Abstract
Metastasis is a complex clinical and biological problem presently under intense study, and several model systems are in use to experimentally recapitulate and dissect the various steps of the metastatic process. Genetically engineered mouse models provide faithful renditions of events in tumor progression, angiogenesis, and local invasion that set the stage for metastasis, whereas engrafting of human or mouse tumor tissues into mouse hosts has been successfully exploited to investigate metastatic dissemination and colonization of distant organs. Real-time, high-resolution microscopy in live animals, and comprehensive genetic and molecular profiling are effective tools to interrogate diverse metastatic cancer cell phenotypes as well as the metastatic tumor microenvironment in different organs. By integrating the information obtained with these complementary approaches the field is currently obtaining an unprecedented level of understanding of the biology, molecular basis, and therapeutic vulnerabilities of metastasis.
Introduction
Metastasis is a challenging clinical problem and the cause of most deaths from cancer. As a biological process, metastasis is quite complex as it reflects the many barriers that cancer cells that leave a primary tumor must overcome to generate aggressive secondary lesions [1]. Depending on the cancer type, metastasis may be achieved by just a rare minority of tumor-initiating cells that reach, survive and eventually overtake a distant tissue microenvironment over a long period of time, as in certain types of breast cancer [2,3], or it may represent a relatively common occurrence among primary tumor cell populations that are primed to perform many of the necessary steps and prone to forming rapidly growing metastatic colonies as, for example, in lung adenocarcinoma [4,5].
Certain fundamental properties of metastatic cells, including migration and invasiveness, have been the subject of many studies using a variety of in vitro model systems. Technological advances such as fluorescent or bioluminescent reporter molecules and sophisticated microscopy have allowed sensitive and accurate analysis of these processes and their molecular underpinnings at the single-cell or cell-cluster levels [6,7]. These experimental systems additionally provide comparatively inexpensive methods of choice to screen RNAi, cDNA or chemical libraries for mediators or inhibitors of these cancer cell functions [8–10]. In vitro model systems have contributed to define the role of candidate metastasis genes in particular steps of the metastatic cascade [11,12]. However, these models can provide surrogate systems for the analysis of only a limited set of events in the metastatic cascade, which in vivo involves multiple steps within specific tissue contexts.
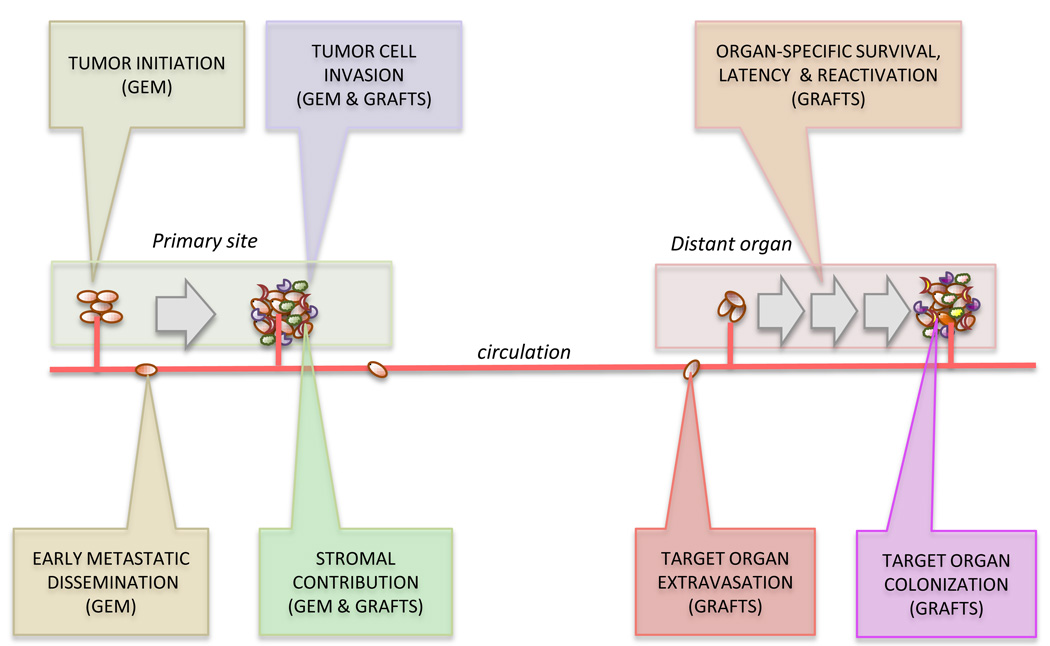
To better understand tumor development and progression in vivo, two general strategies have been pursued in mice: genetically engineered models of cancer, and transplantable tumor model systems. These strategies provide complementary approaches to the dissection of specific metastatic steps. Cancer models driven by the introduction of oncogenic mutations in a tissue-specific manner can faithfully recapitulate important aspects of tumor initiation, local progression, and response to therapy [13,14]. In these models, cancer develops with high penetrance in a stepwise manner, enabling the study of tumor initiation and early steps of metastatic dissemination (Figure 1). However, metastasis in these models is often restricted to lymph nodes and the lungs, or is missing altogether (Table 1). Syngeneic and xenograft models in which mouse or human cancer cells are introduced into immunocompatible or immunocompromised mice provide at present methods of choice to experimentally address metastatic dissemination to, and colonization of relevant organs. Syngeneic models enable the study of the complete microenvironmental interface in the mouse but are limited to study mouse cancer cell metastasis. Conversely, in spite of the caveat of an incomplete immune system, xenograft models provide a superior alternative to the study of metastasis of human cancer cells in vivo.
Figure 1.
Contribution of different mouse models to the various steps of metastatic dissemination. GEM: Genetically engineered mouse models, GRAFTS: xeno- or allograft transplantation.
Table 1.
Metastatic patterns of dissemination
| Tumor Type | Clinical Site Of Relapse | Metastatic pattern observed in GEMs | Metastatic sites in transplantable models |
|---|---|---|---|
| Breast | Bone, lungs, brain, liver, lymph nodes | Lungs, lymph nodes | Bone, lungs, brain, liver, lymph nodes |
| Prostate | Bone, lymph node | Lymph node, lung, rare bone | Bone, lymph node |
| Lung | Lung, brain, bone, lymph node | Lymph node | Lung, brain, bone, lymph node |
| Melanoma | Multiple organs, mostly lymph node, lungs, liver, brain and bone. | Lymph node and lungs | Lymph node, lungs, bone, brain. |
Here we present an overview of the advantages and disadvantages of the principal mouse model systems currently used for the study of metastasis (Table 2), and how the combined use of these systems complemented with in vitro models is yielding an increasingly robust understanding of the multiple modes and steps of metastasis.
Table 2.
Advantages and disadvantages of genetically engineered and transplantable models for the study of metastasis
| Tumor Model | Advantages | Disadvantages |
|---|---|---|
| GEM |
|
|
| Xenografts |
|
|
| Allografts |
|
|
Modeling early metastatic steps
Cancer cells in a primary tumor have already acquired a number of aggressive functions that will remain important throughout the rest of their metastatic progression. These functions generally include motility, invasiveness, resistance to hypoxia and reactive oxygen species, survival after detachment, and evasion of immune surveillance. The accumulation of these properties in human cancer cells is thought to occur in the context of the primary tumor, and genetically engineered tumors in mice provide powerful models to delineate the acquisition of these functions (Figure 1).
It has been proposed that secondary tumor formation involves rare cell variants that have accumulated a complete set of genetic mutations in the primary tumor that enables these cells to grow in a distant organ [15]. However, this hypothesis has been challenged based on the detection of widespread gene expression patterns in primary tumors that strongly predict metastatic competence [16], and the analysis of growth dynamics of human breast primary tumors and metastases [17]. Genetically marked transplantable tumors in mouse mammary carcinoma models were used to demonstrate that cancer cells can disseminate at the premalignant stage [18,19]. Moreover, ex vivo genetic manipulation of tumor-derived cells prior to implantation in cleared mouse mammary fat pads was used to identify genes that influence this process [19]. Other transplantation methods using inducible genetically engineered oncogenes have shown that non-transformed mammary cells introduced in the mouse circulation can extravasate in the lungs, and give rise to tumor foci in the lung parenchyma upon oncogene induction [20]. However, the question of whether early disseminated tumor cells (DTCs) are responsible for the outgrowth of secondary tumors remains a matter of debate. In one approach, early-stage mouse tumors were transplanted for variable time periods, and metastatic outgrowth measured upon resection after a fixed number of weeks. The results suggested that early DTCs are inefficient at initiating secondary tumors [19].
Xenografting of limiting dilutions of cancer cells are standard for evaluating tumor-initiating capacity [21], and has been instrumental in linking epithelial-mesenchymal trans-differentiation (EMT) to tumor-initiating “cancer stem” cell phenotype in breast cancer [22]. A recently introduced interleukin-2 receptor-deficient NOD/SCID mouse strain has been used to demonstrate that one single human melanoma cell can develop into a lethal, metastatic tumor [5]. Tumor engrafting models are extensively used to delineate the role of genes of interest in early metastatic steps at orthotopic sites. Both, xeno- and allo-transplantation have been successfully used to delineate the role of genes and miRNAs in the invasive capacity of tumor cells [12,23–25], and to dissect the distinct contributions of particular genes in the context of mammary tumors versus the context of lung metastasis [11,26].
The importance of the microenvironment in tumor progression is widely recognized, and genetically engineered mouse models have provided key insights into the relevance of the tumor microenvironment in metastasis [27]. Dissenting the contribution of different components of the tumor stroma represent a significant experimental challenge. Nonetheless, progress has been made with the use of bone marrow transplantation from genetically engineered donor mice. Such bone marrow transplants into tumor bearing animals can shed light into the role of a candidate metastasis gene expressed in either the cancer cells or a particular cell type in the tumor microenvironment. Recent examples of this approach have revealed the specific contribution of macrophage-derived cathepsins in pancreatic cancer and breast cancer cell invasion [28]. Similar genetic manipulations in a double transgenic PyMT/RAG1−/− breast cancer model have implicated CD4 T effector cells in tumor cell invasion and metastasis [29]. This methodology is highly promising to define the role of the innate and adaptive immune system in tumor cell dissemination.
Metastatic dissemination and colonization: transplantable tumor models
Establishment of secondary tumors imposes different demands on disseminated cancer cells depending on the target organ. Xenograft models provide an effective system to investigate secondary organ colonization of human cells, and remain the model of choice for pre-clinical studies of human tumor-derived cells (Table 3). Intracardiac inoculation of cancer cells into the arterial circulation of mice allows the systemic distribution of these cells to all organs for the analysis of metastatic functions including organ-specific extravasation, survival in the newly invaded parenchyma, retention of tumor-reinitiating capacity, and overt colonization [30]. In contrast, tail-vein inoculation forces cancer cells to lodge in lung capillaries, which allows an assessment of lung extravasation and colonization functions [31]. Carotid artery inoculation likewise targets cancer cells to the brain [32].
Table 3.
Sources and utilization of clinical material.
| Clinical source | Research use |
|---|---|
| Primary tumor biopsy | Gene validation, xenograft transplantation for stem/progenitor cell capacity, establishment of orthotopic tumors, gene discovery, therapeutic response. |
| Metastatic tumor, lymph node biopsies | Validation of gene expression by IF, establishment of xenograft tumors, gene discovery and functional studies. |
| CTCs (Circulating tumor cells) | Analysis of genetic changes, validation of marker expression, single cell xenograft. |
| DTCs (Disseminated tumor cells from bone marrow aspirates) | Similar as CTCs. In addition, essential contribution to metastatic dormancy and survival studies. |
In vivo selection of organ-specific metastatic variants from human malignant samples and cell lines, coupled with analysis of mRNA and microRNA expression patters has allowed the identification of organ-specific metastasis genes and functions. By comparing the results of this type of analysis with clinical gene expression data sets, it is possible to identify metastasis-associated genes of clinical relevance. Several gene sets have been identified in this manner that are associated with organ-specific relapse in breast cancer patients [33,34]. This information in turn can be used to guide functional studies for the discovery of genes that mediate metastasis, including genes that prime cancer cells for extravasation across the tight endothelial walls in the lungs or the brain [11,34]. Variants of this approach have identified new mediators of circulating cancer cell interaction with vascular capillary walls [35], and genes that support the tumor-reinitiating capacity of disseminated cancer cells in the lung parenchyma [36].
Another approach is based on interrogating clinical gene expression data sets for associations between specific pathways and particular disease outcomes. By combining this information with functional assays it was recently shown that a hyperactive Wnt pathway in lung adenocarcinoma tumors supports aggressive multi-organ metastasis to brain and bones [37], whereas a high level of Src activity in breast tumors endows disseminating cancer cells with an enhanced capacity to survival in the bone marrow microenvironment and may contribute to late-onset bone metastasis [2].
Transplantation studies have also illuminated other aspects of metastasis including the systemic effect of transplanted tumors. Allografted tumors can cause the mobilization of VEGFR1+ bone marrow-derived cells to the lungs for the establishment of a “pre-metastatic niche” to host incoming cancer cells [38]. The use of xenografts has also allowed a demonstration that an indolent tumor can be stimulated to grow by bone marrow-derived cells mobilized by systemic signals from a separate tumor [39]. Inducible RNAi technology in a syngeneic model of lung metastasis has shown that recruitment of endothelial progenitor cells is essential for the angiogenic switch that facilitates macrometastatic growth [40].
A self-seeding mechanism by which circulating cancer cells reinfiltrate and populate the tumor of origin has been proposed to explain certain aspects of tumor growth and metastatic population dynamics [41]. A recent experimental demonstration of this phenomenon employed xenograft and allograft models of orthotopic tumor seeding by cancer cells entering the circulation from a separate tumor mass, from lung metastatic nodules, or from direct intra-arterial inoculation [42].
Although genetically engineered mouse models provide good systems for the pre-clinical evaluation of therapeutic agents [43,44], the response of human cancer cells to therapy in vivo requires the use of xenograft models. Of particular relevance is the xenografting of metastatic cell lines in orthotopic locations, followed by resection of the primary tumors and initiation of therapy. This set up approximates the situation observed in patients with advanced disease [45].
Visualizing metastasis
Tracking cancer cells in real time in whole animals has provided a tremendous advantage in the dynamic monitoring of metastatic development. Of particular relevance, multimodality imaging technology such as the triple fusion protein reporter with herpes simplex virus 1 thymidine kinase (HSV1-TK), fused to enhanced green fluorescent protein (eGFP), and firefly luciferase has enabled the use of nuclear imaging, bioluminiscence and fluorescence imaging in a single experiment. In addition, the same tissues can be further analyzed by histological detection of the fluorescence GFP in frozen sections, or immunohistochemical detection of the reporter [46]. Variants of these reporters with different emission wavelengths, duration of the signal, and split versions for complementation studies exist that are useful in different applications [47]. The development of more advanced reporter systems based on similar technology has also permitted the in vivo monitoring of gene pathway activity and inhibition [48].
Intravital microscopy is a key resource to visualize cancer cells performing various metastatic steps in situ (reviewed in [49]). Macrophage-assisted tumor cell migration involving an autocrine loop has relied on the use of this approach for the analysis of polyoma middle-T transgenic mammary tumors in mice [50]. In this assay, a fine needle containing a chemoattractant is introduced into the tumor, and migrating cells can be recovered for further analysis. The use of this technology has also shed light on the interaction between migrating cancer cells and macrophages (reviewed in [51]).
Long-term spinning disk confocal microscopy represents another application of advanced microscopy and improved tracing techniques to the examination of cellular movements and interactions in the tumor microenvironment. This imaging technique enables the dynamic visualization of stromal cells in defined tumor areas, and its combination with fluorescent reporter knock-in mice in which different immune cell types are marked, or injection of fluorescent antibodies or dextrans, enables the rapid collection of images for studying behavior of moving cells [52]. Rapid and prolonged visualization of tumor growth parameters such as angiogenesis, lymphangiogenesis, tissue viability, and response to therapy in larger tumor areas have also been achieved by the introduction of optical frequency domain imaging. This approach has the advantage of not requiring labeling or contrasting agents [53].
Multiphoton laser scanning microscopy has enabled live imaging of challenging metastatic sites like the brain. The use of cranial windows and fluorescently labeled tumor cells and dextrans has allowed a visualization of brain metastatic cells closely interacting with the microvasculature [54]. A longer term imaging study using genetic labeling of the vasculature has made possible to image several steps in the process of brain metastasis, demonstrating active extravasation of tumors cells into the brain, perivascular growth, and responses to therapy [55]. Advances in the visualization of protease activity such as the use of ACPP (activatable cell penetrating peptide) has also enabled the in vivo labeling of MMP protease activity in xenograft and genetically engineered tumors and small metastatic foci in the lungs [56]. The rapidly evolving field of intravital microscopy is likely to provide in the future new means to analyze metastatic behavior and performing detailed functional analysis of genetically engineered cells.
Conclusions and perspectives
A great deal of progress has been made in the study of the various metastatic steps in the past few years. However, many clinically relevant questions remain unanswered, owing partly to a lack of suitable animal models. The problem of metastatic dormancy and the role of the immune system in metastasis are prominent in this regard. Clinically, metastasis may take decades to become manifest in certain types of cancer [4], and there is a dearth of mouse models to study the biology of the latent metastatic state [2]. Both of these questions would benefit from the development of additional immunocompromised models (rev. in [57]). Progress in this direction is being made with human tissues implanted in the mouse to serve as recipients for human cancer cells [58,59]. Such systems may provide important new models for pre-clinical studies of anti-metastatic agents. Similarly, the ability to uncouple primary tumor growth from specific metastatic steps is a coveted feature of genetically engineered mouse models of cancer. The use of reversibly inducible oncogenic alterations [60–62], and stable [63] or reversible [64] RNAi targeting of genes of interest represents only some of the new tools that can be applied to the analysis of metastatic progression in genetically engineered mouse models. The development of new and improved experimental metastasis methods, and a better integration of the results with clinically data promises to support a sustained expansion of our ability to understand and fight metastasis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 2. Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16(1):67–78. doi: 10.1016/j.ccr.2009.05.017.. ••This article (and ref.37) pioneered the analysis of clinical gene expression analysis with xenograft models of metastasis to identify signaling pathways involved in metastatic dissemination.
- 3.Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4(6):448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 5.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menon MB, Ronkina N, Schwermann J, Kotlyarov A, Gaestel M. Fluorescence-based quantitative scratch wound healing assay demonstrating the role of MAPKAPK-2/3 in fibroblast migration. Cell Motility and the Cytoskeleton. 2009;66(12):1041–1047. doi: 10.1002/cm.20418. 10.1002/cm.20418. [DOI] [PubMed] [Google Scholar]
- 7.Ghajar CM, Bissell MJ. Extracellular matrix control of mammary gland morphogenesis and tumorigenesis: insights from imaging. Histochem Cell Biol. 2008;130(6):1105–1118. doi: 10.1007/s00418-008-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gobeil S, Zhu X, Doillon CJ, Green MR. A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev. 2008;22(21):2932–2940. doi: 10.1101/gad.1714608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 10.Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430(7003):1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 11.Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, Gomis RR, Manova-Todorova K, Massague J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446(7137):765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 12.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nat Rev Cancer. 2002;2(4):251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- 14.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7(9):645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 15.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 16.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418(6900):823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 17.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9(4):302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 18.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, Forni G, Eils R, Fehm T, Riethmuller G, Klein CA. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13(1):58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 19. Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, Pai SY, Ho IC, Werb Z. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13(2):141–152. doi: 10.1016/j.ccr.2008.01.011.. ••Using a mouse mammary tumor model and sofisticated hyperplasia transplantation techniques, the authors demonstrate the timewise dissemination and metastatic development, and demosntrate the role of GATA3 in the process.
- 20.Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321(5897):1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, Coukos G, Zhang L, Huang Q. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11(11):1297–1304. doi: 10.1038/ncb1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massague J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, Berman T, Joyce JA. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24(3):241–255. doi: 10.1101/gad.1874010.. ••See ref.29.
- 29. DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16(2):91–102. doi: 10.1016/j.ccr.2009.06.018.. ••References 28 and 29 demonstrate the power of bone marrow transplantation technology to uncover the role of stromal components in cancer progression.
- 30.Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103(2):197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart IR, Fidler IJ. Role of organ selectivity in the determination of metastatic patterns of B16 melanoma. Cancer Res. 1980;40(7):2281–2287. [PubMed] [Google Scholar]
- 32.Lorger M, Felding-Habermann B. Capturing Changes in the Brain Microenvironment during Initial Steps of Breast Cancer Brain Metastasis. Am J Pathol. 2010 doi: 10.2353/ajpath.2010.090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799.. ••See ref. 34
- 34. Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021.. ••References 33 and 34 have pioneered the combined analysis of xenograft-derived and clinical sample gene expression data to identify organ-specific genes.
- 35.Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5(4):365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- 36.Gupta GP, Perk J, Acharyya S, de Candia P, Mittal V, Todorova-Manova K, Gerald WL, Brogi E, Benezra R, Massague J. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104(49):19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138(1):51–62. doi: 10.1016/j.cell.2009.04.030.. ••See ref.2
- 38.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, Zhu Z, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438(7069):820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McAllister SS, Gifford AM, Greiner AL, Kelleher SP, Saelzler MP, Ince TA, Reinhardt F, Harris LN, Hylander BL, Repasky EA, Weinberg RA. Systemic endocrine instigation of indolent tumor growth requires osteopontin. Cell. 2008;133(6):994–1005. doi: 10.1016/j.cell.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao D, Nolan DJ, Mellick AS, Bambino K, McDonnell K, Mittal V. Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science. 2008;319(5860):195–198. doi: 10.1126/science.1150224. [DOI] [PubMed] [Google Scholar]
- 41.Norton L, Massague J. Is cancer a disease of self-seeding? Nat Med. 2006;12(8):875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- 42.Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, Massague J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139(7):1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Palma M, Mazzieri R, Politi LS, Pucci F, Zonari E, Sitia G, Mazzoleni S, Moi D, Venneri MA, Indraccolo S, Falini A, et al. Tumor-targeted interferon-alpha delivery by Tie2-expressing monocytes inhibits tumor growth and metastasis. Cancer Cell. 2008;14(4):299–311. doi: 10.1016/j.ccr.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15(3):220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009;15(3):232–239. doi: 10.1016/j.ccr.2009.01.021.. ••The authors demonstrate acceleration of metastasis upon anti-angiogenic therapies. Importantly, the authors mimmick clinical situations by removing orthotopically growing tumors before starting the therapy.
- 46.Ponomarev V, Doubrovin M, Serganova I, Vider J, Shavrin A, Beresten T, Ivanova A, Ageyeva L, Tourkova V, Balatoni J, Bornmann W, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging. 2004;31(5):740–751. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 47.Dothager RS, Flentie K, Moss B, Pan MH, Kesarwala A, Piwnica-Worms D. Advances in bioluminescence imaging of live animal models. Curr Opin Biotechnol. 2009;20(1):45–53. doi: 10.1016/j.copbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korpal M, Yan J, Lu X, Xu S, Lerit DA, Kang Y. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nat Med. 2009;15(8):960–966. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- 49.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3(12):921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 50.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67(6):2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 51.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Egeblad M, Ewald AJ, Askautrud HA, Truitt ML, Welm BE, Bainbridge E, Peeters G, Krummel MF, Werb Z. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech. 2008;1(2–3):155–167. doi: 10.1242/dmm.000596. discussion 165.. ••The authors utilize spinning disk confocal microscopy to examine behavior of various genetically labeled cell types in different tumor microenvironments of the same mouse, enabling functional analysis of disctinct cell population in the same tumors.
- 53.Vakoc BJ, Lanning RM, Tyrrell JA, Padera TP, Bartlett LA, Stylianopoulos T, Munn LL, Tearney GJ, Fukumura D, Jain RK, Bouma BE. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat Med. 2009;15(10):1219–1223. doi: 10.1038/nm.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carbonell WS, Ansorge O, Sibson N, Muschel R. The vascular basement membrane as "soil" in brain metastasis. PLoS ONE. 2009;4(6):e5857. doi: 10.1371/journal.pone.0005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16(1):116–122. doi: 10.1038/nm.2072.. ••Through the use of multiphoton laser scaning microscopy, the authors are able to track individual brain metastatic cell fate in relationship to the blood vessels over minutes to months, establishing essential steps in the process of brain metastasis and response to therapy.
- 56.Olson ES, Aguilera TA, Jiang T, Ellies LG, Nguyen QT, Wong EH, Gross LA, Tsien RY. In vivo characterization of activatable cell penetrating peptides for targeting protease activity in cancer. Integr Biol (Camb) 2009;1(5–6):382–393. doi: 10.1039/b904890a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macchiarini F, Manz MG, Palucka AK, Shultz LD. Humanized mice: are we there yet? J Exp Med. 2005;202(10):1307–1311. doi: 10.1084/jem.20051547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Xia TS, Liu XA, Ding Q, Du Q, Yin H, Wang S. A novel orthotopic and metastatic mouse model of breast cancer in human mammary microenvironment. Breast Cancer Res Treat. 2010;120(2):337–344. doi: 10.1007/s10549-009-0391-4. [DOI] [PubMed] [Google Scholar]
- 59.Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101(14):4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2(6):451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 61.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105(13):5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4(2):199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 63.Stern P, Astrof S, Erkeland SJ, Schustak J, Sharp PA, Hynes RO. A system for Creregulated RNA interference in vivo. Proc Natl Acad Sci U S A. 2008;105(37):13895–13900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ, Lowe SW. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39(7):914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]