Abstract
Irreversible inactivation of α-thrombin (T) by the serpin, heparin cofactor II (HCII), is accelerated by ternary complex formation with the glycosaminoglycans (GAGs) heparin and dermatan sulfate (DS). Low expression of human HCII in Escherichia coli was optimized by silent mutation of 27 rare codons and five secondary Shine–Dalgarno sequences in the cDNA. The inhibitory activities of recombinant HCII, and native and deglycosylated plasma HCII, and their affinities for heparin and DS were compared. Recombinant and deglycosylated HCII bound heparin with dissociation constants (KD) of 6 ± 1 and 7 ± 1 μM, respectively, ~6-fold tighter than plasma HCII, with KD 40 ± 4 μM. Binding of recombinant and deglycosylated HCII to DS, both with KD 4 ± 1 μM, was ~4-fold tighter than for plasma HCII, with KD 15 ± 4 μM. Recombinant HCII, lacking N-glycosylation and tyrosine sulfation, inactivated α-thrombin with a 1:1 stoichiometry, similar to plasma HCII. Second-order rate constants for thrombin inactivation by recombinant and deglycosylated HCII were comparable, at optimal GAG concentrations that were lower than those for plasma HCII, consistent with its weaker GAG binding. This weaker binding may be attributed to interference of the Asn169 N-glycan with the HCII heparin-binding site.
Keywords: Recombinant heparin cofactor II, Serpin, Thrombin, Serine protease inactivation
Thrombin, the central proteinase in the coagulation cascade, cleaves soluble fibrinogen to insoluble fibrin which polymerizes to form a blood clot. Thrombin is inactivated by circulating plasma heparin cofactor II (HCII),1 a member of the superfamily of serine proteinase inhibitors (serpins) [1–3]. Serpins possess a reactive center loop (RCL) that is cleaved by their target proteases, with formation of a stabilized, covalent serpin–enzyme complex in which the protease active site is distorted. The thrombin–HCII reaction is accelerated by binding of the sulfated glycosaminoglycans (GAGs), heparin and dermatan sulfate (DS) [4–6], and various other polyanions [7–10] in a ternary complex, and plays an important role in prevention of arterial thrombosis [11]. The thrombin–HCII mechanism utilizes an allosteric interaction of a unique acidic region in the HCII N-terminus with thrombin exosite I [1,12,13], as well as high Mr GAG template formation between thrombin exosite II and the GAG-binding site on helix D of HCII [14,15]. The Glu53-Asp75 sequence in the HCII N-terminus contains two hirudin-like repeats that are absent in antithrombin (AT) [1,13,16,17], and the crystal structure of the HCII·S195A-thrombin Michaelis complex shows direct contact of HCII residues 56–72 with exosite I [18]. This sequence becomes available for allosteric thrombin interaction upon GAG binding of HCII in ternary complexes with thrombin [18,19]. The template mechanism is characterized by bell-shaped logarithmic dependencies of proteinase inactivation rates on the GAG concentration at fixed inhibitory serpin concentrations, and has been demonstrated for GAG-accelerated thrombin inactivation by the serpins AT [20], HCII [4,7,14,21], plasminogen activator inhibitor-1 (PAI-1) [22], and protease nexin-1 [23].
Mutants of HCII are valuable tools for studying its inhibitory mechanism and other functional properties. Previous studies have utilized partially purified Escherichia coli lysates of recombinant human HCII, mutated in the reactive site and the GAG-binding site, respectively, for qualitative characterization of the bifurcated inhibitory and substrate HCII partitioning mechanism [24], and identification of the Arg and Lys residues involved in the partially overlapping binding sites for heparin and DS [21,25–27]. N-terminal deletion mutants were created to probe the role of the HCII Glu53-Asp75 sequence in the interaction with thrombin exosite I [19]. Expression of glycosylated and sulfated human HCII in insect cells [28,29] and Chinese hamster ovary cells [1] has proven useful for studying the biological roles of these posttranslational modifications, and their possible effects on GAG binding. In most of these studies, HCII was detected by immunoblotting and complex formation with radioiodine-labeled thrombin. Quantitative binding and kinetic studies of the thrombin–HCII mechanism require substantial amounts of HCII, and one of the goals of this work was to optimize heterologous expression in E. coli for fast, quantitative production of wild-type and mutant HCII.
Numerous serpins have been expressed in E. coli with varying degrees of success [30]. Low HCII expression levels are in part due to the presence of 27 rare codons [31,32] in the reported HCII sequence [33], as shown in Fig. 1. Of these, 13 are rare Arg codons, with one AGG AGG tandem repeat, and several rare codons in close proximity of one another. AGGAGG is also present in the polypurine sequence (AAGGAGG), known as the Shine–Dalgarno (SD) sequence [34], which is part of the ribosome-binding site (RBS). Four additional secondary SD-like polypurine sequences were identified in the HCII cDNA. Substantial improvement of HCII expression was achieved by substitution of the rare codons with their nonrare counterparts with the highest codon usage in abundantly expressed E. coli genes during the exponential growth phase [35], and silent mutation of the secondary SD-like sequences. The expressed serpin, with an additional Met residue at the N-terminus [36], lacks Tyr sulfation in positions 60 and 74, and N-glycosylation at positions Asn30, Asn169, and Asn368. This study quantitates the dissociation constants (KD) for binding of representative template-forming heparin and DS to HCII expressed in E. coli, in comparison with native and deglycosylated HCII from human plasma, and compares the kinetic profiles of heparin- and DS-accelerated thrombin inactivation by these three HCII forms in a ternary complex model. Our findings indicate that, although recombinant and deglycosylated HCII bind GAGs 4-to 6-fold tighter than plasma HCII, their inhibitory efficiencies at optimal GAG concentrations are essentially comparable, justifying the use of E. coli recombinant HCII for kinetic studies that dissect the encounter, loop insertion, and complex stabilization steps of the thrombin–HCII interaction mechanism.
Fig. 1.
Amino acid sequence of human HCII. HCII expressed in E. coli possesses an N-terminal Met residue, not included in the sequence numbering. Rare codons are indicated in underlined boldface. Positions of Tyr sulfation (SO4) and Asn N-glycosylation (CHO) in plasma HCII are indicated in the sequence. The Leu–Ser scissile bond in the RCL is indicated by a diamond (◇).
Materials and methods
Proteins and materials
The following materials were purchased: oligonucleotide primers (Midland Certified Reagents); Tartoff-Hobbs modified Terrific Broth (TB), MagicMedia, and pfu DNA polymerase (Invitrogen); Luria-Bertani (LB) broth Miller (Fisher Scientific); the restriction enzymes NcoI and SnB1, and DNA markers (New England Biolabs, Beverly, MA); E. coli BL21-CodonPlus (DE3)-RIPL competent cells, DH5α cells, XL1-Blue supercompetent cells, and QuikChange mutagenesis kits (Stratagene);Wizard plus SV Minipreps DNA extraction kits, and Modified Trypsin-Gold (Promega); heparin-Sepharose, Mono S and Mono Q HR 5/50 GL 5 ml prepacked FPLC columns (GE Healthcare); EDTA-free complete protease inhibitor cocktail, and chromogenic substrate Chromozym TH (Roche Applied Science); heparin sodium salt Mr 12,000 from porcine intestinal mucosa, and PNGase F from Elizabethkingia meningoseptica (Sigma–Aldrich); DS Mr 30,000 from porcine intestinal mucosa (Calbiochem); chromogenic substrate CBS 31.39 (Stago); and the fluorescence label TNS (Molecular Probes). The expression vector pET-3d containing the human HCII cDNA was created as published previously [37]. Human prothrombin [38] and HCII [14] were purified from plasma. α-Thrombin, prepared as described [38], was ≥90% active as determined by active site titration [39]. Protein concentrations were determined by absorbance at 280 nm with the absorption coefficients and molecular weights (Mr) of 1.83 (mg/ml)−1 cm−1 and 36,700, for thrombin [40]; 0.59 and 65,600 for plasma HCII [3,5]; and 0.73 and 54,960 for recombinant HCII [41,42]. Active HCII concentrations were determined by stoichiometric titration with thrombin in DS-accelerated reactions. Plasma HCII was deglycosylated by incubation of 2 mg/ml HCII with 15 units of PNGase F in 50 mM Hepes, 0.1 M NaCl, 5 mM CaCl2, 1 mg/ml PEG 8000 buffer, pH 7.4 (reaction buffer) for 24 h at room temperature, under nondenaturing conditions, and glycan fragments were removed by desalting on a 1-ml G25 column. DS was treated with nitrous acid to remove traces of heparin [12].
Site-directed mutagenesis and expression of recombinant human HCII
Silent mutation of 27 rare codons and 5 secondary SD-like sequences in the HCII cDNA was performed at the positions listed in Tables 1 and 2, using the Stratagene QuikChange site-directed mutagenesis kit. Human HCII cDNA, cloned into the pET3d - derived vector, using 5′ NcoI and 3′ SnB1 restriction sites, was used to transform DH5α E. coli cells. Mini DNA preps were made from 5 ml overnight cultures, incubated at 37 °C. The DNA was digested with the restriction enzymes, and DNA quality and restriction sites were verified by 0.7% agarose gel electrophoresis. The HCII constructs were amplified by PCR using the forward and reverse sets of the primers listed in Tables 1 and 2. The reaction (50 μl), containing 50 ng pET-3d HCII, 125 ng each of forward and reverse primer, and 1 μl of dNTP mix, was initiated with Pfu-Turbo DNA polymerase (2.5 U/μl). Cycling parameters were 95 °C, 60 s (denaturation); 55 °C, 60 s (annealing); 68 °C, 15 min (extension); 16 cycles, followed by cooling at 4 °C. Amplification was verified by 0.7% agarose gel electrophoresis. Parental, methylated DNA in the amplification mixture was digested immediately with the restriction enzyme DpnI (10 U/μl) for 1 h at 37 °C, and the mutagenized pET-3d plasmid was used to transform XL1-Blue supercompetent cells by the heat-shock method [43]. Transformed cells were grown on agar plates containing 50 μg/ml ampicillin for 16 h at 37 °C. LB broth (10 ml) containing ampicillin was inoculated with selected colonies, and incubated overnight at 37 °C for plasmid isolation, using the SV Minipreps DNA isolation kit. The plasmids were sequenced to verify the accuracy of each mutation. XL1-Blue glycerol stocks of correctly mutated clones were stored at −80 °C.
Table 1.
Silent mutations of rare codons in the HCII cDNA.
| Position | Primer sequence | Rare codon |
|---|---|---|
| 9 | 5′GAT CAG CTG GAG AAA GGC GGG GAA ACT GCT CAG 3′ | CTA |
| 21 | 5′ GCT CAG TCT GCA GAT CCG CAG TGG GAG CAG TTA 3′ | CCC |
| 66 | 5′ CTG GAC CTG GAG AAG ATC TTC AGT GAA GAC GAC 3′ | ATA |
| 120 | 5′ GCT TTC AAC CTC TAC CGT GTG CTG AAA GAC CAG 3′ | CGA |
| 134/136 | 5′ TTC GAT AAC ATC TTC ATC GCA CCG GTC GGC ATT TCT ACT 3′ | ATA, CCC |
| 192/193 | 5′ ACT CAT CGC CTC TTC CGC CGC AAT TTT GGG TAC 3′ | AGG, AGG |
| 218/222 | 5′ATC CTG CTT GAC TTC CGC ACT AAA GTA CGC GAG TAT TAC TTT GCT 3′ | AGA, AGA |
| 231 | 5′ TTT GCT GAG GCC CAG ATC GCT GAC TTC TCA GAC CCT GCC TTC ATA TCA 3′ | ATA |
| 240 | 5′ TCA GAC CCT GCC TTC ATC TCA AAA ACC AAC AAC 3′ | ATA |
| 255/262 | 5′ GGC CTC ATC AAA GAT GCT CTG GAG AAT ATC GAC CCT GCT ACC CAG 3′ | ATA, ATA |
| 299 | 5′ TTC CGG CTG AAT GAG CGT GAG GTA GTT AAG GTT 3′ | AGA |
| 337 | 5′ GGG GGC ATC AGC ATG CTG ATT GTG GTC CCA CAC 3′ | CTA |
| 356 | 5′ GAA GCG CAA CTG ACA CCG CGG GTG GTG GAG 3′ | CCC |
| 361/369/371 | 5′ GTG GTG GAG CGC TGG CAA AAA AGC ATG ACA AAC CGC ACT CGC GAA GTG CTT CTG CCG 3′ | AGA, AGA, CGA |
| 386 | 5′ GAG AAG AAC TAC AAT CTG GTG GAG TCC CTG AAG 3′ | CTA |
| 396 | 5′ AAG TTG ATG GGG ATC CGT ATG CTG TTT GAC AAA 3′ | AGG |
| 412 | 5′ GGC ATC TCA GAC CAA CGT ATC GCC ATC GAC CTG 3′ | AGG |
| 455 | 5′ TTC ACT GTC GAC CGC CCG TTT CTT TTC CTC ATC 3′ | CCC |
| 473/477/479 | 5′C TTC ATG GGA CGT GTG GCC AAC CCG AGC CGT TCC TAG AGG TGG 3′ | AGA, CCC, AGG |
Forward primer sequences, with rare codons corrected at the positions in boldface. Original rare codon sequences are listed in the third column.
Table 2.
Silent mutations of SD-like sequences in the HCII cDNA.
| Position | Primer sequence | Silent mutation |
|---|---|---|
| 9/12 | 5′ GAT CAG CTG GAG AAA GGC GGG GAA ACT GCT CAG 3′ | CTA, GGA |
| 42 | 5′ CCT GCC GAC TTC CAC AAA GAA AAC ACC GTC ACC 3′ | AAG |
| 55/56 | 5′ TGG ATT CCA GAG GGG GAA GAA GAC GAC GAC TAT CTG 3′ | GAG, GAG |
| 192/193 | 5′ ACT CAT CGC CTC TTC CGC CGC AAT TTT GGG TAC 3′ | AGG, AGG |
| 310 | 5′ TCC ATG ATG CAG ACC AAA GGG AAC TTC CTC GCA 3′ | AAG |
Forward primer sequences, with silent mutations in the Shine–Dalgarno-like sequences introduced at the positions in boldface. Original sequences are listed in the third column.
BL21-CodonPlus (DE3)-RIPL competent cells were transformed with modified pET-3dHCII by heat shock. Transformed cells were plated onto agar plates containing 50 μg/ml ampicillin, and colonies (>20/plate) were grown for 16 h at 37 °C. LB media (5 ml) containing 50 μg/ml ampicillin was inoculated with several colonies and incubated overnight at 37 °C for preparation of glycerol stocks. Starter cultures (100 ml) of TB, MagicMedia, and LB media containing 50 μg/ml ampicillin, inoculated with transformed BL21-Codon-Plus (DE3)-RIPL cells, were grown overnight at 37 °C. Ten milliliters of this starter culture was used to inoculate 1-L media flasks, with a total prep volume of 10 L, and the cultures were grown under constant shaking at 37 °C. Cultures in LB and TB media, grown to A600 0.6, were induced with 0.3 mM IPTG, after a 1 h cool down to 25 °C, and grown overnight under constant shaking at 25 °C. MagicMedia, utilizing the T7/lacUV5 promoter mechanism, required no IPTG induction, and cultures were grown overnight at 25 °C. The cell pellets were harvested by centrifugation and stored at −80 °C until further use.
Purification of recombinant human HCII
Cells (100 g pellets) were resuspended in 200 ml of 50 mM Hepes, 50 mM NaCl, 0.1 mM EDTA, and 0.02% NaN3, pH 7.5 (start buffer). Protease inhibitor cocktail (2 tablets), 6 mg of lysozyme powder, and 1 mM PMSF (final) were added to the buffer prior to resuspension of the cell pellet. Cells were sonicated in an ice bath, for 5 cycles of 45 s, with 10 min intervals, and cell debris was removed by centrifugation. The crude lysate was loaded onto a heparin-Sepharose column (2.5 × 23 cm) in start buffer at room temperature, and the column was washed until the absorbance at 280 nm (A280) was ≤0.08. Protein was eluted using a 250 × 250 ml linear gradient of 0.05–0.50 M NaCl in start buffer, and 5.0 ml fractions were collected. HCII activity was measured spectrophotometrically by reacting 10–20 nM α-thrombin with 100 μl fraction sample in 1 ml 50 mM Hepes, 0.1 M NaCl, 5 mM CaCl2, 1 mg/ml PEG 8000 buffer, pH 7.4 (reaction buffer), in the presence of 1.5 μM heparin (Mr 12,000), and 100 μM of competing chromogenic substrate CBS31.39, and continuously monitoring the inhibition of pNA production at 405 nm. Active fractions were pooled, dialyzed against Mono Q and S HR 5/50 start buffer, 20 mM potassium phosphate buffer, pH 7.4, and applied to the Mono Q FPLC column equilibrated in start buffer. The column was washed until A280 ≤ 0.05, and eluted with 35 column volumes of 0.05–0.50 M NaCl gradient in start buffer, in 1.5 ml fractions. Active HCII fractions were pooled, dialyzed against start buffer, and applied to the Mono S FPLC column. After washing with 5 column volumes of start buffer, elution was performed with a 30 column volume 0.05–0.50 M NaCl gradient in start buffer, in 1.5 ml fractions. Active fractions were analyzed by 4–20% SDS–PAGE. Purified HCII was concentrated, dialyzed against 50 mM Hepes, 0.1 M NaCl, pH 7.4, storage buffer, and quick-frozen at −80 °C.
Mass spectrometry of native and recombinant HCII
Plasma and recombinant HCII were resolved by SDS–PAGE and visualized by Coomassie Blue R250 staining. Gel-excised protein bands of interest were equilibrated in 100 mM NH4HCO3 and then reduced with dithiothreitol (3 mM in 100 mM NH4HCO3, 37 °C for 15 min) and alkylated with iodoacetamide (6 mM in 100 mM NH4HCO3, 15 min). Gel slices were dehydrated with acetonitrile and then rehydrated with 15 μl of 25mM NH4HCO3 containing 0.01 μg/μl Modified Trypsin-Gold protease, and digestion was carried out for >2 h at 37 °C. Peptides were extracted with 60% acetonitrile, 0.1% trifluoroacetic acid, dried by vacuum centrifugation, and reconstituted in 10 μl of 60% acetonitrile, 0.1% trifluoroacetic acid. Aliquots (0.4 μl) were applied to target plates and overlayed with 0.4 μl of α-cyano-4-hydroxycinnamic acid matrix (5 mg/ml, supplemented with 1 mg/ml ammonium citrate). Matrix-assisted laser desorption/ionization, time-of-flight (MALDI-TOF) MS, and tandem TOF/TOF MS/MS was carried out using a Voyager 4700 mass spectrometer (Applied Biosystems), operated in positive-ion reflectron mode. Each MALDI-TOF mass spectrum (representing a peptide mass map of the digested protein) was calibrated to within 10 ppm using trypsin autolytic peptides present in the sample (m/z 842.50, 1045.56, and 2211.10). Peptide masses were then manually compared to the list of predicted peptides for HCII from each digested protein (either isolated from plasma or expressed/purified from E. coli), as well as to each other.
Fluorescence equilibrium binding of GAGs to recombinant, native, and deglycosylated HCII
Binding of heparin and DS to native, deglycosylated, and recombinant HCII (500, 480, and 405 nM, respectively) was quantitated in reaction buffer, pH 7.4, at 25 °C, by measuring the change in fluorescence of the reversible probe TNS (12 μM final concentration) in the HCII·TNS complex upon GAG binding to HCII [44,45]. Titrations were performed with an SLM 8100 spectrofluorometer in the ratio mode as described previously [46], at 330 and 428 nm excitation and emission wavelengths, with 8 nm band passes. Blank titrations with TNS and GAGs were performed in parallel to correct for background fluorescence. Data were expressed as the fractional change in the initial fluorescence ((Fobs − Fo)/Fo = ΔF/Fo) as a function of total titrant concentration, and were fit by the quadratic binding equation for binding of a single ligand [47]. The fitted parameters were the maximum fluorescence intensity changes ΔFmax,HCII,GAG/Fo and the dissociation constants KHCII,GAG for heparin and DS binding to HCII.
Kinetics of GAG-accelerated thrombin inactivation by recombinant, native, and deglycosylated HCII
Kinetic experiments and stoichiometric titrations were performed at 25 °C in pH 7.4 reaction buffer. The stoichiometries of thrombin reacting with native, deglycosylated, and recombinant HCII were determined in the absence of GAGs, and in the presence of 5 μM heparin or DS. Incubations were performed in PEG 20,000-coated 1 ml Eppendorf tubes containing 0.5–1 μM active-site titrated thrombin and increasing HCII concentrations. Reactions were incubated overnight, and thrombin activity was assayed with the chromogenic substrate CBS31.39 (100 μM) at 405 nm with a Shimadzu 2401 spectrophotometer. Residual rates for thrombin–HCII incubations were compared to thrombin rates in the absence of HCII, and the stoichiometries were determined from the equivalence points at which complete thrombin neutralization was observed. The second-order rate constants of thrombin inactivation by the three HCII forms were measured from the loss of thrombin chromogenic substrate activity, under pseudo-first-order conditions (~500 nM [HCII]o, 50 nM [T]o), in the absence of GAGs. Residual thrombin activity was measured at discrete time intervals, using discontinuous sampling into cuvettes containing 150 μM S2238 or 120 μM Chromozym TH, and expressed as the fractional residual thrombin activity, [T]t/[T]o. The first-order time courses of the fractional residual thrombin activity were analyzed by nonlinear least-squares fitting of
| (1) |
with the first-order rate constant k = k′[HCII], the product of the second-order rate constant k′ for the HCII–T reaction and the HCII concentration. Thrombin inactivation by HCII in the presence of GAGs was monitored by continuous competitive chromogenic substrate hydrolysis, under first-order conditions of chromogenic substrate and inhibitor, as described previously [14,48–50]. Km and kcat for hydrolysis of Chromozym TH and CBS31.39 were 11 μM and 167 s−1, and 240 μM, and 49 s−1, respectively [50]. Thrombin was added to a mixture of HCII, GAG, and chromogenic substrate at time zero, and pNA formation was monitored continuously at 405 nm. Time courses were analyzed by a nonlinear least squares fit by
| (2) |
with [pNA]t the concentration of pNA formed during the reaction, A the amplitude of the progress curve, and the observed pseudo-first-order rate constant kobs = k/(1 + [S]o/Km). Km and [S]o are the Michaelis constant and concentration of chromogenic substrate, and k is the first-order rate constant without chromogenic substrate competition as described above. X is a slow, second exponential to account for the combined effects of the uncatalyzed reaction and slow reacting thrombin forms [51].
GAG and HCII dependencies of kobs were established as described previously [14,50], and analyzed by Eq. (3), using the mass conservation equations for thrombin, HCII and GAG:
| (3) |
The fixed parameters were Km and [S]o, the Michaelis constant and total concentration of the chromogenic substrate; and KHCII,GAG, the dissociation constant for GAG binding to HCII, appearing in the mass conservation equation for HCII, and determined independently by equilibrium binding. The fitted parameters were klim, the limiting first-order rate constant for GAG-accelerated thrombin inactivation; KT,GAG for binding of GAG to thrombin; and Kcomplex the apparent Michaelis constant for assembly of the ternary T·GAG·HCII complex. [GAG]free and HCIIfree were calculated from the mass balances. Least-squares fitting of binding and kinetic data were performed with SCIENTIST Software (MicroMath). All reported estimates of error represent ± 2 SD.
Results
Site-directed mutagenesis and expression of recombinant human HCII
Of the 27 rare codons, 11 were clustered in positions Arg192-Arg193; Ile134-Pro136; Arg218-Arg222; Arg369-Arg371; and Arg473-Pro477-Arg479. Initial silent mutation of these clusters, and positions Pro9 and Arg361, combined with the use of the BL21-CodonPlus (DE3)-RIPL cells, containing extra copies of the E. coli argU, ileY, leuW, and proL tRNA genes, improved expression levels in TB and LB media, with higher expression at 25 °C compared to 30 and 37 °C. Expression in MagicMedia, utilizing auto-induction, was very low, and the use of this medium was discontinued. Silent mutation of the remaining rare codons, and in the additional SD-like sequences AAGGAGG, AAGGAA, AGGAGG, and AAGGGG, respectively, spanning residues Lys11-Gly13, Lys42-Glu43, Glu55-Asp57, and Lys310-Gly311 (Tables 1 and 2) further substantially improved expression levels, with comparable yields in TB and LB media. The IPTG induction concentration for optimal expression in these systems was determined to be 0.3 mM.
Purification of recombinant human HCII
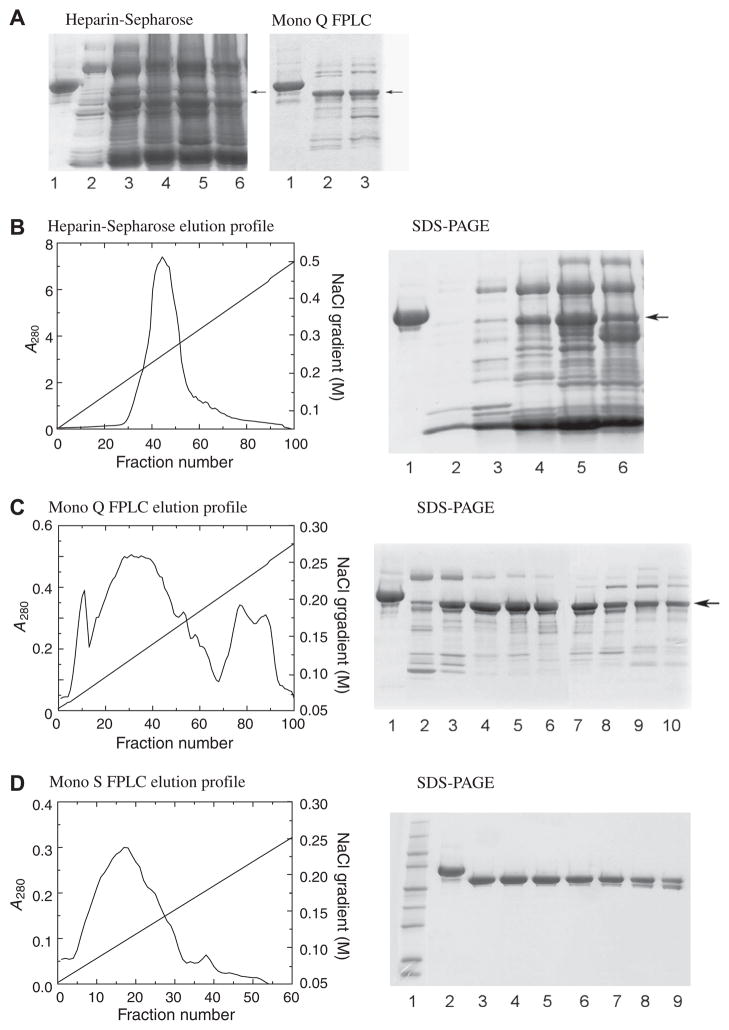
HCII eluted from the heparin-Sepharose, Mono Q and Mono S columns between 0.20 and 0.30, 0.10 and 0.20, and 0.08 and 0.20 M NaCl, respectively. Fig. 2A shows the SDS–PAGE of recombinant HCII, expressed from the construct bearing 13 initial mutations, after heparin-Sepharose chromatography, and after partial purification on Mono Q FPLC. Yields of these preps were typically ~3 mg from 10 L starting media. Fig. 2B–D show the elution profiles and SDS–PAGE of HCII expressed from the final construct containing all silent mutations, after consecutive heparin-Sepharose, and Mono Q and Mono S FPLC purification. Shallow FPLC gradients were necessary to resolve the HCII peak. HCII yields of 10 L preps after Mono Q were up to 35 mg, with one major HCII peak, and several side peaks containing HCII as well as impurities and minor bands of cleaved HCII. These side pools were saved for rechromatography on Mono Q. Impurities and cleaved protein in the major HCII peak were removed by Mono S FPLC. Final yields after Mono S FPLC were typically 10–20 mg.
Fig. 2.
Purification of recombinant HCII. (A) SDS–PAGE of recombinant HCII (active fractions) after partial purification on heparin-Sepharose (left panel) and Mono Q FPLC (right panel) of HCII expressed from a construct with 13 initial mutations, with plasma HCII (lanes 1) as a standard. Arrows indicate the HCII bands. The electrophoretic mobility of recombinant HCII is slightly faster than that of plasma HCII due to the lack of glycosylation. (B) Elution profile on heparin-Sepharose of HCII expressed from the construct containing all silent mutations, and SDS–PAGE of the active fractions (lane 1, plasma HCII; lanes 2–6 are fractions 32, 35, 40, 45, 50). (C) Elution profile of recombinant HCII on a 5 ml Mono Q FPLC column, with SDS–PAGE of active fractions (lane 1, plasma HCII; lanes 2–10 are every 5th fraction, from 20 to 60). (D) Elution profile of recombinant HCII on a 5 ml Mono S FPLC column, with SDS–PAGE of active fractions (lane 1, Mr markers; lane 2, plasma HCII; lanes 3–9 are every 5th fraction, from 10 to 40). A280 values of the fractions were measured in 1 cm cuvettes. The yield of this prep was 14 mg.
Mass spectrometry of native and recombinant HCII
The plasma and recombinant HCII spectra showed complete similarity and accounted for all major ions observed (Fig. 3), except for one peptide with mass/charge ratio m/z of 1120.63, identified as QFPILLDFK in plasma HCII, as predicted from the genomic sequence [52], and QFPILLDFR with m/z 1148.64 in recombinant HCII [33]. The recombinant HCII spectrum also showed a peptide with m/z ratio 1131.62, with expected loss of 17 Da due to the cyclization of the N-terminal Q to pyroglutamate on the QFPILLDFR peptide. Sequence confirmation was provided by tandem TOF/TOF mass spectrometry on these ions, where the fragmentation pattern was consistent with the expected amino acid sequence (data not shown). The QFPILLDFR peptide ionized much more efficiently due to the presence of arginine, and altered the fragmentation pattern in a predictable way. The K-R substitution at position 218 in human HCII has been reported as a polymorphism [33,53,54].
Fig. 3.
MALDI-TOF peptide mass map of plasma and recombinant HCII. The ionization patterns of plasma HCII and recombinant HCII are shown in the top and bottom panels, respectively. Ionization of plasma HCII yields the QFPILLDFK peptide with m/z ratio 1120.63. The corresponding peptide in recombinant HCII is QFPILLDFR with m/z ratio of 1148.64, resulting from a polymorphism. An additional peptide is present, with m/z ratio 1131.62, and expected loss of 17 Da due to the cyclization of the N-terminal Q to pyroglutamate on the QFPILLDFR peptide.
Fluorescence equilibrium binding of GAGs to recombinant, native, and deglycosylated HCII
GAG binding to HCII was measured by the fluorescence change of the reversible TNS·HCII complex, specifically reporting GAG binding to the serpin heparin-binding site [44,45]. The analysis for binding of heparin with Mr 12,000 gave apparent dissociation constants KHCII,H 6 ± 1 μM for recombinant HCII, 7 ± 1 μM for deglycosylated plasma HCII, and 40 ± 4 μM for native plasma HCII; and respective fluorescence amplitudes ΔFmax,HCII,H/Fo −60 ± 1, −31 ± 1, and −47 ± 2% (Fig. 4A). Binding of DS with Mr 30,000 was characterized by KHCII,DS 4 ± 1 μM for recombinant HCII, 4 ± 1 μM for deglycosylated plasma HCII, and 15 ± 4 μM for native plasma HCII; and respective fluorescence amplitudes ΔFmax,HCII,DS/Fo −68 ± 4, −32 ± 1, and −42 ± 4% (Fig. 4B), with one GAG-binding site assumed on HCII. Fluorescence amplitudes for heparin and DS binding were similar for each HCII form, with consistently larger amplitudes for GAG binding to recombinant HCII.
Fig. 4.
Fluorescence equilibrium binding of heparin and DS to HCII. Fractional change in fluorescence (ΔF/Fo) of 500 nM plasma HCII (●), 480 nM deglycosylated plasma HCII (○) and 405 nM recombinant HCII (■) in the presence of 12 μM TNS as a function of total heparin concentration ([Heparin]o) (A) and total dermatan sulfate concentration ([DS]o) (B). Solid lines represent least-squares fitting of the data by the quadratic equation for binding of a single ligand [47], with the fitted parameters given in the text. Binding experiments were performed and analyzed as described under Materials and methods.
Kinetics of GAG-accelerated thrombin inactivation by recombinant, native, and deglycosylated HCII
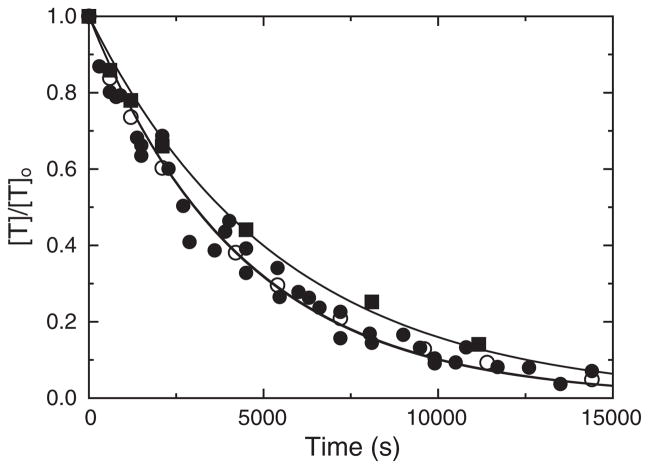
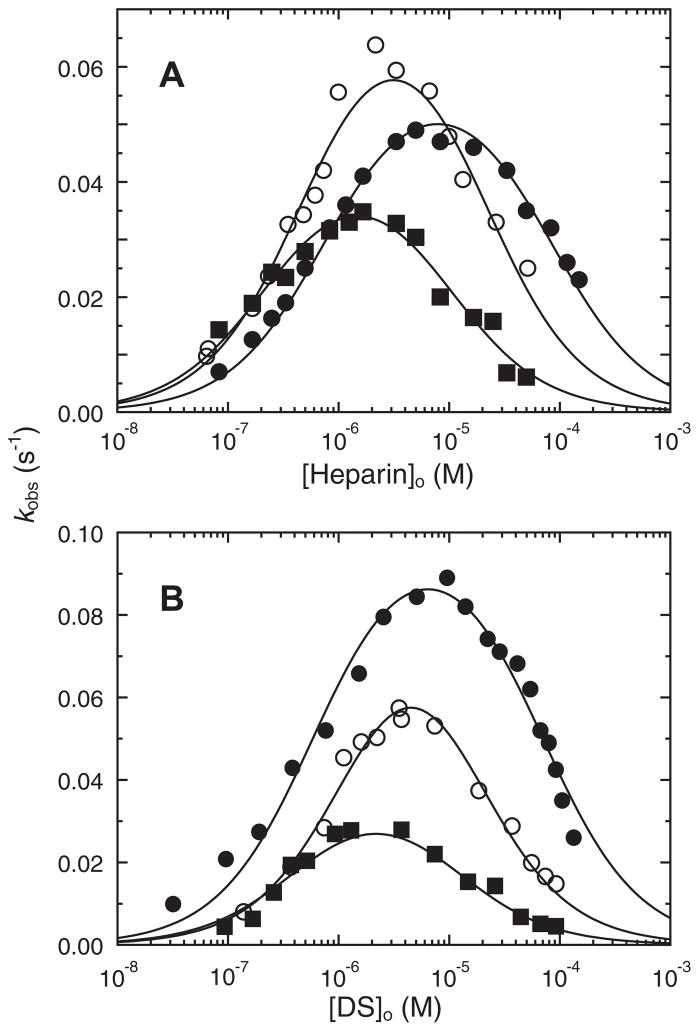
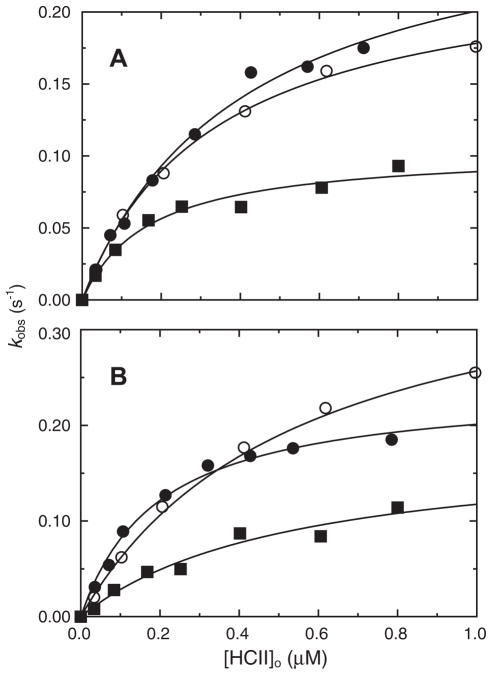
Stoichiometric titration of recombinant HCII with thrombin demonstrated a 1:1 thrombin:HCII stoichiometry for the uncatalyzed and DS-catalyzed reactions, and a 1:1.3 stoichiometry for the heparin-catalyzed reaction, in good agreement with the results for native and deglycosylated HCII. Rates of uncatalyzed inactivation of thrombin by recombinant, native, and deglycosylated HCII were indistinguishable, with second-order rate constants of 460 ± 30, 460 ± 20, and 420 ± 20 M−1 s−1, respectively, as shown in Fig. 5. The GAG and HCII dependencies of the first-order inactivation rate constant kobs for recombinant, native, and deglycosylated plasma HCII are shown in Figs. 6A and B and 7A and B, respectively. Simultaneous analysis of the GAG and HCII concentration dependencies by Eq. (3) and the mass conservation equations for thrombin, HCII, and GAG gave similar second-order rate constants ranging from 9 to 11 μM−1 s−1 for the heparin- and DS-catalyzed reactions of recombinant and native HCII, and 13–16 μM−1 s−1 for deglycosylated HCII; however, these differences were not significant. The equilibrium and rate constants for all interactions are given in Table 3. The KD values for heparin and DS binding to recombinant, plasma, and deglycosylated HCII, obtained by equilibrium binding, and fitted from the kinetic data, were in good agreement.
Fig. 5.
Inhibitory activity of recombinant, native, and deglycosylated HCII in the absence of GAGs. Thrombin activity, expressed as a fraction of the initial concentration ([T]t/[T]o), is shown as a function of time, for reactions of 402 nM recombinant HCII (■), 500 nM plasma HCII (●), and 538 nM deglycosylated HCII (○) with 50 nM thrombin, in the absence of GAGs. Solid lines represent the fit of the data by Eq. (3). Experiments were performed and analyzed as described under Materials and methods.
Fig. 6.
Heparin and DS concentration dependencies of kobs. The first-order inactivation rate constant (kobs), measured in the presence of 120 μM Chromozym TH, as a function of total heparin (A) or DS (B) concentration. Recombinant (■), plasma (●), and deglycosylated (○) HCII concentrations were 84, 107, and 103 nM, respectively, and thrombin concentrations ranged from 0.6 to 2.7 nM. Solid lines represent global least-squares fitting by Eq. (5), with the fitted parameters given in the text. Experiments were performed and analyzed as described under Materials and methods.
Fig. 7.
HCII concentration dependencies of kobs. The first-order inactivation rate constant (kobs), measured in the presence of 120 μM Chromozym TH, as a function of total recombinant (■), plasma (●), or deglycosylated (○) HCII concentration ([HCII]o), in the presence of and 1.66 μM heparin (A) or 3.7 μM DS (B). Thrombin concentrations ranged from 0.6 to 6.9 nM. Solid lines represent global least-squares fitting by Eq. (5), with the fitted parameters given in the text. Experiments were performed and analyzed as described under Materials and methods.
Table 3.
Equilibrium binding and kinetic parameters for GAG-accelerated thrombin inactivation by HCII.
| Recombinant HCII | Deglycosylated HCII | Native HCII | |
|---|---|---|---|
| KHCII,H | 6 ± 1 μMa | 7 ± 1 μMa | 40 ± 4 μMa |
| 5 ± 2 μMb | 13 ± 5 μMb | 60 ± 10 μMb | |
| KHCII,DS | 4 ±1 μMa | 4 ± 1 μMa | 15 ± 4 μMa |
| 8 ± 5 μMb | 12 ± 5 μMb | 40 ± 10 μMb | |
| KT,H | 0.4 ± 0.3 μM | 0.8 ± 0.2 μM | 1.0 ± 0.5 μM |
| KT,DS | 0.6 ± 0.3 μM | 1.0 ± 0.4 μM | 1.0 ± 0.3 μM |
| KTH,HCII | 0.010 ± 0.004 μM | 0.018 ± 0.003 μM | 0.030 ± 0.010 μM |
| KTDS,HCII | 0.020 ± 0.006 μM | 0.020 ± 0.003 μM | 0.022 ± 0.002 μM |
| klim,H | 0.11 ± 0.01 s−1 | 0.24 ± 0.01 s−1 | 0.28 ± 0.04 s−1 |
| klim,DS | 0.17 ± 0.03 s−1 | 0.32 ± 0.03 s−1 | 0.24 ± 0.04 s−1 |
| k′H | 11 ± 4 μM−1 s−1 | 13 ± 4 μM−1 s−1 | 9 ± 3 μM−1 s−1 |
| k′DS | 9 ± 3 μM−1 s−1 | 16 ± 3 μM−1 s−1 | 11 ± 2 μM−1 s−1 |
Dissociation constants for heparin (H) and dermatan sulfate (DS) binding to native, deglycosylated, and recombinant HCII were determined by equilibrium bindinga and nonlinear least-squares fitting of the inactivation rates by Eq. (3) and the mass conservation equation for HCIIb. Dissociation constants for heparin and DS binding to thrombin (KT,H, KT,DS), and ternary complex formation (KTH,HCII, KTDS,HCII), and limiting rates (klim,H, klim,DS) for thrombin inactivation were determined by kinetic analysis of the HCII and GAG concentration dependencies by Eq. (3). Second-order rate constants k′H and k′DS were calculated as klim,H/KTH,HCII and klim,DS/KTDS,HCII, respectively. Experiments were performed and analyzed as described under Materials and methods. Errors represent ± 2 SD.
Discussion
Quantitative expression of wild-type HCII in E. coli has to date been performed mainly by semilarge scale incubation [19], or high cell density fermentation [37]. In our initial studies, expression of wild-type HCII in BL21-CodonPlus (DE3)-RIPL cells at 37 °C, induced by 0.3 mM IPTG, did not significantly improve the yield, although these cells contain a plasmid encoding extra copies of rare E. coli tRNA genes. Variation of cell lines, media, expression temperature, duration of incubation, and of type and concentration of induction agent resulted only in marginal improvement in some cases, whereas other conditions caused complete arrest. Expression of HCII in S. cerevisiae W303-1a, using the p423-MET25 construct [55], although readily detectable by immunoblotting, was insufficient for production of mg quantities of HCII, mainly due to low codon usage of the Arg CGG, CGA, and CGC, Ser TCG, and Leu CTC codons in S. cerevisiae (0.4–0.9% fraction, GCG Codon-Frequency Program, Accelrys).
Heterologous expression of human HCII in E. coli is negatively affected by the presence of 27 rare codons, some of them clustered. The codon usage in E. coli is the lowest for the Arg codons AGG and AGA, 0.3% and 0.6%, respectively [56], and the abundance of their cognate tRNAs is similarly very low [57]. Other codons with low usage are Ile ATA (0.6%), Leu CTA (0.8%), Arg CGA (1.1%), and Pro CCC (1.6%). Consecutive AGG codons in various heterologous genes are known to cause ribosome stalling, resulting in frameshift, hopping, or chain termination, depending on the location of the AGG AGG sequence [58,59]. Similarly, other secondary SD-like sequences in the cDNA of certain proteins were reported to cause significant decrease or even arrest of expression [60,61]. Both in-frame and out-of-frame AGG clusters may result in inhibition, suggesting that reduced expression is caused by the SD-like sequence binding to 16S RNA [62]. Gene optimization was performed in two stages. Initial silent mutation of 13 rare codons including the tandem AGG repeat and several clustered sequences significantly improved expression, and a final optimization stage including the remaining rare codons and secondary SD-like sequences resulted in vastly improved yields, up to 35 mg/10 L media in the second purification step, and 10–20 mg/10 L after final purification. Optimal conditions of expression temperature (25 °C) and induction conditions (0.3 mM IPTG) were established. Yields in LB and enriched TB [63] were comparable, in contrast to Magic-Media utilizing auto-induction [64], in which no expression was observed. Cell densities (OD600nm) after overnight IPTG induction in LB and TB at 25 °C were between 3.5 and 4, below those typically obtained in high-density fermentation at 37 °C. Optimized high-density methods with controlled pH and oxygen availability, utilizing colony selection, have been described [65]; however, high cell density may result in plasmid loss from E. coli [66]. Expression at 25 °C decreases protein misfolding and processing in inclusion bodies, sometimes encountered during 37 °C incubations [67]. In endogenous E. coli proteins, codon usage in the second triplet, following the N-terminal Met, is selective [68], and further improvement of HCII expression may be obtained by silent mutation of the GGG, the codon for HCII N-terminal Gly, to GGT.
Both recombinant and deglycosylated HCII exhibited similar, increased GAG-binding affinities compared to native plasma HCII, indicating that the presence of the Arg218 polymorphism in the recombinant protein did not affect GAG binding. Steric hindrance and charge effects of the N-glycan chain at Asn169, in the vicinity of the HCII heparin-binding site, may be responsible for the reduced GAG affinity of plasma HCII. Isoforms of plasma HCII lacking Asn169 N-glycosylation, similar to AT-β, lacking N-glycosylation at Asn135, have not yet been reported. AT-β, representing 5–10% of the total circulating antithrombin, binds heparin 3.5-fold tighter than fully glycosylated AT [69], and exerts its antithrombotic function at the endothelial and subendothelial cell surface [70,71]. Native and deglycosylated HCII possess two sulfated Tyr residues at positions 60 and 73, increasing the net negative charge of the protein compared to E. coli recombinant HCII. Expression of HCII in CHO cells treated with tyrosine sulfation inhibitors reportedly resulted in production of nonsulfated, glycosylated HCII with increased affinity for heparin-Sepharose [52]. During purification we did not observe significant differences in chromatographic behavior of plasma HCII and HCII from E. coli lysates on heparin-Sepharose [14], possibly due to the presence of large concentrations of other heparin-binding proteins and compounds in the first purification step, and differences in heparin-Sepharose matrix-binding behavior may become apparent for the purified HCII forms. Whereas our fluorescence equilibrium binding results demonstrated similar GAG affinities of recombinant and deglycosylated HCII in a solution system, the resulting quenches in fluorescence intensity were much more pronounced for recombinant HCII, with ~2-fold larger amplitudes for recombinant HCII, compared to the Tyr-sulfated native and deglycosylated HCII. These results suggest that the increased affinity for GAGs may at least in part be due to the absence of N-glycosylation, and that the sulfated Tyr residues in HCII may affect the fluorescence of the TNS probe bound to the serpin, but not necessarily the affinity of GAGs in solution. Recombinant deletion of HCII residues 1–74, with both sulfated Tyr residues contained in the acidic region spanning residues 53–74, was shown to result in a 2-fold increase in affinity for the heparin-Sepharose matrix, and a drastic reduction of GAG-accelerated reactivity toward α-thrombin possessing an intact exosite I [19]. Binding of the hirudin-like acidic N-terminal region in HCII to thrombin exosite I is an essential part of the inactivation mechanism [18,19], and our kinetic studies with recombinant HCII proposed to investigate whether the sulfation status of both tyrosines in the acidic region is obligatory for proper interaction with thrombin exosite I. If this were the case, we would expect to observe substantial differences in the kinetics of GAG-accelerated thrombin inactivation by recombinant and deglycosylated HCII.
The HCII dependencies of the inactivation rate, measured at the respective optimal GAG concentrations, displayed hyperbolic saturation, and the logarithmic GAG dependencies were bell-shaped, consistent with a mechanism of ternary thrombin·GAG·HCII complex formation. The amplitudes of the GAG dependencies for recombinant HCII were lower due to the lower fixed HCII concentrations in these experiments. The HCII dependencies defined the limiting rates, klim, the ternary complex affinities, Kcomplex, and the second-order rate constants k′ expressed as the ratio klim/Kcomplex, whereas the GAG dependencies defined the GAG affinities for thrombin (KT,GAG), HCII (KHCII,GAG), and also the ternary complex affinities, Kcomplex. The maxima of the logarithmic GAG concentration dependencies of the inactivation rate were shifted toward lower GAG concentrations for recombinant and deglycosylated HCII compared to native HCII, consistent with tighter GAG binding of these forms. Whereas the limiting rates for the recombinant HCII reactions were typically lower than those for deglycosylated and native HCII, the second-order rate constants k′ were largely comparable, indicating equally efficient ternary complex formation and inhibitory behavior. The fitted dissociation constants for GAG binding to HCII in the kinetic profiles were generally higher than those measured independently by equilibrium binding, and were similarly lower for recombinant and deglycosylated HCII, compared to native HCII. The fits of the GAG profiles, spanning concentrations that differ by 3–4 orders of magnitude, with equal weighting of data, gave comparable values for GAG binding to thrombin, from 0.4 to 1 μM, validating the data analysis employing a linkage mechanism of ternary complex formation [72]. The affinities of these GAGs in binary and ternary complexes with thrombin and HCII, although representative, are expected to vary with the type, degree of sulfation, and molecular weight of the GAG.
In conclusion, the present work demonstrates that the increased GAG affinity of E. coli recombinant HCII is largely caused by the lack of N-glycosylation. Tyr sulfation in the acidic N-terminal HCII sequence does not appear to play a major role in GAG binding, and is not required for efficient GAG-accelerated thrombin inactivation. This study reports for the first time a quantitative analysis of the binding and kinetic properties of recombinant human HCII expressed in E. coli, of importance in comparative interpretation of the kinetic behavior of HCII variants engineered as functional probes of the thrombin inactivation mechanism.
Acknowledgments
The project described was supported by Grants R01 HL 080018 (I.M.V) and R01 HL 55520 (D.M.T.) from the National Heart, Lung, and Blood Institute of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health. We are grateful to Ms. Sarah R. Stuart for excellent technical assistance with the mass spectrometry experiments.
Footnotes
Abbreviations used: serpin, serine proteinase inhibitor; HCII, heparin cofactor II; AT, antithrombin; RCL, reactive center loop; SDS–PAGE, sodium dodecyl sulfate polyacrylamide electrophoresis; T, human α-thrombin; GAG, glycosaminoglycan; IPTG, isopropyl-1-thio-β-D-galactopyranoside; TNS, 2-(p-toluidinyl)naphthalene-6-sulfonic acid; DS, dermatan sulfate; CBS31.39, CH3SO2-D-Leu-Gly-Arg-p-nitroanilide; Chromozym TH, Tos-Gly-Pro-Arg-p-nitroanilide; pNA, p-nitroaniline; Hepes, N-(2-hydroxyethyl)-N′-2-piperazine-ethanesulfonic acid; PEG, polyethylene glycol 8000.
References
- 1.Ragg H, Ulshofer T, Gerewitz J. Glycosaminoglycan-mediated leuserpin-2/thrombin interaction, Structure–function relationships. J Biol Chem. 1990;265:22386–22391. [PubMed] [Google Scholar]
- 2.Tollefsen DM. Insight into the mechanism of action of heparin cofactor II. Thromb Haemost. 1995;74:1209–1214. [PubMed] [Google Scholar]
- 3.Church FC, Meade JB, Pratt CW. Structure–function relationships in heparin cofactor II: spectral analysis of aromatic residues and absence of a role for sulfhydryl groups in thrombin inhibition. Arch Biochem Biophys. 1987;259:331–340. doi: 10.1016/0003-9861(87)90499-1. [DOI] [PubMed] [Google Scholar]
- 4.Tollefsen DM, Pestka CA, Monafo WJ. Activation of heparin cofactor II by dermatan sulfate. J Biol Chem. 1983;258:6713–6716. [PubMed] [Google Scholar]
- 5.Tollefsen DM, Majerus DW, Blank MK. Heparin cofactor II. Purification and properties of a heparin-dependent inhibitor of thrombin in human plasma. J Biol Chem. 1982;257:2162–2169. [PubMed] [Google Scholar]
- 6.Pratt CW, Whinna HC, Church FC. A comparison of three heparin-binding serine proteinase inhibitors. J Biol Chem. 1992;267:8795–8801. [PubMed] [Google Scholar]
- 7.Church FC, Meade JB, Treanor RE, Whinna HC. Antithrombin activity of fucoidan. The interaction of fucoidan with heparin cofactor II, antithrombin III, and thrombin. J Biol Chem. 1989;264:3618–3623. [PubMed] [Google Scholar]
- 8.Church FC, Pratt CW, Treanor RE, Whinna HC. Antithrombin action of phosvitin and other phosphate-containing polyanions is mediated by heparin cofactor II. FEBS Lett. 1988;237:26–30. doi: 10.1016/0014-5793(88)80164-9. [DOI] [PubMed] [Google Scholar]
- 9.Church FC, Treanor RE, Sherrill GB, Whinna HC. Carboxylate polyanions accelerate inhibition of thrombin by heparin cofactor II. Biochem Biophys Res Commun. 1987;148:362–368. doi: 10.1016/0006-291x(87)91119-3. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi T, Hayakawa Y, Hayashi T, Sasaki H, Sakuragawa N. Sulfated polysaccharide from the leaves of Artemisia Princeps activates heparin cofactor II independently of the Lys173 and Arg189 residues of heparin cofactor II. Thromb Res. 1997;87:105–112. doi: 10.1016/s0049-3848(97)00109-6. [DOI] [PubMed] [Google Scholar]
- 11.He L, Vicente CP, Westrick RJ, Eitzman DT, Tollefsen DM. Heparin cofactor II inhibits arterial thrombosis after endothelial injury. J Clin Invest. 2002;109:213–219. doi: 10.1172/JCI13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maimone MM, Tollefsen DM. Structure of a dermatan sulfate hexasaccharide that binds to heparin cofactor II with high affinity. J Biol Chem. 1990;265:18263–18271. [PubMed] [Google Scholar]
- 13.Mitchell JW, Church FC. Aspartic acid residues 72 and 75 and tyrosine-sulfate 73 of heparin cofactor II promote intramolecular interactions during glycosaminoglycan binding and thrombin inhibition. J Biol Chem. 2002;277:19823–19830. doi: 10.1074/jbc.M200630200. [DOI] [PubMed] [Google Scholar]
- 14.Verhamme IM, Bock PE, Jackson CM. The preferred pathway of glycosaminoglycan-accelerated inactivation of thrombin by heparin cofactor II. J Biol Chem. 2004;279:9785–9795. doi: 10.1074/jbc.M313962200. [DOI] [PubMed] [Google Scholar]
- 15.Fortenberry YM, Whinna HC, Gentry HR, Myles T, Leung LL, Church FC. Molecular mapping of the thrombin–heparin cofactor II complex. J Biol Chem. 2004;279:43237–43244. doi: 10.1074/jbc.M406716200. [DOI] [PubMed] [Google Scholar]
- 16.Hortin GL, Tollefsen DM, Benutto BM. Antithrombin activity of a peptide corresponding to residues 54–75 of heparin cofactor II. J Biol Chem. 1989;264:13979–13982. [PubMed] [Google Scholar]
- 17.Myles T, Church FC, Whinna HC, Monard D, Stone SR. Role of thrombin anion-binding exosite-I in the formation of thrombin–serpin complexes. J Biol Chem. 1998;273:31203–31208. doi: 10.1074/jbc.273.47.31203. [DOI] [PubMed] [Google Scholar]
- 18.Baglin TP, Carrell RW, Church FC, Esmon CT, Huntington JA. Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multi-step allosteric mechanism. Proc Natl Acad Sci USA. 2002;99:11079–11084. doi: 10.1073/pnas.162232399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Deerlin VM, Tollefsen DM. The N-terminal acidic domain of heparin cofactor II mediates the inhibition of alpha-thrombin in the presence of glycosaminoglycans. J Biol Chem. 1991;266:20223–20231. [PubMed] [Google Scholar]
- 20.Streusand VJ, Björk I, Gettins PG, Petitou M, Olson ST. Mechanism of acceleration of antithrombin-proteinase reactions by low affinity heparin. Role of the antithrombin binding pentasaccharide in heparin rate enhancement. J Biol Chem. 1995;270:9043–9051. doi: 10.1074/jbc.270.16.9043. [DOI] [PubMed] [Google Scholar]
- 21.Colwell NS, Grupe MJ, Tollefsen DM. Amino acid residues of heparin cofactor II required for stimulation of thrombin inhibition by sulphated polyanions. Biochim Biophys Acta. 1999;1431:148–156. doi: 10.1016/s0167-4838(99)00051-5. [DOI] [PubMed] [Google Scholar]
- 22.Rezaie AR. Role of exosites 1 and 2 in thrombin reaction with plasminogen activator inhibitor-1 in the absence and presence of cofactors. Biochemistry. 1999;38:14592–14599. doi: 10.1021/bi9913303. [DOI] [PubMed] [Google Scholar]
- 23.Stone SR, Brown-Luedi ML, Rovelli G, Guidolin A, McGlynn E, Monard D. Localization of the heparin-binding site of glia-derived nexin/protease nexin-1 by site-directed mutagenesis. Biochemistry. 1994;33:7731–7735. doi: 10.1021/bi00190a028. [DOI] [PubMed] [Google Scholar]
- 24.Derechin VM, Blinder MA, Tollefsen DM. Substitution of arginine for Leu444 in the reactive site of heparin cofactor II enhances the rate of thrombin inhibition. J Biol Chem. 1990;265:5623–5628. [PubMed] [Google Scholar]
- 25.Blinder MA, Andersson TR, Abildgaard U, Tollefsen DM. Heparin cofactor II Oslo. Mutation of Arg-189 to His decreases the affinity for dermatan sulfate. J Biol Chem. 1989;264:5128–5133. [PubMed] [Google Scholar]
- 26.Blinder MA, Tollefsen DM. Site-directed mutagenesis of arginine 103 and lysine 185 in the proposed glycosaminoglycan-binding site of heparin cofactor II. J Biol Chem. 1990;265:286–291. [PubMed] [Google Scholar]
- 27.Whinna HC, Blinder MA, Szewczyk M, Tollefsen DM, Church FC. Role of lysine 173 in heparin binding to heparin cofactor II. J Biol Chem. 1991;266:8129–8135. [PubMed] [Google Scholar]
- 28.Bauman SJ, Church FC. Enhancement of heparin cofactor II anticoagulant activity. J Biol Chem. 1999;274:34556–34565. doi: 10.1074/jbc.274.49.34556. [DOI] [PubMed] [Google Scholar]
- 29.Ciaccia AV, Cunningham EL, Church FC. Characterization of recombinant heparin cofactor II expressed in insect cells. Protein Expr Purif. 1995;6:806–812. doi: 10.1006/prep.1995.0012. [DOI] [PubMed] [Google Scholar]
- 30.Bird PI, Pak SC, Worrall DM, Bottomley SP. Production of recombinant serpins in Escherichia coli. Methods. 2004;32:169–176. doi: 10.1016/s1046-2023(03)00208-1. [DOI] [PubMed] [Google Scholar]
- 31.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 32.Kurland C, Gallant J. Errors of heterologous protein expression. Curr Opin Biotechnol. 1996;7:489–493. doi: 10.1016/s0958-1669(96)80050-4. [DOI] [PubMed] [Google Scholar]
- 33.Blinder MA, Marasa JC, Reynolds CH, Deaven LL, Tollefsen DM. Heparin cofactor II: cDNA sequence, chromosome localization, restriction fragment length polymorphism, and expression in Escherichia coli. Biochemistry. 1988;27:752–759. doi: 10.1021/bi00402a039. [DOI] [PubMed] [Google Scholar]
- 34.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henaut A, Danchin A. Analysis and predictions from Escherichia coli sequences, or E. coli in silico. In: Neidhardt FC, editor. Escherichia coli and Salmonella Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 2047–2066. [Google Scholar]
- 36.Burgess AW, Begley CG, Johnson GR, Lopez AF, Williamson DJ, Mermod JJ, Simpson RJ, Schmitz A, DeLamarter JF. Purification and properties of bacterially synthesized human granulocyte-macrophage colony stimulating factor. Blood. 1987;69:43–51. [PubMed] [Google Scholar]
- 37.Han JH, Van Deerlin VM, Tollefsen DM. Heparin facilitates dissociation of complexes between thrombin and a reactive site mutant (L444R) of heparin cofactor II. J Biol Chem. 1997;272:8243–8249. doi: 10.1074/jbc.272.13.8243. [DOI] [PubMed] [Google Scholar]
- 38.Bock PE. Active-site-selective labeling of blood coagulation proteinases with fluorescence probes by the use of thioester peptide chloromethyl ketones. I. Specificity of thrombin labeling. J Biol Chem. 1992;267:14963–14973. [PubMed] [Google Scholar]
- 39.Chase T, Jr, Shaw E. Comparison of the esterase activities of trypsin, plasmin, and thrombin on guanidinobenzoate esters. Titration of the enzymes. Biochemistry. 1969;8:2212–2224. doi: 10.1021/bi00833a063. [DOI] [PubMed] [Google Scholar]
- 40.Fenton JW, 2nd, Fasco MJ, Stackrow AB. Human thrombins. Production, evaluation, and properties of alpha-thrombin. J Biol Chem. 1977;252:3587–3598. [PubMed] [Google Scholar]
- 41.Creighton TE. Protein Structure, a Practical Approach. Oxford University Press; 1997. [Google Scholar]
- 42.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson M, McClelland M. Use of DNA methyltransferase/endonuclease enzyme combinations for megabase mapping of chromosomes. Methods Enzymol. 1992;216:279–303. doi: 10.1016/0076-6879(92)16027-h. [DOI] [PubMed] [Google Scholar]
- 44.Meagher JL, Olson ST, Gettins PG. Critical role of the linker region between helix D and strand 2A in heparin activation of antithrombin. J Biol Chem. 2000;275:2698–2704. doi: 10.1074/jbc.275.4.2698. [DOI] [PubMed] [Google Scholar]
- 45.O’Keeffe D, Olson ST, Gasiunas N, Gallagher J, Baglin TP, Huntington JA. The heparin binding properties of heparin cofactor II suggest an antithrombin-like activation mechanism. J Biol Chem. 2004;279:50267–50273. doi: 10.1074/jbc.M408774200. [DOI] [PubMed] [Google Scholar]
- 46.Verhamme IM, Olson ST, Tollefsen DM, Bock PE. Binding of exosite ligands to human thrombin. Re-evaluation of allosteric linkage between thrombin exosites I and II. J Biol Chem. 2002;277:6788–6798. doi: 10.1074/jbc.M110257200. [DOI] [PubMed] [Google Scholar]
- 47.Bock PE. Active-site-selective labeling of blood coagulation proteinases with fluorescence probes by the use of thioester peptide chloromethyl ketones. II. Properties of thrombin derivatives as reporters of prothrombin fragment 2 binding and specificity of the labeling approach for other proteinases. J Biol Chem. 1992;267:14974–14981. [PubMed] [Google Scholar]
- 48.Tian WX, Tsou CL. Determination of the rate constant of enzyme modification by measuring the substrate reaction in the presence of the modifier. Biochemistry. 1982;21:1028–1032. doi: 10.1021/bi00534a031. [DOI] [PubMed] [Google Scholar]
- 49.Hogg PJ, Jackson CM. Fibrin monomer protects thrombin from inactivation by heparin–antithrombin III: implications for heparin efficacy. Proc Natl Acad Sci USA. 1989;86:3619–3623. doi: 10.1073/pnas.86.10.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarilla S, Habib SY, Kravtsov DV, Matafonov A, Gailani D, Verhamme IM. Sucrose octasulfate selectively accelerates thrombin inactivation by heparin cofactor II. J Biol Chem. 2010;285:8278–8289. doi: 10.1074/jbc.M109.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Cera E. Thrombin. Mol Aspects Med. 2008;29:203–254. doi: 10.1016/j.mam.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Böhme C, Nimtz M, Grabenhorst E, Conradt HS, Strathmann A, Ragg H. Tyrosine sulfation and N-glycosylation of human heparin cofactor II from plasma and recombinant Chinese hamster ovary cells and their effects on heparin binding. Eur J Biochem. 2002;269:977–988. doi: 10.1046/j.0014-2956.2001.02732.x. [DOI] [PubMed] [Google Scholar]
- 53.Herzog R, Lutz S, Blin N, Marasa JC, Blinder MA, Tollefsen DM. Complete nucleotide sequence of the gene for human heparin cofactor II and mapping to chromosomal band 22q11. Biochemistry. 1991;30:1350–1357. doi: 10.1021/bi00219a027. [DOI] [PubMed] [Google Scholar]
- 54.Ragg H. A new member of the plasma protease inhibitor gene family. Nucleic Acids Res. 1986;14:1073–1088. doi: 10.1093/nar/14.2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang SP, Zubay G, Goldman E. Low-usage codons in Escherichia coli, yeast, fruit fly and primates. Gene. 1991;105:61–72. doi: 10.1016/0378-1119(91)90514-c. [DOI] [PubMed] [Google Scholar]
- 57.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 58.Spanjaard RA, Chen K, Walker JR, van Duin J. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNA(Arg) Nucleic Acids Res. 1990;18:5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenberg AH, Goldman E, Dunn JJ, Studier FW, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hrzenjak A, Artl A, Knipping G, Kostner G, Sattler W, Malle E. Silent mutations in secondary Shine–Dalgarno sequences in the cDNA of human serum amyloid A4 promotes expression of recombinant protein in Escherichia coli. Protein Eng. 2001;14:949–952. doi: 10.1093/protein/14.12.949. [DOI] [PubMed] [Google Scholar]
- 61.Jin H, Zhao Q, Gonzalez de Valdivia EI, Ardell DH, Stenström M, Isaksson LA. Influences on gene expression in vivo by a Shine–Dalgarno sequence. Mol Microbiol. 2006;60:480–492. doi: 10.1111/j.1365-2958.2006.05110.x. [DOI] [PubMed] [Google Scholar]
- 62.Ivanov I, Alexandrova R, Dragulev B, Saraffova A, AbouHaidar MG. Effect of tandemly repeated AGG triplets on the translation of CAT-mRNA in E. coli. FEBS Lett. 1992;307:173–176. doi: 10.1016/0014-5793(92)80761-5. [DOI] [PubMed] [Google Scholar]
- 63.Tartoff KD, Hobbs CA. Improved media for growing plasmid and cosmid clones. Bethesda Res Lab Focus. 1987;9:12. [Google Scholar]
- 64.Attrill H, Harding PJ, Smith E, Ross S, Watts A. Improved yield of a ligand-binding GPCR expressed in E. coli for structural studies. Protein Expr Purif. 2009;64:32–38. doi: 10.1016/j.pep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 65.Sivashanmugam A, Murray V, Cui C, Zhang Y, Wang J, Li Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 2009;18:936–948. doi: 10.1002/pro.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baneyx F. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 1999;10:411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 67.Sim BJ, Tan DSH, Liu X, Sim TS. Production of high levels of soluble recombinant Streptomyces clavuligerus isopenicillin N synthase in Escherichia coli. J Mol Catal B: Enzyme. 1996;2:71–83. [Google Scholar]
- 68.Looman AC, Bodlaender J, Comstock LJ, Eaton D, Jhurani P, de Boer HA, van Knippenberg PH. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J. 1987;6:2489–2492. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson CB, Blackburn MN. Isolation and characterization of an antithrombin III variant with reduced carbohydrate content and enhanced heparin binding. J Biol Chem. 1985;260:610–615. [PubMed] [Google Scholar]
- 70.Frebelius S, Isaksson S, Swedenborg J. Thrombin inhibition by antithrombin III on the subendothelium is explained by the isoform AT beta. Arterioscler Thromb Vasc Biol. 1996;16:1292–1297. doi: 10.1161/01.atv.16.10.1292. [DOI] [PubMed] [Google Scholar]
- 71.Swedenborg J. The mechanisms of action of alpha- and beta-isoforms of antithrombin. Blood Coagul Fibrinolysis. 1998;9(Suppl 3):S7–S10. [PubMed] [Google Scholar]
- 72.Olson ST. Transient kinetics of heparin-catalyzed protease inactivation by antithrombin III. Linkage of protease-inhibitor-heparin interactions in the reaction with thrombin. J Biol Chem. 1988;263:1698–1708. [PubMed] [Google Scholar]