buffer, 0.4 μ HotStarTaq DNA polymerase (Qiagen Inc., Valencia CA, USA), 250 μM dNTPs, 10 μM forward and reverse primers, and 50 ng genomic DNA. Reactions were carried out using the following conditions: 95°C/5 min (1 cycle); 95°C/30 s, 55°C/30 s, 72°C/1 min (35 cycles); and a final extension at 72°C/5 min (1 cycle). To detect the presence of Zmetr2b, a forward primer, ZmETR2-F1, 5′-ATGGTGGTGGGAACGGCGCCGTGCGGGG-3′, and a reverse primer, ZmETR2-R1, 5′-TGCAGTCTGGAAGGAATTCCGAGCTTCC-3′ were used. To detect the presence of Zmers1b, a forward primer, ZmERS1-F1, 5′-ATGGACGGATGTGATTGCATCGA-3′, and a reverse primer, ZmERS1-R1, 5′-AACAGCTAGAAAATCATTGCGAGCACG-3′ were used. To detect the presence of AtETR1, a forward primer, AtETR1-F1, 5′-GCGGTTGTTAAGAAATTACCCATCACACT-3′, and a reverse primer, AtETR1-R1, 5′-ATCCAAATGTTACCCTCCATCAGATTCAC-3′ were used. To detect the presence of the T-DNA insertion in the etr1-9 mutant, a forward primer, AtETR1-F1, and a reverse primer, T_DNA-L, 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′ were used. To detect the presence of wild-type AtERS1, a forward primer, AtERS1-F1, 5′-CAGAGAGTTCTGTCACTCCTGGAAATGGT-3′, and a reverse primer, AtERS1-R1, 5′-CACAACCGCGCAAGAGACTTTAGCAATAGT-3′ were used. To detect the presence of the T-DNA insertion in the ers1-3 mutant, a forward primer, AtERS1-F2, 5′-GAACAGGGAATTGTTTCTCAAGAAGAAAGC-3′, and the reverse primer, T_DNA-L were used.
qPCR analysis
Plant material was frozen in liquid nitrogen, ground to a fine powder, and 100 mg was resuspended in 1 ml TRIZOL® Reagent (Invitrogen, Carlsbad CA, USA). Following centrifugation, the supernatant was extracted with 200 μl chloroform and centrifuged to separate the phases. RNA was precipitated from the aqueous phase using isopropyl alcohol, the RNA pellet washed with 75% ethanol and resuspended in RNase-free H2O. 1 μg RNA was used to obtain the first-strand cDNA by Omniscript RT Kit (Qiagen, Valencia CA, USA) in a 20 μl reaction. The qPCR analysis was performed using a iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules CA, USA) in 25 μl reactions containing 1× SYBR Green SuperMix 500 nM forward and reverse primers and 10 ng cDNA. Reactions were carried out using the following conditions: 95°C/5 min (1 cycle); 95°C/30 s, 55°C/30 s, 72°C/30 s (35 cycles). To detect the presence of Zmetr2, a forward primer, ZmETR2-F5, 5′-GAGTTCAAACGGTCAGATAAGTCATGG-3′, and a reverse primer, ZmETR2-R8, 5′-TCGGCTCAAGTCTGAAGACGCC-3′ were used. To detect the presence of Zmers1, a forward primer, ZmERS1-F3, 5′-GGCTGCAGTCCGTGTACCTCTTC-3′, and a reverse primer, ZmERS1-R4, 5′-AACAGCTAGAAAATCATTGCGAGCACG were used. Protein phosphatase PP2A (At1g13320) was used as the reference gene for the quantitation of Zmetr2 and Zmers1 expression in Arabidopsis leaves and roots. To detect the expression of PP2A, a forward primer, PP2A-FW, 5′-AGTATCGCTTCTCGCTCCAG-3′ and a reverse primer, PP2A-RV, 5′-GTTCTCCACAACCGCTTGGT-3′ were used. The efficiency of PCR was determined by five 10-fold serial dilutions of the template DNAs in triplicate.
Analysis of etr1-9;ers1-3 double mutants
Seeds of a homozygous etr1-9; heterozygous ers1-3 (i.e., etr1-9/etr1-9;ers1-3/+) plant were surface sterilized, cold treated, and germinated on 0.25× MS medium. etr1-9;ers1-3 double null plants are significantly smaller than plants segregating for the ers1-3 locus and usually die before flowering. Consequently, these seedlings were removed from the segregating population at this stage. The remaining plants were transferred to soil and grew to flowering under a 24 h light cycle at 20°C. The presence of the ers1-3 allele was identified by PCR genotyping the adult plants, and etr1-9/etr1-9;ers1-3/+ plants were crossed with either T:Zmetr2b line L9 or line T:Zmers1b line L11. Screening for the presence of the ers1-3 allele in F1 progeny from each cross was performed by PCR, and plants that were T:Zmetr2b;etr1-9/+;ers1-3/+ or T:Zmers1b;etr1-9/+;ers1-3/+ were allowed to self pollinate. F2 seeds were germinated on 0.25× MS medium and plants exhibiting the small growth phenotype characteristic of etr1-9;ers1-3 double null plants were genotyped by PCR analysis to determine the presence of each maize transgene as well as the presence of the ETR1, etr1-9, ERS1, and ers1-3 loci.
Results
Dominant negative Zmetr2b and Zmers1b mutant receptors confer ethylene insensitivity in Arabidopsis
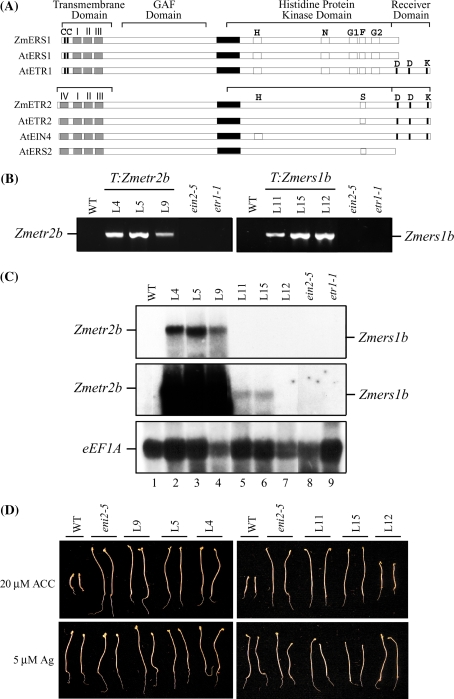
ZmERS1a and ZmERS1b, encoded by ZmERS1a and ZmERS1b (originally designated ZmERS1-14 and ZmERS1-25, respectively), are 96% identical at the amino acid level (Gallie and Young 2004). ZmETR2a and ZmETR2b, encoded by ZmETR2a and ZmETR2b (originally designated ZmETR2-9 and ZmETR2-40, respectively), are 92% identical at the amino acid level of the mature protein (Gallie and Young 2004). The ZmERS1 receptors are similar to Arabidopsis ERS1 in that they contain an N-terminal domain composed of three transmembrane spanning regions, followed by a GAF domain and a His-kinase domain that possesses the amino acid sequences and motifs required for His-kinase activity, but lack a C-terminal receiver domain (Fig. 1(A)). ZmETR2 receptors are similar to Arabidopsis ETR2 in that they contain an N-terminal domain composed of four transmembrane spanning regions, followed by a GAF domain, a His-kinase domain that lacks several of the amino acid sequences and motifs required for His-kinase activity, and a C-terminal receiver domain (Fig. 1(A)).
Fig. 1.
Zmetr2b and Zmers1b function as dominant negative mutants in Arabidopsis. (A) Comparison of ZmERS1 with Arabidopsis subfamily I receptors, i.e., AtETR1 and AtERS1, and comparison of ZmETR2 with Arabidopsis subfamily II receptors, i.e., AtETR2, AtEIN4, and AtERS2. The N-terminal, hydrophobic, transmembrane domains are indicated by gray boxes. Cys-4 and Cys-6, are indicated by the Cs at the left end of the proteins. The five consensus motifs (H, N, G1, F, and G2) within the histidine protein kinase domain (Hua et al. 1998) are indicated, and the aspartate and lysine residues conserved in the receiver domain of ETR1 are indicated. The serine-rich domain (S) is also indicated. The proposed coiled-coil region is indicated by the black box. (B) PCR amplification of the Zmetr2b and Zmers1b transgenes from three lines containing Zmetr2b (i.e., L4, L5, L9) and three lines containing Zmers1b (i.e., L11, L15, L12) confirming the presence of the transgene in the transformants. Wild-type (WT), ein2-5, and etr1-1 plants were included as negative controls. (C) Northern analysis of seedlings of the same Zmetr2b or Zmers1b lines germinated in the dark for 10 days. The level of Zmetr2b and Zmers1b expression was measured using a mixture of Zmetr2b and Zmers1b probes after a 24 h (top panel) or 2 week (middle panel) exposure of the membrane to film. Expression of the translation elongation factor 1A (eEF1A) mRNA was determined as an RNA loading control from the same membrane after it had been stripped (bottom panel). (D) Seeds from the same lines were germinated in the dark for 5 days on media containing either 20 μM ACC or 5 μM AgNO3 to assay for their triple response. Two representative seedlings are shown for each line. Quantitative measurements for hypocotyl and root lengths with standard deviations are shown in Table 1
In order to determine the extent to which the maize ethylene receptors are functionally conserved with those of Arabidopsis, the sequences of ZmETR2b and ZmERS1b were used to amplify the open reading frame of each gene from the inbred B73. The sequence of each was then mutated at the codon corresponding to Cys65 in Arabidopsis ETR1 to change the specified amino acid from Cys to Tyr, as in the etr1-1 mutant receptor, resulting in the generation of the Zmetr2b and Zmers1b mutants. The Zmetr2b and Zmers1b coding regions were placed under the control of the 35S promoter in pBI121 for Agrobacterium-mediated transformation of Arabidopsis, from which several independent transformants homozygous for each transgene were isolated. The presence of the Zmetr2b or Zmers1b transgene in candidate transformant lines was confirmed by PCR (Fig. 1(B)).
To investigate the induction of Zmetr2b or Zmers1b expression and ethylene insensitivity, three independent transformant lines homozygous for either Zmetr2b (i.e., lines L4, L5, L9) or Zmers1b (i.e., lines L11, L12, L15) were germinated and RNA extracted from 10 day old seedlings for Northern analysis. Expression of Zmetr2b was readily observed in each of the lines (lanes 2–4, top panel, Fig. 1(C)). Expression of Zmers1b in lines L11 and L15 appeared lower than Zmetr2b expression and was observed only upon longer exposure (lanes 5–7, middle panel, Fig. 1(C)), despite similar levels of total RNA loaded as determined by the transcript abundance of translation elongation factor 1A (eEF1A) mRNA, which was used as the internal control (bottom panel, Fig. 1(C)). Little to no Zmers1b expression was observed in line L12 (lane 7, middle panel, Fig. 1(C)). The Zmetr2b or Zmers1b probes used for the Northern analysis did not detect the expression of Arabidopsis ethylene receptors (lanes 1, 8–9, middle panel, Fig. 1(C)).
To determine whether expression of Zmetr2b or Zmers1b could repress ethylene responses in Arabidopsis, the triple response of transgenic seedlings was examined when grown on medium containing ACC, the precursor to ethylene. The triple response in Arabidopsis is an ethylene-mediated response of dark-grown seedlings characterized by the radial expansion of the hypocotyl, inhibition of root and hypocotyl elongation, and the presence of an exaggerated apical hook (Neljubow 1901). Wild-type (WT) seedlings grown in the dark on 20 μM ACC exhibited these characteristics (top panel, Fig. 1(D)). Hypocotyl growth in seedlings expressing Zmetr2b, however, was substantially greater than WT seedlings and was similar to the growth of the ethylene insensitive mutant, ein2-5 (top panel, Fig. 1(D); Table 1). Root growth in lines expressing Zmetr2b was greater than WT seedlings but slightly reduced relative to ein2-5 roots (Table 1). No difference in the growth of WT seedlings, the ein2-5 mutant, or lines expressing Zmetr2b was observed during growth on 5 μM Ag2+, which inhibits ethylene perception by likely replacing the copper cofactor in receptors and uncoupling ethylene binding from signal output (Rodríguez et al. 1999). Similar results were obtained for lines expressing Zmers1b in that a lack of a triple response was observed for lines expressing Zmers1b (i.e., L11 and L15) when grown in the dark on 20 μM ACC and that root growth was slightly reduced relative to that observed for the ein2-5 mutant (top panel, Fig. 1(D); Table 1). Ethylene insensitivity in the Zmers1b-expressing lines was observed despite the apparent lower level of Zmers1b expression relative to that of Zmetr2b expression in Zmetr2b-expressing lines, suggesting that the amount of Zmers1b receptor produced was sufficient to confer ethylene insensitivity. In Zmers1b L12, however, only a slight reduction in ethylene sensitivity was observed (top panel, Fig. 1(D); Table 1), which correlated with the lower level of Zmers1b expression than in the other lines, i.e., L11 and L15.
Table 1.
Expression of mutant Zmetr2b and Zmerslb receptors confers ethylene insensitivity in Arabidopsis
| 20 μM ACC | 5 μM Ag2+ | |||||||
|---|---|---|---|---|---|---|---|---|
| Hypocotyl length (mm)a | t-Test | Root length (mm)a | t-Test | Hypocotyl length (mm)a | t-Test | Root length (mm)a | t-Test | |
| WT | 5.0 ± 0.71 | 3.00 ± 1.03 | 14.7 ± 1.12 | 6.69 ± 1.41 | ||||
| ein2-5 | 13.9 ± 1.71 | P < 0.001 | 7.43 ± 2.63 | P < 0.001 | 11.0 ± 3.13 | P < 0.001 | 6.27 ± 1.93 | P = 0.371 |
| T:Zmetr2b L4 | 14.2 ± 1.61 | P < 0.001 | 6.05 ± 1.36 | P < 0.001 | 13.0 ± 3.20 | P < 0.001 | 7.33 ± 1.81 | P = 0.122 |
| T:Zmetr2b L5 | 14.4 ± 1.97 | P < 0.001 | 4.87 ± 1.16 | P < 0.001 | 11.5 ± 2.87 | P < 0.001 | 5.39 ± 1.27 | P < 0.0001 |
| T:Zmetr2b L9 | 13.8 ± 2.36 | P < 0.001 | 6.27 ± 1.76 | P < 0.001 | 12.1 ± 2.40 | P < 0.001 | 7.07 ± 1.58 | P = 0.354 |
| WT | 6.6 ± 1.33 | 3.10 ± 0.92 | 16.9 ± 2.41 | 5.48 ± 0.94 | ||||
| ein2-5 | 17.0 ± 2.98 | P < 0.001 | 6.51 ± 1.56 | P < 0.001 | 14.7 ± 3.78 | P < 0.001 | 5.97 ± 1.56 | P = 0.349 |
| T:Zmerslb L11 | 17.6 ± 3.10 | P < 0.001 | 4.60 ± 0.88 | P < 0.001 | 10.9 ± 3.68 | P < 0.001 | 6.35 ± 1.87 | P = 0.086 |
| T:Zmerslb L15 | 18.0 ± 1.38 | P < 0.001 | 5.88 ± 1.40 | P < 0.001 | 11.7 ± 5.01 | P < 0.001 | 5.51 ± 1.45 | P = 0.921 |
| T:Zmers1b L12 | 8.8 ± 1.37 | P < 0.001 | 3.33 ± 0.97 | P = 0.406 | 17.3 ± 1.95 | P = 0.561 | 7.14 ± 1.97 | P < 0.001 |
aMeasurements taken from 5 day old seedlings
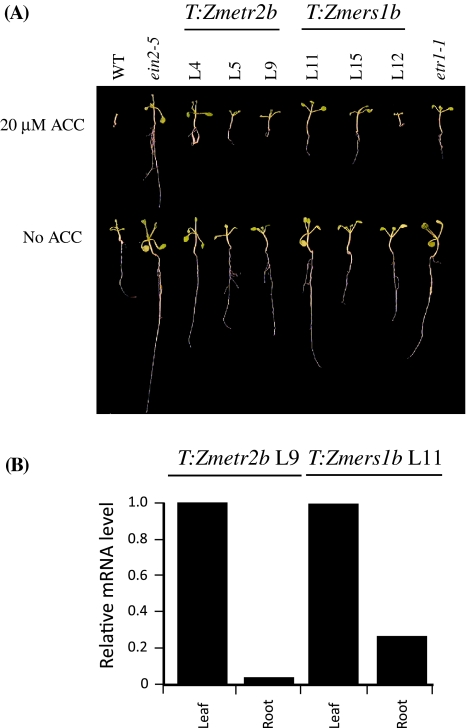
In light-grown seedlings, exposure to elevated levels of ethylene inhibits cotyledon expansion, represses root growth, and delays the emergence of true leaves (Smalle et al. 1997). Light-grown WT seedlings exhibited these characteristics in the presence of 20 μM ACC whereas growth was normal in the absence of ACC (Fig. 2(A)). In Zmetr2b-expressing seedlings, cotyledon expansion and the emergence of the first true leaves were not as inhibited by growth on 20 μM ACC as in WT seedlings and was similar to that observed in ein2-5 and etr1-1 seedlings (Fig. 2(A)). Root growth in Zmetr2b-expressing seedlings grown in the presence of 20 μM ACC was greater than in WT seedlings but less than in ein2-5 and etr1-1 seedlings. qPCR analysis of light-grown, T:Zmetr2b seedlings (line L9) demonstrated that Zmetr2b expression in roots was 1.7% of the level in leaves (Fig. 2(B)), correlating with the observed lower level of ethylene insensitivity in its roots. Cotyledon expansion and the emergence of the first true leaves in Zmers1b-expressing seedlings was also greater than that of WT seedlings in the presence of 20 μM ACC (Fig. 2(A)). Root growth was also greater than in WT seedlings but less than in ein2-5 seedlings and was greater than in Zmetr2b-expressing seedlings (Fig. 2(A)). qPCR analysis of light-grown, Zmers1b-expressing seedlings (line L11) demonstrated that Zmers1b expression in roots was 23.2% of the level in leaves (Fig. 2(B)), consistent with the greater level of root growth observed in Zmers1b seedlings relative to Zmetr2b seedlings (Fig. 2(A)). In line L12, seedling growth was similar to WT seedlings in the presence of 20 μM ACC, supporting the conclusion that there was, at best, a slight reduction in ethylene sensitivity in this line, correlating with its low level of Zmers1b expression.
Fig. 2.
Expression of Zmetr2b and Zmers1b confers ethylene insensitivity in light-grown Arabidopsis seedlings. In (A), the same lines used in Fig. 1 were germinated in the light for 10 days on media in the presence or absence of 20 μM ACC. One representative seedling is shown for each line. In (B), qPCR analysis of leaf and root tissue of light-grown seedlings of lines T:Zmetr2b L9 and T:Zmers1b L11. The level of expression of each transgene in leaves was set to a value of one and transgene expression in roots is shown relative to that in leaves
Growth of the Zmetr2b and Zmers1b-expressing lines was followed throughout their life cycle. A delay in flowering and an increase in rosette size have been reported for ein2-5 (Guzmán and Ecker 1990). An increase in leaf number and a delay in flowering were observed for Zmetr2b and Zmers1b-expressing lines, especially for T:Zmetr2b line L9 (Table 2). No significant change in chlorophyll content or in the chlorophyll a/b ratio was observed for the Zmetr2b and Zmers1b-expressing lines or the ein2-5 mutant for 3 week-old plants (Table 2) but an increase in leaf size (Fig. 3(A)) and a delay in leaf senescence (Fig. 3(B)) was observed for Zmetr2b and Zmers1b-expressing lines as was observed for the ein2-5 and etr1-1 mutants. The increase in leaf size and delay in leaf senescence correlated with the degree of ethylene insensitivity exhibited by each line. For example, T:Zmetr2b line L9 and T:Zmers1b line L11, both of which were highly ethylene insensitive, had substantially larger adult leaves (Fig. 3(C)) and a pronounced delay in leaf senescence whereas leaf size and rate of leaf senescence in T:Zmers1b line L12, which had exhibited only a slight reduction in ethylene sensitivity, was similar to WT plants (Fig. 3).
Table 2.
Phenotypes of Arabidopsis transformants expressing maize Zmetr2b and Zmers1b receptors
| Flowering timea (days) | Leaf number | Chlorophyll a (ng/mg FW) | Chlorophyll b (ng/mg FW) | Chlorophyll a/b ratio | |
|---|---|---|---|---|---|
| WT | 22.0 | 11.1 ± 1.7 | 960 ± 92 | 281 ± 21 | 3.42 |
| ein2-5 | 22.5 | 12.1 ± 2.0 | 923 ± 67 | 296 ± 21 | 3.11 |
| T:Zmetr2b L4 | 22.5 | 12.9 ± 1.9 | 988 ± 86 | 298 ± 27 | 3.32 |
| T:Zmetr2b L5 | 22.5 | 13.8 ± 2.0 | 903 ± 93 | 271 ± 16 | 3.33 |
| T:Zmetr2b L9 | 28.0 | 16.2 ± 1.2 | 965 ± 104 | 304 ± 39 | 3.18 |
aMeasurements taken from plants grown under 100 μmol m−2 s−1
Fig. 3.
Arabidopsis expressing Zmetr2b or Zmers1b exhibit a larger leaf size and a delay in senescence. The same lines used in Fig. 1 were grown under 100 μmol m−2 s−1 for 4 (A) or 7 (B) weeks. Wild-type (WT) plants were included as an ethylene sensitive control. ein2-5 and etr1-1 were included as ethylene insensitive controls. In (C), the size of leaf 6 from 4 week-old plants is compared for each line
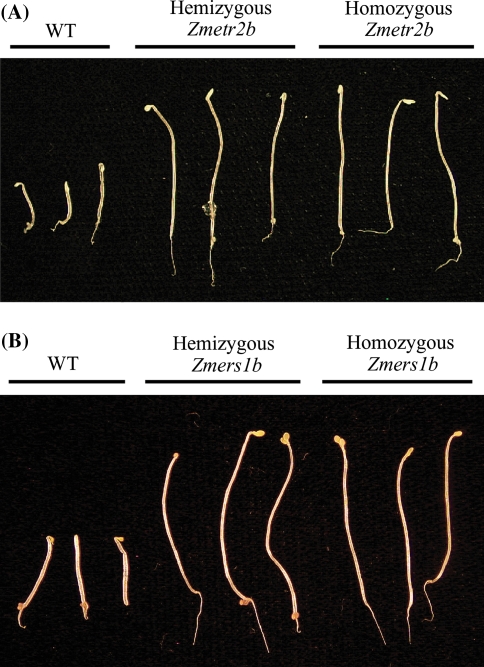
To determine whether expression of the Zmetr2b and Zmers1b transgenes would exert dominance when present in a hemizygous state, T:Zmetr2b line L9 and T:Zmers1b line L11 were crossed with WT plants to generate seed in which each transgene was present in a hemizygous state and their level of ethylene insensitivity in the triple response assay compared to lines homozygous for Zmetr2b or Zmers1b. When grown in the presence of 20 μM ACC, hemizygous Zmetr2b or Zmers1b seedlings exhibited a similar degree of ethylene insensitivity as seedlings homozygous for Zmetr2b or Zmers1b as determined by growth of the hypocotyl and root and the lack of an apical hook (Fig. 4; Table 3). A slight reduction in root growth in hemizygous Zmetr2b seedlings relative to roots of homozygous Zmetr2b seedlings was observed (Table 3). These results suggest that expression of Zmetr2b and Zmers1b can exert dominance when present in a hemizygous state.
Fig. 4.
Zmetr2b and Zmers1b exert dominance in Arabidopsis in a hemizygous state. Line T:Zmetr2b L9 (A) and T:Zmers1b L11 (B) were crossed with wild-type (WT) Arabidopsis to generate seed hemizygous for each transgene. Seed containing each transgene in a hemizygous or homozygous state were germinated in the dark for 5 days on media with 20 μM ACC to assay for their triple response. Three representative seedlings are shown for each line. Quantitative measurements for hypocotyl and root lengths with standard deviations are shown in Table 3. Wild-type (WT) plants were included as an ethylene sensitive control
Table 3.
Zmetr2b and Zmers1b are dominant when present in a hemizygous state in Arabidopsis
| Hypocotyl length (mm)a | t-Test | Root length (mm)a | t-Test | |
|---|---|---|---|---|
| WT | 6.37 ± 1.00 | 3.20 ± 1.14 | ||
| Hemizygous Zmetr2b | 14.0 ± 2.04 | P < 0.001 | 6.01 ± 1.64 | P < 0.001 |
| Homozygous Zmetr2b | 17.2 ± 1.58 | P < 0.001 | 5.83 ± 1.18 | P < 0.001 |
| WT | 8.52 ± 0.65 | 2.78 ± 0.83 | ||
| Hemizygous Zmerslb | 16.5 ± 4.98 | P < 0.001 | 4.78 ± 1.60 | P < 0.001 |
| Homozygous Zmerslb | 17.3 ± 2.74 | P < 0.001 | 4.77 ± 0.92 | P < 0.001 |
aMeasurements for lines containing the Zmetr2b or Zmers1b transgenes were taken from plants grown for 4 or 5 days, respectively, on 20 μM ACC in the dark
Segregation of the ethylene insensitivity phenotype in F2 progeny from a hemizygous Zmetr2b or a hemizygous Zmers1b parent was also performed to determine genetically the copy number of each transgene. Of 156 Zmetr2b progeny analyzed, 123 were ethylene insensitive and 33 were ethylene sensitive as determined by grown in the light in the presence of 20 μM ACC. Of 149 Zmers1b progeny analyzed, 114 were ethylene insensitive and 35 were ethylene sensitive. This represents a segregation 3.7:1 ratio for the Zmetr2b transgene and a 3.26 ratio for the Zmers1b transgene which, for a dominant phenotype, suggests that each transgene is present as a single copy.
Expression of Zmetr2b or Zmers1b confers a high level of insensitivity to ethylene
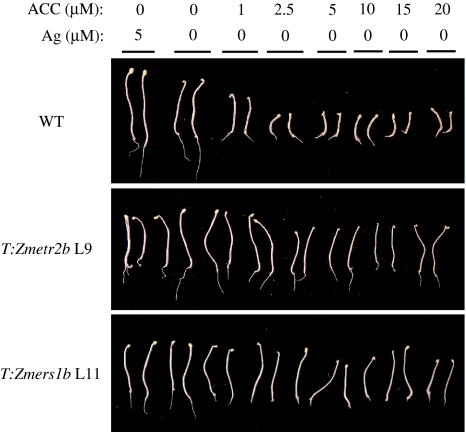
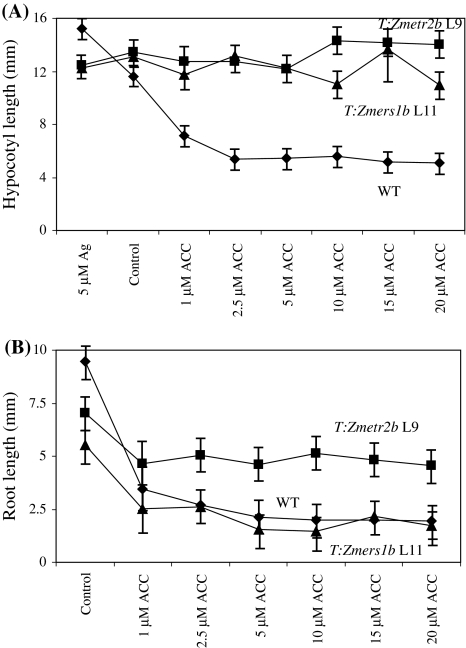
To determine the degree to which expression of Zmetr2b and Zmers1b confers a state of ethylene insensitivity, the growth of T:Zmetr2b line L9 and T:Zmers1b line L11 seedlings was compared to WT seedlings in the triple response assay on medium containing different levels of ACC. Hypocotyl growth was greatest for WT seedlings grown in the presence of Ag2+ (Fig. 5), which was confirmed by quantitative measurements (Fig. 6). In the absence of Ag2+, hypocotyl growth of WT seedlings was reduced relative to growth on Ag2+. Significant inhibition of hypocotyl and root growth and a prominent apical hook in WT seedlings was observed at 1 μM ACC (Fig. 5) and full inhibition was achieved by approximately 2.5 μM ACC (Fig. 6).
Fig. 5.
Zmetr2b and Zmers1b expression confers ethylene insensitivity over a range of ACC concentrations. Seeds of T:Zmetr2b L9 and T:Zmers1b L11 were germinated in the presence of either 5 μM AgNO3 or ACC at the concentrations indicated and grown in the dark for 5 days. Wild-type (WT) plants were included as an ethylene sensitive control. Two representative seedlings are shown for each line
Fig. 6.
Quantitative measurement of the dominance of Zmetr2b and Zmers1b expression in Arabidopsis. Measurements were made of hypocotyl (A) and root (B) lengths of the T:Zmetr2b L9 (squares), T:Zmers1b L11 (triangles), and WT (diamonds) seedlings presented in Fig. 5
Hypocotyl growth of T:Zmetr2b line L9 and T:Zmers1b line L11 seedlings was slightly less than that in WT seedlings in the presence of Ag2+ but greater than that in WT seedlings in the absence of Ag2+ (Fig. 6). The growth of the hypocotyl in T:Zmetr2b line L9 and T:Zmers1b line L11 seedlings was largely unaffected by increasing ACC concentrations up to 5 μM and no apical hook was evident (Fig. 5). A slight reduction in hypocotyl growth was observed in T:Zmers1b line L11 seedlings at higher ACC concentrations (Fig. 6) with a few seedlings exhibiting an apical hook (Fig. 5). T:Zmetr2b line L9 roots were slightly shorter than WT roots when grown in the absence of ACC but their growth was not inhibited to same extent as in WT seedlings by increasing concentrations of ACC (Fig. 6). In contrast, T:Zmers1b line L11 root growth was inhibited to a similar extent as WT roots in the presence of increasing concentrations of ACC (Fig. 6). These results demonstrate that the state of ethylene insensitivity in the hypocotyl conferred by Zmetr2b and Zmers1b expression is maintained over a wide range of ACC concentrations with a reduction in sensitivity to ethylene in roots.
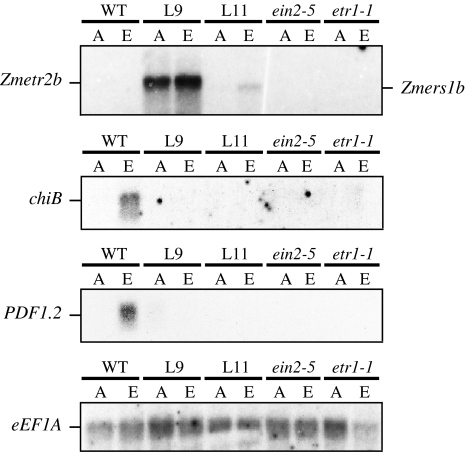
Expression of Zmetr2b and Zmers1b in Arabidopsis conferred insensitivity to ethylene as measured by the lack of ethylene-mediated hypocotyl growth inhibition. To examine whether the dominance of Zmetr2b and Zmers1b mutant expression also inhibited ethylene responses at the molecular level, the expression of genes known to be ethylene inducible was examined. Light-grown T:Zmetr2b line L9 and T:Zmers1b line L11 plants were treated with 100 ppm ethylene for 24 h while additional plants were maintained in air for the same period to serve as an air-treated control. Total RNA was extracted from both the ethylene and air-treated plants for Northern analysis. Expression of Zmetr2b and Zmers1b mRNA was somewhat higher in ethylene-treated plants relative to air-treated plants when normalized to eEF1A mRNA, which was used as the RNA loading control (Fig. 7). As observed in Fig. 1, the Zmetr2b or Zmers1b probes did not cross react with Arabidopsis ethylene receptor mRNA as demonstrated in WT plants (Fig. 7). Expression from chiB and PDF1.2, two ethylene-inducible genes in adult Arabidopsis leaves, was absent in air-treated WT plants but was induced in ethylene-treated plants (Fig. 7). No expression of either gene was observed in Zmetr2b L9 or Zmers1b L11 plants in the presence or absence of ethylene or in ein2-5 or etr1-1 plants as would be expected for these ethylene insensitive mutants (Fig. 7). These results suggest that Zmetr2b or Zmers1b expression represses the induction of ethylene-regulated genes.
Fig. 7.
Induction of ethylene regulated gene expression in Arabidopsis expressing Zmetr2b or Zmers1b. Northern analysis was performed on total RNA from light-grown T:Zmetr2b line L9 and T:Zmers1b line L11 plants that were treated either with 100 ppm ethylene (E) or air (A) for 24 h. Following resolution and transfer of the RNA to membrane, the membrane was probed for the presence of Zmetr2b or Zmers1b mRNA using a combination of both probes (top panel). Northern analysis was also performed for chiB (basic chitinase) or PDF1.2 (plant defensin 1.2) mRNAs which are ethylene regulated. Northern analysis was also performed for the translation elongation factor 1A (eEF1A) which served as an RNA loading control. Wild-type (WT) plants were included as an ethylene sensitive control. ein2-5 and etr1-1 were included as ethylene insensitive controls
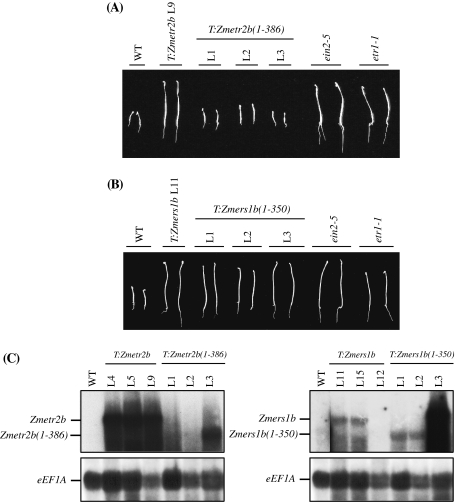
The N-terminal domain of Zmers1b but not Zmetr2b is sufficient to confer ethylene insensitivity
Expression of the N-terminal 349 amino acids of etr1-1, i.e., etr1-1(1-349) can confer ethylene insensitivity in Arabidopsis (Gamble et al. 2002). To determine whether the N-terminal domain of maize ethylene receptors containing the C65Y mutation is sufficient to exert dominance over the Arabidopsis receptors, the portion of the Zmetr2b and Zmers1b coding region corresponding to etr1-1(1-349) was placed under the control of the 35S promoter in pBI121 for Agrobacterium-mediated transformation of Arabidopsis, from which several independent transformants homozygous for each transgene were isolated. The presence of the Zmetr2b(1-386) or Zmers1b(1-350) in the candidate transformants was confirmed by PCR (data not shown). To determine whether expression of Zmetr2b(1-386) or Zmers1b(1-350) was able to confer ethylene insensitivity, seeds from three independent transformed lines for each transgene were germinated in the presence of 20 μM ACC to examine the degree of their triple response. Expression of full-length Zmetr2b resulted in a level of ethylene insensitivity similar to that in ein2-5 or etr1-1 seedlings (Fig. 8(A)) as observed previously. Expression of Zmetr2b(1-386) was unable to confer ethylene insensitivity in the three independent transformed lines examined. This failure was not a result of a lack of Zmetr2b(1-386) expression as its expression was easily detected in at least one of the three lines (Fig. 8(C)). In contrast, expression of Zmers1b(1-350) was sufficient to exert dominance over the Arabidopsis receptors in all three of the transformed lines tested (Fig. 8(B)) and in which Zmers1b(1-350) expression was easily detected (Fig. 8(C)). These results demonstrate that the N-terminal domain of the Zmers1 receptor but not the Zmetr2 receptor is sufficient to confer ethylene insensitivity in Arabidopsis.
Table 4.
Analysis of the ability of Zmetr2b(1-386) or Zmers1b(1-350) to confer ethylene insensitivity
| Hypocotyl length (mm)a | t-Test | Root length (mm)a | t-Test | |
|---|---|---|---|---|
| WT | 5.25 ± 0.39 | 2.81 ± 0.78 | ||
| ein2-5 | 16.0 ± 1.55 | P < 0.001 | 9.44 ± 1.66 | P < 0.001 |
| etrl-l | 15.0 ± 1.35 | P < 0.001 | 7.27 ± 1.83 | P < 0.001 |
| T:Zmetr2b L9 | 16.6 ± 1.26 | P < 0.001 | 5.64 ± 1.27 | P < 0.001 |
| T:Zmetr2b(1-386) L1 | 5.91 ± 0.83 | P < 0.001 | 2.66 ± 0.93 | P = 0.534 |
| T:Zmetr2b(1-386) L2 | 6.17 ± 0.89 | P < 0.001 | 3.05 ± 0.91 | P = 0.321 |
| T:Zmetr2b(1-386) L3 | 5.25 ± 0.59 | P = 0.977 | 2.84 ± 0.83 | P = 0.889 |
| T:Zmers1b L11 | 19.4 ± 1.86 | P < 0.001 | 4.82 ± 1.56 | P < 0.001 |
| T:Zmers1b(1-350) L1 | 16.4 ± 1.31 | P < 0.001 | 3.63 ± 0.97 | P < 0.005 |
| T:Zmers1b(1-350) L2 | 12.4 ± 0.64 | P < 0.001 | 3.07 ± 0.56 | P < 0.05 |
| T:Zmers1b(1-350) L3 | 18.6 ± 0.84 | P < 0.001 | 5.51 ± 0.86 | P < 0.001 |
aMeasurements were taken from 4 day old seedlings grown on 20 μM ACC in the dark
Fig. 8.
The N-terminal domain of Zmers1b but not Zmetr2b is sufficient to exert dominance in Arabidopsis. Arabidopsis was transformed with the portion of the Zmetr2b coding region representing the N-terminal domain, i.e., Zmetr2b(1-386), to generate lines T:Zmetr2b(1-386) L1, L2, and L3 or transformed with the portion of the Zmers1b coding region representing the N-terminal domain, i.e., Zmers1b(1-350), to generate lines T:Zmers1b(1-350) L1, L2, and L3. Seeds from lines homozygous for Zmetr2b(1-386) (A) or Zmers1b(1-350) (B) were germinated in the dark for 5 days on medium containing 20 μM ACC to assay for their triple response. T:Zmetr2b L9 and T:Zmers1b L11 were included as full-length controls. Wild-type (WT) plants were included as an ethylene sensitive control. ein2-5 and etr1-1 were included as ethylene insensitive controls. Two representative seedlings are shown for each line. Quantitative measurements for hypocotyl and root lengths with standard deviations are shown in Table 4. In (C), RNA was extracted from the same seedlings and Northern analysis performed to detect expression of full-length Zmetr2b or Zmetr2b(1-386) (left top panel), full-length Zmers1b or Zmers1b(1-350) (right top panel), or eEF1A (bottom panels) as an RNA loading control from the same membrane after it had been stripped
Zmetr2b and Zmers1b function is dependent on subfamily 1 expression in Arabidopsis
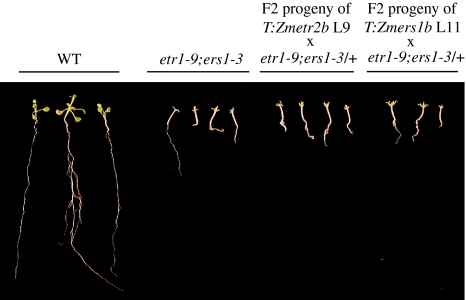
The function of etr1-1 as a dominant negative regulator of ethylene signaling is dependent on the expression of subfamily 1 receptors, which, in Arabidopsis, includes ETR1 and ERS1 (Xie et al. 2006). To examine whether Zmetr2b or Zmers1b exert their dominance through subfamily 1 receptors, the ability of each mutant maize receptor to confer ethylene insensitivity was tested in etr1-9;ers1-3, a double knockout mutant that does not express ETR1 or ERS1 (Qu et al. 2007). Loss of ETR1 and ERS1 expression results in growth phenotypes more severe than those observed for ctr1, which itself is characterized by constitutive ethylene signaling (Kieber et al. 1993). The etr1-9;ers1-3 double mutant plant is extremely small and typically dies before flowering (Qu et al. 2007), precluding crosses with this mutant. As a consequence, the two mutations are typically maintained in plants containing the etr1-9 mutation in a homozygous state and the ers1-3 mutation in a heterozygous state, which are viable and fertile (Qu et al. 2007). The Zmetr2b or Zmers1b transgene was introduced into the etr1-9;ers1-3/+ mutant through crosses with T:Zmetr2b line L9 or T:Zmers1b line L11, generating F1 progeny that were hemizygous for either Zmetr2b or Zmers1b, heterozygous for etr1-9, and either heterozygous for ers1-3 or homozygous for ERS1. F1 progeny identified as ers1-3/+ by PCR genotyping were selfed and F2 seed germinated in the light. Those F2 progeny exhibiting the extremely small growth phenotype typical of etr1-9;ers1-3 plants were genotyped by PCR to verify the presence of the etr1-9 and ers1-3 mutations as well as the absence of the corresponding wild-type loci. The same plants were also genotyped to determine the presence of either the Zmetr2b or Zmers1b transgene. If either Zmetr2b or Zmers1b functioned to confer a state of ethylene insensitivity to etr1-9;ers1-3 plants, then neither the Zmetr2b nor the Zmers1b transgene would be detected in etr1-9;ers1-3 F2 progeny exhibiting the extremely small growth phenotype. If, however, Zmetr2b or Zmers1b failed to function in etr1-9;ers1-3 plants (i.e., in the absence of ETR1 and ERS1 expression), the Zmetr2b or Zmers1b transgene would be expected to segregate in etr1-9;ers1-3 F2 progeny exhibiting the extremely small growth phenotype. Analysis of small F2 progeny from crosses between T:Zmetr2b line L9 and etr1-9;ers1-3/+ plants revealed the presence of the Zmetr2b transgene (Table 5). F2 progeny containing Zmetr2b included plants that were etr1-9;ers1-3 (Table 5) and such plants were substantially smaller than plants exhibiting wild type growth (Fig. 9). Similar results were obtained for the Zmers1b transgene in that Zmers1b was present in several small F2 progeny from crosses between T:Zmers1b line L11 and etr1-9;ers1-3/+ plants and that F2 progeny containing the Zmers1b transgene included plants that were etr1-9;ers1-3 (Table 5). Such plants were substantially smaller than plants exhibiting wild type growth (Fig. 9). These results demonstrate that Zmetr2b and Zmers1b fail to rescue the small growth phenotype of the etr1-9;ers1-3 double mutant, indicating that the function of Zmetr2b and Zmers1b receptors is dependent on the expression of subfamily 1 members.
Table 5.
Zmetr2b and Zmers1b require subfamily 1 receptors to confer ethylene insensitivity in Arabidopsis
| Genotype | Number of small F2 progeny with or without Zmetr2b from a T:Zmetr2b L9 × etr1-9;ers1-3/+ cross | Number of small F2 progeny with or without Zmers1b from a T:Zmers1b L11 × etr1-9;ers1-3/+ cross | ||
|---|---|---|---|---|
| −Zmetr2b | +Zmetr2b | −Zmers1b | +Zmers1b | |
| etr1-9/etr1-9;ers1-3/ers1-3 | 5 | 11 | 0 | 7 |
| etr1-9/etr1-9;ERS1/ers1-3 | 1 | 1 | 1 | 0 |
| etr1-9/etr1-9;ERS1/ERS1 | 0 | 1 | 0 | 0 |
| ETR1/etr1-9;ers1-3/ers1-3 | 0 | 1 | 0 | 0 |
| ETR1/etr1-9;ERS1/ers1-3 | 1 | 1 | 0 | 2 |
| ETR1/etr1-9;ERS1/ERS1 | 0 | 0 | 0 | 0 |
| ETR1/ETR1;ers1-3/ers1-3 | 0 | 0 | 0 | 0 |
| ETR1/ETR1;ERS1/ers1-3 | 0 | 0 | 0 | 0 |
| ETR1/ETR1;ERS1/ERS1 | 0 | 0 | 0 | 0 |
Fig. 9.
Zmers1b and Zmetr2b require expression of subfamily I receptors for their function. T:Zmetr2b line L9 or T:Zmers1b line L11 was crossed with etr1-9;ers1-3/+ plants and F1 progeny that were T:Zmetr2b;etr1-9/+;ers1-3/+ or T:Zmers1b;etr1-9/+;ers1-3/+ were selfed. F2 progeny were germinated in the light for 2 weeks on medium. Examples of plants exhibiting the extremely small growth phenotype typical of etr1-9;ers1-3 plants are shown and were genotyped by PCR analysis to determine the presence of each maize transgene as well as the presence of the ETR1, etr1-9, ERS1, and ers1-3 loci as summarized in Table 5. Also shown are etr1-9;ers1-3 and WT plants
Discussion
Based on sequence conservation and domain structure, maize expresses only two types of ethylene receptors, i.e., ZmERS1 and ZmETR2, in contrast to the five types of receptors expressed in Arabidopsis. In this study, we show that, despite the difference in sequence and types of receptors in the two species, maize receptor function is conserved in Arabidopsis. Introducing the same C65Y mutation into ZmERS1b and ZmETR2b that is present in the etr1-1 dominant negative mutant resulted in dominant negative mutant receptors that conferred ethylene insensitivity in Arabidopsis. Plants expressing Zmers1b or Zmetr2b exhibited many of the phenotypes associated with ethylene insensitive Arabidopsis mutants, including a lack of a triple response when dark-grown seedlings were germinated in the presence of ACC, a larger leaf size and a delay in leaf senescence in light-grown plants, and repression of ethylene-inducible gene expression. Zmers1b and Zmetr2b conferred a state of ethylene insensitivity in Arabidopsis seedlings when present in either a hemizygous or a homozygous state. Rice also expresses only ERS1-like and ETR2-like receptors (Yau et al. 2004), suggesting that the perception of ethylene in monocots may be limited to these two receptor types. Thus, the observations made with the maize ethylene receptors in this study may have broad applicability to monocots in general.
Expression of Zmers1b and Zmetr2b in Arabidopsis resulted in a level of ethylene insensitivity in the hypocotyl of dark-grown seedlings or in leaves of light-grown seedlings comparable to that in ein2-5 or etr1-1 mutants, but in roots, they conferred only partial insensitivity, correlating with their lower expression in roots relative to leaves. The native maize promoter was not used to express Zmers1b and Zmetr2b as there was no assurance that either monocot promoter would function appropriately in Arabidopsis to provide a “native” level of expression. Moreover, the use of an Arabidopsis ethylene receptor promoter to express the maize receptors would not assure a “native” level of expression as the stability and translational efficiency of maize receptor mRNAs and their protein stability will contribute to their steady state level of expression. Using the 35S promoter to express wild-type ZmERS1b and ZmETR2b receptors did not result in an observable phenotype in Arabidopsis (data not shown), suggesting that the level of expression of Zmers1b and Zmetr2b in Arabidopsis is not due to unusual expression patterns or levels. These observations are consistent with the conclusion that maize Zmers1b or Zmetr2b receptors function in Arabidopsis and that the C65Y mutation has a similar effect in both maize receptors.
How such C65Y mutants, best studied in etr1-1, exert dominance over endogenous receptors is not fully understood. The C65Y mutation in etr1-1 perturbs binding of the Cu cofactor at this site, preventing binding of ethylene (Hall et al. 1999; Rodríguez et al. 1999). This is thought to maintain the mutant receptor in a state that constitutively represses activation of the downstream components of the signaling pathway, e.g., EIN2 and EIN3 (Gamble et al. 2002; Xie et al. 2006). Thus, binding of ethylene to wild type receptors fails to activate an ethylene response in the presence of etr1-1 that constitutively represses ethylene responses. This suggests that the dominance of etr1-1 resides in its ability to constitutively repress ethylene responses whether or not ethylene is present by maintaining output signaling to the downstream components of the signaling pathway.
Expression of etr1-1(1-349), which lacks the His-kinase and receiver domains, is sufficient to repress ethylene responses (Gamble et al. 2002), suggesting that this region is responsible for output signaling itself or that it exerts its dominance through interaction with wild-type receptors. The observation that etr1-1(1-349) functions to repress ethylene responses in an etr1-7;ers1-2 mutant, in which a low level of ERS1 expression remains, but not in an etr1-7;ers1-3 mutant, in which no ERS1 expression is detectable, supports the notion that etr1-1(1-349) requires expression of subfamily I members (Xie et al. 2006). Expression of Zmers1b(1-350) was sufficient to cause ethylene insensitivity in Arabidopsis, demonstrating that the putative His-kinase domain is not required for Zmers1b function. Although the ability of the N-terminal region of an Arabidopsis ers1(C65Y) mutant to function in ethylene signaling has not been reported, given the similarity between ETR1 and ERS1 and that the major difference between the two, i.e., the presence of the receiver domain in ETR1 and its absence in ERS1, is lacking in etr1-1(1-349), it is possible that an N-terminal ers1(C65Y) mutant may be capable of repressing ethylene signaling. The observation that Zmers1b(1-350) was sufficient to cause ethylene insensitivity in Arabidopsis indicates that the N-terminal region of the ERS1 class of receptors is capable of output signaling, either directly or through interactions with endogenous receptors, when the C65Y mutation is present. Interestingly, expression of Zmetr2b(1-386) was unable to cause ethylene insensitivity, suggesting that the truncated peptide was not capable of ethylene output signaling, either directly or indirectly, despite the fact that full-length Zmetr2b provided strong ethylene signaling. The failure of Zmetr2b(1-386) to repress ethylene responses was not due to a lack of expression as it was readily detected by Northern analysis. If the function of the N-terminal region of receptor mutants, such as etr1-1(1-349) or Zmers1b(1-350), is mediated through interactions with endogenous receptors, the extent of their conservation with endogenous receptors may determine their interaction and therefore their effectiveness. ZmERS1 is approximately 73% identical with Arabidopsis ERS1 but ZmETR2 is only 45% identical with Arabidopsis ETR2 (Gallie and Young 2004). The difference in conservation with their respective Arabidopsis subfamily receptors may account for the difference in the ability of the Zmers1b(1-350) and Zmetr2b(1-386) peptides to exert dominance. It is possible, however, that the inability of Zmetr2b(1-386) to repress ethylene responses is a result of the instability of the peptide or its inability to fold correctly. It is also possible the C-proximal sequence missing in Zmetr2b(1-386) is required for the dominant function exhibited by the full-length Zmetr2 mutant receptor.
Zmers1b and Zmetr2b failed to confer ethylene insensitivity in the etr1-9;ers1-3 mutant, demonstrating that the function of both mutant receptors requires expression of subfamily I members in Arabidopsis. These observations suggest that Zmers1b(1-350) may function through an interaction with subfamily I members to exert its dominance as proposed for etr1-1(1-349) (Xie et al. 2006), indicating that the N-terminal region of Zmers1 is sufficiently conserved with Arabidopsis subfamily I receptors to permit the functional interaction needed to repress ethylene responses. Such a functional interaction for etr1-1(1-349) may involve maintaining subfamily I receptors in a signaling state or that the interaction with subfamily I receptors permits signaling from the etr1-1(1-349) truncated protein itself (Xie et al. 2006).
ETR1 in Arabidopsis can form covalently linked dimers through a disulfide bond formed between Cys-4 and Cys-6 which may be involved in ethylene signaling (Schaller et al. 1995; Gamble et al. 2002; Qu and Schaller 2004; Chen et al. 2010). etr1-1(1-349) covalently dimerizes with ETR1 (Gamble et al. 2002), demonstrating that the C65Y mutation does not disrupt its interaction with the wild-type receptor. Although mutation of Cys-4 and Cys-6 in etr1-1(1-349) did not abolish N-terminal signaling, the dominant signaling from the truncated receptor was reduced (Xie et al. 2006), raising the possibility that the interaction mediated through the disulfide bonds may contribute to signaling. The maize ZmERS1 and ZmETR2 receptors share structural similarity with the Arabidopsis subfamily I and II receptors, respectively. The Cys-4 and Cys-6 present in ETR1 and ERS1 are conserved in ZmERS1b and in ZmETR2b (Gallie and Young 2004). In ZmETR2b, the cysteines are C-proximal to a putative signal peptide as they are in Arabidopsis subfamily II receptors. Whether any interaction between ZmERS1b and subfamily I receptors requires the formation of disulfide bonds between maize and Arabidopsis receptors remains to be determined. The observation that mutation of Cys-4 and Cys-6 in etr1-1(1-349) did not abolish N-terminal signaling (Xie et al. 2006), however, suggests that the interaction between receptors may also be facilitated by non-covalent interactions.
An observed interaction between ETR1 and ERS2 in Arabidopsis was largely disrupted by SDS treatment, indicating their association is maintained by higher order interactions although 20% of the heterodimers was resistant to the treatment suggesting that the formation of disulfide bonds may contribute to receptor association (Gao et al. 2008; Chen et al. 2010). These findings suggest that receptor interactions are largely maintained through higher order interactions that may include non-covalent interactions between GAF domains, which in other two-component regulators, can dimerize (Aravind and Ponting 1997; Ho et al. 2000; Martinez et al. 2002). The GAF domain may function similarly in ethylene receptors as the GAF domain is sufficient to mediate the interaction between Arabidopsis ETR1 and ETR2 (Gao et al. 2008). These results also demonstrate cross interactions between ethylene receptor subfamilies. The GAF domain is present in the Zmers1b(1-350) and Zmetr2b(1-386) peptides (Fig. 1(A)). The ZmERS1b GAF domain exhibits a high level of conservation with Arabidopsis subfamily I receptor GAF domains whereas the conservation between the GAF domain of ZmETR2b and Arabidopsis subfamily II receptor GAF domains is considerably lower (Gallie and Young 2004). An investigation into the extent to which the GAF domain determines interactions between ethylene receptors may provide greater insight into receptor function, particularly for dominant receptor mutations.
In conclusion, the analysis of maize ethylene receptors in Arabidopsis has revealed considerable functional conservation in the role that Cys65 plays in ethylene signaling, the ability of the full-length Zmers1b and Zmetr2b receptors or Zmers1b(1-350) to repress ethylene responses, and the dependence of Zmers1b and Zmetr2b on subfamily 1 ethylene receptors for their function.
Acknowledgments
The authors thank Dr. G. Eric Schaller for providing the etr1-9;ers1-3 mutant. This work was supported by AgroFresh, Inc., and by the University of California Agricultural Experiment Station.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ACC
1-Aminocyclopropane-1-carboxylic acid
- ACS
ACC synthase
- ACO
ACC oxidase
- β-ME
β-Mercaptoethanol
- EDTA
Ethylenediaminetetraacetic acid
- EtOH
Ethanol
- MS
Murishige and Skoog
- PFD
Photon flux density
- SDS
Sodium dodecyl sulfate
- SSPE
150 mM NaCl, 10 mM NaH2PO4 · H2O, 1 mM EDTA
- TE
Tris–EDTA
References
- Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in plant biology, 2nd edn. Academic Press, Inc., San Diego, 414 pp
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Anderson E. What is Zea mays-a report of progress. Chron Bot. 1945;9:88–92. [Google Scholar]
- Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/S0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Esch JJ, Hall AE, Rodríguez FI, Binder BM. The ethylene-receptor family from Arabidopsis: structure and function. Philos Trans R Soc Lond B Biol Sci. 1998;353:1405–1412. doi: 10.1098/rstb.1998.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Bleecker AB. Ethylene biology. More than a gas. Plant Physiol. 2004;136:2895–2899. doi: 10.1104/pp.104.900122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–358. doi: 10.1016/S1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- Chang C, Stadler R. Ethylene hormone receptor action in Arabidopsis. Bioessays. 2001;23:619–627. doi: 10.1002/bies.1087. [DOI] [PubMed] [Google Scholar]
- Chang C, Stewart RC. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117:723–731. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/S0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Chen QG, Bleecker AB. Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol. 1995;108:597–607. doi: 10.1104/pp.108.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem. 2002;277:19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- Chen YF, Gao Z, Kerris RJ, III, Wang W, Binder BM, Schaller GE. Ethylene receptors function as components of high-molecular-mass protein complexes in Arabidopsis. PLoS One. 2010;5:e8640. doi: 10.1371/journal.pone.0008640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L. Pointing roots in the right direction: the role of auxin transport in response to gravity. Genes Dev. 1998;12:2091–2095. doi: 10.1101/gad.12.14.2091. [DOI] [PubMed] [Google Scholar]
- Ecker JR, Davis RW. Plant defense genes are regulated by ethylene. Proc Natl Acad Sci USA. 1987;84:5202–5206. doi: 10.1073/pnas.84.15.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman LJ. Regulation of root development. Annu Rev Plant Physiol. 1984;35:223–242. doi: 10.1146/annurev.pp.35.060184.001255. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Young TE. The ethylene biosynthetic and perception machinery is differentially expressed during endosperm and embryo development in maize. Mol Genet Genomics. 2004;271:267–281. doi: 10.1007/s00438-004-0977-9. [DOI] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE. Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol. 2002;128:1428–1438. doi: 10.1104/pp.010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Wen CK, Binder BM, Chen YF, Chang J, Chiang YH, Kerris RJ, III, Chang C, Schaller GE. Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J Biol Chem. 2008;283:23801–23810. doi: 10.1074/jbc.M800641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Bleecker AB. Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell. 2003;15:2032–2041. doi: 10.1105/tpc.013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Chen QG, Findell JL, Schaller GE, Bleecker AB. The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 1999;121:291–300. doi: 10.1104/pp.121.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YS, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/S0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldman KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-B. [DOI] [PubMed] [Google Scholar]
- Klee HJ. Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiol. 2004;135:660–667. doi: 10.1104/pp.104.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Abel S, Keller JA, Shen NF, Theologis A. The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1992;89:11046–11050. doi: 10.1073/pnas.89.22.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60:3311–3336. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Harter K. Plant two-component signaling systems and the role of response regulators. Plant Physiol. 2002;128:363–369. doi: 10.1104/pp.010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, Hol WG, Beavo JA. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Natl Acad Sci USA. 2002;99:13260–13265. doi: 10.1073/pnas.192374899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Suttle JC (1991) The plant hormone ethylene. CRC Press, Boca Raton, 337 pp
- Morgan PG, Drew MC. Ethylene and plant responses to stress. Physiol Plant. 1997;100:620–630. doi: 10.1111/j.1399-3054.1997.tb03068.x. [DOI] [Google Scholar]
- Moussatche P, Klee HJ. Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem. 2004;279:48734–48741. doi: 10.1074/jbc.M403100200. [DOI] [PubMed] [Google Scholar]
- Neljubow DN. Uber die horizontale Nutation der Stengel von Pisum sativum und einiger Anderen Pflanzen. Beih Bot Zentralbl. 1901;10:128–139. [Google Scholar]
- O’Malley RC, Rodriguez FI, Esch JJ, Binder BM, O’Donnell P, Klee HJ, Bleecker AB. Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J. 2005;41:651–659. doi: 10.1111/j.1365-313X.2004.02331.x. [DOI] [PubMed] [Google Scholar]
- Qu X, Schaller GE. Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol. 2004;136:2961–2970. doi: 10.1104/pp.104.047126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Hall BP, Gao Z, Schaller GE. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 2007;7:3. doi: 10.1186/1471-2229-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MM. Duplicate genes in maize. Am Nat. 1951;85:105–110. doi: 10.1086/281658. [DOI] [Google Scholar]
- Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE. Ethylene and cytokinin signalling in plants: the role of two-component systems. Essays Biochem. 1997;32:101–111. [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem. 1995;270:12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW. Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 2000;124:1525–1531. doi: 10.1104/pp.124.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Van Montagu M, Straeten DV. Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA. 1997;94:2756–2761. doi: 10.1073/pnas.94.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. Genes nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM (2005) Arabidopsis ethylene signaling pathway. Sci STKE 276, cm4 [DOI] [PubMed]
- Tsuchisaka A, Theologis A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 2004;136:2982–3000. doi: 10.1104/pp.104.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics. 2009;183:979–1003. doi: 10.1534/genetics.109.107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hall AE, O’Malley R, Bleecker AB. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA. 2003;100:352–357. doi: 10.1073/pnas.0237085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Liu Q, Wen CK. Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiol. 2006;142:492–508. doi: 10.1104/pp.106.082628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Ann Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- Yau CP, Wang L, Yu M, Zee SY, Yip WK. Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions. J Exp Bot. 2004;55:547–556. doi: 10.1093/jxb/erh055. [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Programmed cell death during endosperm development. Plant Mol Biol. 2000;44:283–301. doi: 10.1023/A:1026588408152. [DOI] [PubMed] [Google Scholar]
- Zarembinski T, Theologis A. Ethylene biosynthesis and action: a case of conservation. Plant Mol Biol. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]