Abstract
Methylthiotransferases (MTTases) are a closely related family of proteins that perform both radical-S-adenosylmethionine (SAM) mediated sulfur insertion and SAM-dependent methylation to modify nucleic acid or protein targets with a methyl thioether group (–SCH3). Members of two of the four known subgroups of MTTases have been characterized, typified by MiaB, which modifies N6-isopentenyladenosine (i6A) to 2-methylthio-N6-isopentenyladenosine (ms2i6A) in tRNA, and RimO, which modifies a specific aspartate residue in ribosomal protein S12. In this work, we have characterized the two MTTases encoded by Bacillus subtilis 168 and find that, consistent with bioinformatic predictions, ymcB is required for ms2i6A formation (MiaB activity), and yqeV is required for modification of N6-threonylcarbamoyladenosine (t6A) to 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A) in tRNA. The enzyme responsible for the latter activity belongs to a third MTTase subgroup, no member of which has previously been characterized. We performed domain-swapping experiments between YmcB and YqeV to narrow down the protein domain(s) responsible for distinguishing i6A from t6A and found that the C-terminal TRAM domain, putatively involved with RNA binding, is likely not involved with this discrimination. Finally, we performed a computational analysis to identify candidate residues outside the TRAM domain that may be involved with substrate recognition. These residues represent interesting targets for further analysis.
INTRODUCTION
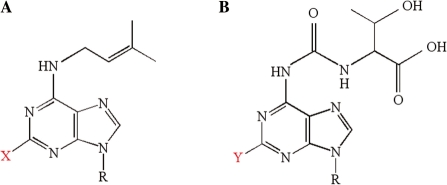
Transfer RNA molecules from all three domains of life undergo numerous, and often complex, post-transcriptional modifications in the course of maturation. Residue 37, which is 3′-adjacent to the anticodon and almost invariably purine, is a frequent target of modification, the specific type of which appears to vary with the identity of residue 36 (the third residue of the anticodon) (1). Among the modified residues found exclusively at this position are N6-isopentenyladenosine (i6A) and N6-threonylcarbamoyladenosine (t6A), and their methylthiolated derivatives 2-methylthio-N6-isopentenyladenosine (ms2i6A) and 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A) (Figure 1). In Escherichia coli and Bacillus subtilis, i6A or ms2i6A occur in most tRNAs with A36 (reading UNN codons), while t6A or ms2t6A occur in most tRNAs with U36 (reading ANN codons) (1,2). Experimental evidence supports the hypothesis that these hydrophobic modifications stabilize the relatively weak A : U and U : A base pairs formed by the third base of the anticodons of these tRNAs with the first base of their complementary codons, thereby improving translational fidelity (1,3,4).
Figure 1.
Schematic structures of the methylthiolated nucleic acid residues from B. subtilis. (A) i6A when X = H and ms2i6A when X = SCH3. (B) t6A when Y = H and ms2t6A when Y = SCH3.
To the extent that the pathways are known, these modifications are constructed in a stepwise manner, and some of the enzymes involved have been discovered and characterized. Modification of adenosine to i6A is carried out by the gene products of miaA in E. coli and MOD5 in yeast, which transfer the Δ2-isopentenyl group from dimethylallyl diphosphate, a mevalonic acid derivative, to A37 N6 (5–8). Mutations in miaA (originally called trpX) result in the accumulation of tRNA with strictly unmodified A37, indicating the formation of i6A is a prerequisite for any further modification (9). Formation of t6A from adenosine is less well characterized but is known to be an ATP-dependent reaction requiring threonine and carbonate (10). Recent genetic evidence shows that E. coli yrdC and yeast SUA5 are required for this modification, but the involvement of other proteins in the reaction cannot yet be ruled out (11). Although not yet demonstrated, it seems reasonable to expect that t6A formation is a requisite first step for further modification based on analogy to the i6A pathway.
Methylthiolation (–SCH3 addition) at C2 of the adenosine ring forms ms2i6A and ms2t6A from i6A and t6A, respectively (Figure 1). The miaB gene product has been shown to catalyze the formation of ms2i6A in both E. coli and Thermotoga maritima (12–14). This reaction was originally hypothesized to occur by sequential steps, possibly catalyzed by separate enzymes: sulfur insertion by a miaB activity, followed by S-adenosylmethionine (SAM)-dependent methylation of a thiolated intermediate by a miaC activity (15,16). This was seemingly borne out by experiments using an E. coli rel met cys mutant (auxotrophic for methionine and cysteine yet capable of RNA synthesis in their absence). Under conditions of methionine starvation, such cells accumulated an uncharacterized i6A derivative that was capable of subsequent labeling using 14C-SAM, presumably by methylation (15). It was speculated that this uncharacterized nucleoside was the intermediate s2i6A, but this has yet to be confirmed, and this intermediate has not been observed elsewhere. Furthermore, recent results with the closely related enzyme RimO contradict this, suggesting instead that methylation of the sulfur atom occurs on an enzyme-bound FeS cluster prior to insertion (17). In any case, once discovered, purified MiaB was shown to be a methylthiotransferase (MTTase), responsible for both thiolation and methylation of i6A (18).
Phylogenetic analysis of the MTTase family shows that it consists of four clades, members of two of which have been characterized (19,20). One clade includes MiaB and its homologs, and is found exclusively in bacteria and eukaryotic organelles. The second characterized clade, also exclusively bacterial, includes RimO, a MTTase that modifies D88 of ribosomal protein S12 in E. coli (19). A third bacterial clade, typified by B. subtilis YqeV, and a fourth clade, found exclusively in archaea and eukaryotes and typified by Methanocaldococcus jannaschii Mj0867, remain uncharacterized. Given that the nucleoside ms2t6A has been observed both in bacteria [including B. subtilis (21) but not E. coli] and in archaea (22), it seems reasonable to expect that members of both of these clades are responsible for methylthiolating t6A to ms2t6A, with the phylogenetic distinction reflecting the ancient split of the bacterial domain from archaea and eukaryotes rather than a functional differentiation within the protein family.
MTTases are members of the so-called ‘radical-SAM’ superfamily of proteins, which use a reducing equivalent from a prosthetic [4Fe–4S]1+ cluster to cleave SAM, generating methionine and a reactive 5′-dA• radical (23). In the case of MiaB and RimO, this radical facilitates the difficult C–H to C–S bond conversion in the nucleoside or amino acid substrate, respectively, by abstracting the hydrogen atom and creating a reactive substrate radical that is amenable to sulfur insertion. All MTTases share a common tripartite domain structure, with the central domain responsible for this radical-SAM chemistry. The N-terminal domain has been shown in MiaB and RimO to harbor a second FeS cluster (17,24), a feature common to other radical-SAM proteins that catalyze sulfur insertion reactions such as BioB and LipA (25); this cluster is speculated to serve as the immediate sulfur donor in the thiolation reaction. The C-terminal TRAM domain has been shown to bind RNA in the context of other proteins (26), suggesting a similar function in tRNA-modifying MTTases. Its presence in the protein-modifying RimO is less obvious, but a recently published structure suggests that in this particular protein it has adapted to bind the protein substrate rather than RNA (27).
In this work, we identify and characterize the two MTTases encoded in the genome of B. subtilis str. 168, products of the ymcB and yqeV genes. We confirm the earlier prediction that YqeV performs the novel t6A methylthiolation function, and verify that YmcB, a MiaB ortholog, methylthiolates i6A. We then went on to construct several chimeric proteins derived from these two closely-related enzymes and show that, RNA-binding function of the TRAM domain notwithstanding, the ability of these two enzymes to discriminate between i6A and t6A resides in the N-terminal and/or radical-SAM domains, and not in the TRAM domain. Finally, we were unable to complement the loss of yqeV in B. subtilis with either the mj0867 gene or a mesophilic ortholog in trans. We suggest that yqeV be renamed tmtB, for the second step in tRNA-methylthiolation, reserving tmtA for the t6A modifying activity.
MATERIALS AND METHODS
Enzymes
All restriction enzymes and T4 DNA ligase were from New England Biolabs (Ipswich, MA, USA). All PCR reactions were performed using Phusion DNA polymerase (New England Biolabs) in the HF buffer, unless otherwise noted.
Bacterial strains and media
Table 1 describes the bacterial strains and plasmids used in this study. Bacillus subtilis strains BSF2608 and YQEVd were derived from strain 168 by single-crossover (Campbell type) insertion of the plasmid pMUTIN (28). The insertion in BSF2608 is after ymcB nt 533, and that in YQEVd is after yqeV nt 280 (29). Insertions were confirmed by PCR amplification (data not shown).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Bacillus subtilis | ||
| 168 | trpC2 | (52) |
| BSF2608 | 168, ymcB::pMUTIN | (29) |
| YQEVd | 168, yqeV::pMUTIN | (29) |
| B(124) | BSF2608 × pDM124c7 | This work |
| V(124) | YQEVd × pDM124c7 | This work |
| B(B) | BSF2608 × pDMymcB | This work |
| B(MM) | BSF2608 × pDMmmar | This work |
| V(V) | YQEVd × pDMyqeV | This work |
| V(MJ) | YQEVd × pDMmj0867 | This work |
| V(MM) | YQEVd × pDMmmar | This work |
| B(B1V) | BSF2608 × pDM-B1V | This work |
| B(B2V) | BSF2608 × pDM-B2V | This work |
| B(V1B) | BSF2608 × pDM-V1B | This work |
| B(V2B) | BSF2608 × pDM-V2B | This work |
| V(B1V) | YQEVd × pDM-B1V | This work |
| V(B2V) | YQEVd × pDM-B2V | This work |
| V(V1B) | YQEVd × pDM-V1B | This work |
| V(V2B) | YQEVd × pDM-V2B | This work |
| Plasmids | ||
| pDM124c7 | E. coli/B. subtilis shuttle plasmid with the promoter region from the B. amyloliquefaciens α-amylase gene followed by a multiple cloning site | J. Benner and D. Martin (unpublished) |
| pDMymcB | pDM124c7 with ymcB cloned between NdeI and XhoI | This work |
| pDMyqeV | pDM124c7 with yqeV cloned between NdeI and XhoI | This work |
| pDMmj0867 | pDM124c7 with mj0867 cloned between NdeI and XhoI | This work |
| pDMmmar | pDM124c7 with mmar cloned between NdeI and XhoI | This work |
| pDM-B1V | pDM124c7 with ymcB/yqeV chimera B1V between NdeI and XhoI | This work |
| pDM-B2V | pDM124c7 with ymcB/yqeV chimera B2V between NdeI and XhoI | This work |
| pDM-V1B | pDM124c7 with ymcB/yqeV chimera V1B between NdeI and XhoI | This work |
| pDM-V2B | pDM124c7 with ymcB/yqeV chimera V2B between NdeI and XhoI | This work |
To make B. subtilis competent cells, 5 ml Rich medium supplemented with 3 mM MgSO4 was inoculated with a single colony and grown at 37°C with vigorous aeration to OD600 ∼1.0. Then, 0.5 ml of this culture was used to inoculate 10 ml minimal medium (0.9× PC buffer, 2% glucose, 3 mM MgSO4, 2.5 mg/ml potassium aspartate, 11 µg/ml ferric ammonium citrate, 50 µg/ml phenylalanine, 50 µg/ml tryptophan), which was then grown 4 h at 37°C with vigorous aeration. Three milliliters of 50% glycerol was then added to the culture, which was frozen at −80°C in 1 ml aliquots until ready for use. The composition of 1× PC buffer was 0.1 M potassium phosphate, 3 mM trisodium citrate, pH adjusted to 7.5 with KOH.
Transformation of B. subtilis was accomplished by mixing 1.5 µg plasmid DNA with 200 µl competent cells, growing 37°C 1 h with vigorous aeration, and plating on Rich medium supplemented with 12.5 µg/ml chloramphenicol as required. Plasmids used for B. subtilis transformation were purified from E. coli NEBTurbo (New England Biolabs), a recA+ strain.
Plasmid construction
The shuttle plasmid used for complementation, pDM124c7 (J. Benner and D. Martin, unpublished data), contains a ColE1 origin of replication and ampicillin-resistance marker for propagation in E. coli and a chloramphenicol-resistance marker for selection in B. subtilis. Genes were cloned at the NdeI-XhoI sites, with expression in B. subtilis driven constitutively by the B. amyloliquefaciens α-amylase promoter and expressed proteins targeted to the cytoplasm.
Genes encoding MTTases were cloned by PCR amplification, with ymcB and yqeV from B. subtilis 168 genomic DNA, gene mj0867 from M. jannaschii genomic DNA, and gene mmar from Methanococcus maripaludis genomic DNA, using primers described in Table 2. All four genes were digested at the NdeI and XhoI sites (underlined in Table 2) and inserted at the same sites in pDM124c7. Cloning steps were performed in E. coli, and constructs verified by DNA sequencing.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence | Target sitea |
|---|---|---|
| ymcB_F | ATAAAACATATGAATGAAAAACAAAAATTAGAGAG | ymcB nt 1–26, forward |
| ymcB_R | ATAAAACTCGAGTCATTTCACCTCGATTGCTTCTCCTACC | ymcB nt 1530–1503, reverse |
| yqeV_F | TATTTTCATATGGCAACTGTTGCTTTCCATACGCTTG | yqeV nt 1–28, forward |
| yqeV_R | TACCCCCTCGAGTTAAGAAGACAAACGCATGTGTTCAG | yqeV nt 1356–1331, reverse |
| mj0867_F | ATGGCGCATATGTGGTTATATTATTTACAAGTGGTG | mj0867 nt 1–27, forward |
| mj0867_R | ATGCCGCTCGAGTTAAAGGATAAGCTCCCCTTTCAATCC | mj0867 nt 1284–1258, reverse |
| mmar_F | ATGGCGCATATGAAAATTTACATTGAAGGATACGG | mmar nt 1–26, forward |
| mmar_R | ATGCCGCTCGAGTTAATTTATCAGTTTACCCGAAAGTCC | mmar nt 1278–1252, reverse |
| mmar_I | CCTTCACATATGGGTAGTGCGGTAATTAATCC | mmar nt 446–421, reverse |
| ymcBch_F1 | AACACCAGCUGCTAAGATGAAAGATAATG | ymcB nt 1224–1243, forward |
| ymcBch_R1 | AGCTGGTGTUCCTTCACGCGGAGAGTAAATG | ymcB nt 1214–1194, reverse |
| ymcBch_F2 | AGAATACGCUAAGGAATACGAAGGCAAGG | ymcB nt 1318–1336, forward |
| ymcBch_R2 | AGCGTATTCUGCAGAAATTTCATTCACCAG | ymcB nt 1307–1288, reverse |
| yqeVch_F1 | AACACCAGCUGCACGAATGGAAGACCAAG | yqeV nt 1011–1039, forward |
| yqeVch_R1 | AGCTGGTGTUCCTGTACGCTTACTGTAAGG | yqeV nt 1020–991, reverse |
| yqeVch_F2 | AGAATACGCUTCTCAGTATGAAAATGAAG | yqeV nt 1104–1132, forward |
| yqeVch_R2 | AGCGTATTCUTTTGCAAGCTGGTCAGAAAG | yqeV nt 1113–1084, reverse |
| bla_F | AGTTACATGAUCCCCCATGTTGTGCAAAAAAG | bla nt 474–443, reverse |
| bla_R | ATCATGTAACUCGCCTTGATCGTTGGGAAC | bla nt 464–493, forward |
aRegions of hybridization are: for forward cloning primers, from the ATG within the NdeI site (underlined); for reverse cloning primers, 3′ of the XhoI site (underlined); for the internal mmar cloning primer (mmar_I), from the NdeI site (underlined) to the 3′ end; for ymcBch USER primers, 3′ of the uracil used for nicking (underlined); for yqeVch and bla USER primers, the entire primer. bla is oriented counter-clockwise in pDM124c7, so bla_F is forward/clockwise with respect to the plasmid but reverse/bottom-strand with respect to the gene sequence, and vice versa for bla_R.
An internal NdeI site in mmar necessitated a second step in the cloning of that gene, as follows. The cloning steps above yielded a construct containing a truncated gene, with the 450 bp at the 5′ end of the gene missing. The missing fragment was amplified using primers mmar_F and mmar_I (Table 2), digested with NdeI and inserted at the NdeI site of the truncated construct, thus reconstructing the intact gene. Our strain of M. maripaludis is distinct from the four strains whose genome sequences have been published to date, so we use the generic name mmar to refer to the mj0867 ortholog from our strain. The mmar gene sequence and a protein sequence alignment of Mmar with the four published M. maripaludis orthologs are shown in Supplementary Figure S1.
Plasmids containing ymcB/yqeV chimeric genes were constructed using the USER Friendly Cloning Kit (New England Biolabs). Two fragments (designated ‘N’ and ‘C’), which together encompassed the entirety of the plasmid template, were amplified from each of pDMymcB and pDMyqeV using PfuCx Hotstart polymerase (Stratagene, Cedar Creek, TX, USA), with primers described in Table 2. Junctions between the two fragments were (i) within the ymcB or yqeV gene, as described in ‘Results’ section, and (ii) within the ampicillin-resistance marker. Following treatment with the USER enzyme, fragments were annealed such that fragment N derived from one plasmid was paired with fragment C derived from the other. Constructs were verified by DNA sequencing.
tRNA purification
tRNA was purified from 500 ml B. subtilis cultures grown at 37°C in Rich medium and harvested at OD600 between 0.8 and 1.0. Cell pellets were washed with 4 ml TE (10 mM Tris–HCl pH 8.0, 1 mM EDTA), then incubated 37°C 3 h in 4 ml TE with 40 mg/ml lysozyme. Samples were then vigorously mixed with 16 ml TRI Reagent (Sigma-Aldrich, St Louis, MO, USA) and left 10 min at room temperature. Chloroform (3.2 ml) was added, and samples were left for 5 min at room temperature before centrifuging at 8000 r.p.m. for 10 min. Total RNA was precipitated from the aqueous phase with 0.3 M sodium acetate and 70% ethanol and collected by centrifugation at 12 000 r.p.m. for 20 min. tRNA was solubilized by vortexing pellets in 3.2 ml TE with 2 M LiCl and recentrifuging at 8000 r.p.m. for 10 min, then precipitated from the supernatant with 70% ethanol and resuspended in 450 µl TE.
tRNA was cleaned for LC/MS by three rounds of precipitation with 0.3 M ammonium acetate and 70% ethanol, and finally resuspended in 450 µl TE. Typical yield was roughly 1 mg. Eighty micrograms of this was then purified over Nucleobond AX-R 80 ion exchange columns (Macherey-Nagel, Bethlehem, PA, USA), collecting 1.2 ml eluate. tRNA was precipitated from the eluate with 45% isopropanol and resuspended in 40 µl H2O. Typical yield was 65 µg tRNA.
tRNA was digested to nucleosides as follows. Forty microliters (roughly 65 µg) of tRNA was denatured 3 min at 100°C, then rapidly chilled in an ice-H2O slurry. Four microliter 0.1 M ammonium acetate pH 5.3 and 8 U Nuclease P1 (Sigma-Aldrich) were added, and samples were incubated at 45°C for 2 h. Four microliters of 0.1 M ammonium bicarbonate and 0.1 U Phosphodiesterase I (Worthington Biochemical, Lakewood, NJ, USA) were added, and samples were incubated at 37°C for 2 h. Five units Antarctic phosphatase (New England Biolabs) were added, and samples were incubated at 37°C for 1 h.
Liquid chromatography and ESI-MS analysis
Nucleosides were separated, at room temperature, on a Hitachi HPLC system (L-7100 pump) with UV detection at 260 nm (L-7400 UV detector). The column used was a Supelcosil LC-18-S (25 cm × 2.1 mm, 5 μm diameter particles, with a 2 cm × 2.1 mm guard column), which was run at a flow rate of 0.3 ml min−1. The mobile phases used were (i) 5 mM ammonium acetate pH 5.3, and (ii) acetonitrile/water (40:60, v/v) with the gradient described by Pomerantz and McCloskey (30) with minor alterations. The column effluent was split, with one-third directed to a Thermo LTQ-XL (or LTQ-FT) mass spectrometer and two-thirds to the UV detector. Mass spectra were recorded in the positive ion mode from m/z 103 to 510. Electrospray and MS conditions were optimized using adenosine, introduced post-column. The capillary temperature used was 275°C, source voltage 3.7–5 kV, sheath gas flow 45 arbitrary units, auxiliary gas flow 25 arbitrary units and sweep gas flow 10 arbitrary units.
RESULTS
Identification of B. subtilis MTTase genes
A previous BLASTP search of the translated B. subtilis 168 genome using E. coli MiaB as the query revealed two putative MTTases, products of ymcB (BSU17010) and yqeV (BSU25430). Based on phylogenetic considerations, ymcB was predicted to be a true miaB ortholog, with its product responsible for the ms2i6A modification observed in B. subtilis (31), while yqeV belonged to a novel subfamily predicted (19) to be responsible for the observed ms2t6A modification (32). ymcB appears to be a part of bicistronic operon with the gene ymcA, with potential −35 and −10 elements upstream of ymcB and a putative rho-independent terminator downstream of ymcA (33). yqeV is the terminal gene of the heptacistronic dnaK operon, whose transcripts have been mapped and regulation described by Homuth and coworkers (who refer to yqeV as orf50) (34,35). It is expressed from both a heat-inducible promoter upstream of hrcA and from a constitutive promoter upstream of dnaJ. The tetracistronic constitutive transcript encodes, between dnaJ and yqeV, two other methyltransferases involved with modifying the translational machinery: yqeT, a probable homolog of the ribosomal protein L11 MTase prmA, and yqeU, a probable homolog of the 16S rRNA MTase rsmE.
Characterization of mutant phenotypes
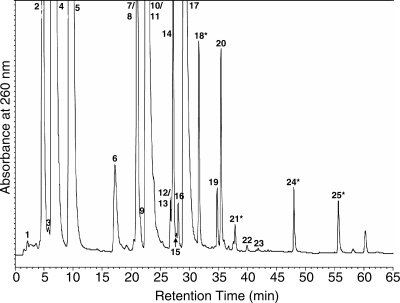
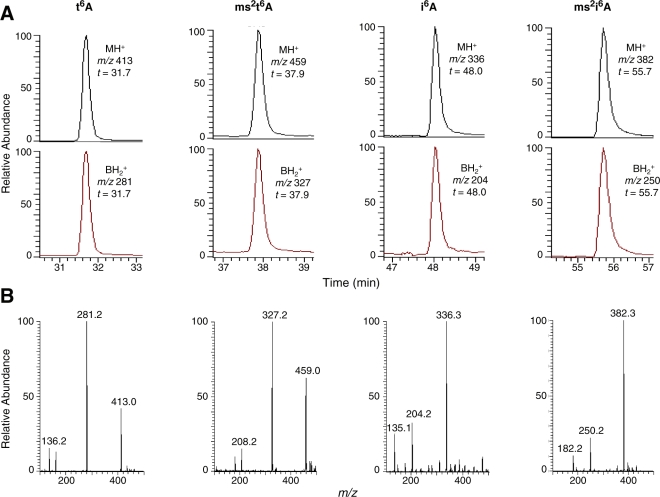
Mutant strains with insertions in both ymcB and yqeV have been constructed (29), indicating that neither gene is essential under normal growth conditions. We obtained these mutant strains as well as the parental strain 168 from the B. subtilis sequencing consortium (http://bacillus.genome.ad.jp). To investigate the effect of these insertion mutants on tRNA modification, we purified and digested to nucleosides total tRNA from the parental and two mutant strains. These nucleoside digests were analyzed by LC/UV and LC/MS to determine the presence or absence of post-transcriptional modifications, specifically t6A, ms2t6A, i6A and ms2i6A. Figure 2 shows the UV chromatogram of the nucleoside digest of tRNA from wild-type strain 168, indicating the presence of peaks at retention times expected for all four modifications (highlighted with ‘asterisks’). The identities of these peaks were confirmed by LC/MS, as shown in Figure 3. The characteristic collision-induced fragmentation pattern of modified nucleosides involves the cleavage of the N-glycosidic bond and the neutral loss of ribose (132 mass units when the 2′-OH is unmethylated, as is the case for the four modified adenosines in question). Thus, the major ions detected by MS are the protonated molecular ion (MH+) and the protonated base ion (BH2+). The corresponding mass spectra in Figure 3 reveal that in each case, the MH+ and BH2+ ions characteristic of the expected nucleoside are present and track to the equivalent retention time as the UV peak.
Figure 2.
UV chromatogram of wild-type B. subtilis 168 total tRNA digested to nucleosides. Numbered peaks are as follows: (1) dihydrouridine (D), (2) pseudouridine (Ψ), (3) 5-carboxymethylaminomethyluridine (cmnm5U), (4) cytidine, (5) uridine, (6) 1-methyladenosine (m1A), (7) 5-methyluridine (m5U), (8) 5-methoxyuridine (mo5U), (9) inosine (I), (10) guanosine, (11) 7-methylguanosine (m7G), (12) 2′-O-methylguanosine (Gm), (13) queuosine (Q), (14) 1-methylguanosine (m1G), (15) lysidine (k2C), (16) N2-methylguanosine (m2G), (17) adenosine, (18) t6A, (19) 2-methyladenosine (m2A), (20) N6-methyladenosine (m6A), (21) ms2t6A, (22) 2-methylthioadenosine (ms2A), (23) N6,N6-methyladenosine (m62A), (24) i6A, (25) ms2i6A. The four peaks relevant to this work are marked with asterisks.
Figure 3.
LC/MS spectra of the adenosine derivatives t6A, ms2t6A, i6A and ms2i6A from digested wild-type B. subtilis 168 total tRNA. (A) Selected ion chromatograms of molecular and base ions. (B) Mass spectra.
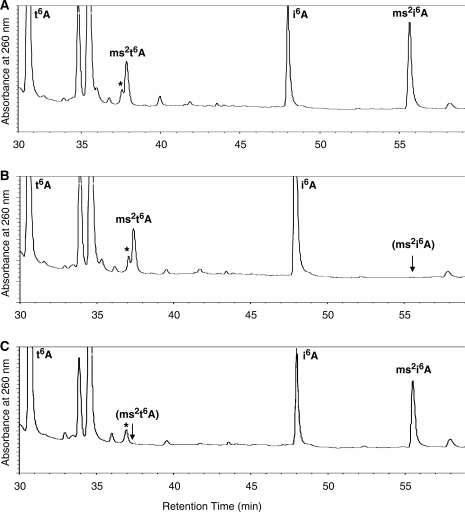
UV chromatograms of nucleoside digests from the two mutant strains are shown in Figure 4. Strain BSF2608 (ymcB) shows specific loss of the ms2i6A peak (Figure 4B), and strain YQEVd (yqeV) shows specific loss of the ms2t6A peak (Figure 4C). Both genes were separately subcloned into the plasmid pDM124c7 (‘Materials and methods’ section) and reintroduced in trans into the respective mutant strains to complement the disrupted alleles. In both cases, the intact plasmid-encoded gene rescued the modification-deficient phenotype, whereas the empty plasmid vector did not, indicating that ymcB and yqeV are required for ms2i6A and ms2t6A modification, respectively, confirming previous phylogenomic predictions (19). These results are summarized in Table 3, and complete LC/UV and LC/MS data for all samples are included as Supplementary Figure S2. Although we will continue to refer to these genes and their products here as ymcB and yqeV for clarity, we suggest that ymcB be renamed miaB and suggest the name tmtB for yqeV.
Figure 4.
UV chromatograms of B. subtilis total tRNA nucleoside digests from the wild-type and two mutant strains, focused on later eluting species. Peaks representing t6A, ms2t6A, i6A and ms2i6A are labeled, with parentheses indicating loss of the expected peak. A non-nucleoside peak eluting close to ms2t6A is indicated by an asterisk. (A) Wild-type strain 168, (B) strain BSF2608 and (C) strain YQEVd.
Table 3.
Modified nucleosides observed in B. subtilis strains
| Strain | Genotypea |
Phenotypeb |
||||
|---|---|---|---|---|---|---|
| ymcB | yqeV | i6A | ms2i6A | t6A | ms2t6A | |
| 168 | + | + | + | + | + | + |
| BSF2608 | − | + | + | −c | + | + |
| B(124) | − | + | + | −c | + | + |
| B(B) | +P | + | + | + | + | + |
| B(MM) | (?)P | + | + | − | + | + |
| B(B1V) | (?)P | + | + | + | + | + |
| B(B2V) | (?)P | + | + | + | + | + |
| B(V1B) | (?)P | + | + | − | + | + |
| B(V2B) | (?)P | + | + | − | + | + |
| YQEVd | + | − | + | + | + | − |
| V(124) | + | − | + | + | + | − |
| V(V) | +P | + | + | + | + | + |
| V(MJ) | + | (?)P | + | + | + | − |
| V(MM) | + | (?)P | + | + | + | − |
| V(B1V) | + | (?)P | + | + | + | − |
| V(B2V) | + | (?)P | + | + | + | − |
| V(V1B) | + | (?)P | + | + | + | − |
| V(V2B) | + | (?)P | + | + | + | − |
aIntact genes indicated by ‘plus’; insertion mutants indicated by ‘minus’; question marks indicate the presence of a heterologous gene that may or may not complement the gene in question; superscript ‘P’ indicates plasmid-encoded copy.
bPhenotypes indicate the presence (plus) or absence (minus) of an LC/UV peak indicative of this modified nucleoside. MS results were consistent with the UV results except where indicated (footnote c).
cWeak MS signal detected in these samples (Supplementary Figure S2).
The ms2i6A- and ms2t6A-deficient B. subtilis strains identified above are, in addition, suitable for characterizing heterologous MTTases of unknown substrate specificity. We cloned mj0867 from M. jannaschii (a hyperthermophile), and the orthologous gene mmar from M. maripaludis (a mesophilic relative of M. jannaschii), into pDM124c7 and introduced them separately into the ms2t6A− strain YQEVd as above. However, despite being predicted to be ms2t6A MTTases, neither gene was able to rescue the modification-deficient phenotype (Table 3 and Supplementary Figure S2). In addition, mmar failed to rescue the ms2i6A deficiency in strain BF2608. Both archaeal proteins appeared largely insoluble in the B. subtilis extracts (data not shown), suggesting that these proteins may be misfolded, and therefore inactive, in this heterologous context.
Construction and characterization of YmcB/YqeV chimeric proteins
We further used the availability of the modification-deficient strains to explore the determinants of substrate recognition in YmcB and YqeV by constructing chimeric proteins. Specifically, we sought to determine whether the TRAM domain, believed to bind RNA, also conferred the recognition of the N6 moiety that differentiates the substrates of these two proteins. YmcB and YqeV share significant sequence similarity across all three domains (Figure 5). We wished to generate a breakpoint between the radical-SAM and TRAM domains at which the TRAM domains could be swapped. However, we noted significant sequence conservation in the region between these two domains as defined by Pfam (Figure 5), and it was thus unclear where the functionally required elements of one domain ended and the other began. Accordingly, we generated two alternative breakpoints for each construct, one close to the Pfam-defined boundary of the radical-SAM domain (breakpoint 1, Figure 5 red arrows) and the other close to the boundary of the TRAM domain (breakpoint 2, Figure 5 green arrows). Four constructs were generated in all, designated B1V, B2V, V1B and V2B, with the first letter (‘B’ for YmcB and ‘V’ for YqeV) describing the source of the N-terminal and radical-SAM domains, the second letter describing the source of the TRAM domain, and the number between them indicating the location of the breakpoint. Note that, due to the requirements of the cloning methodology, breakpoint 2 differs by three residues between B2V and V2B.
Figure 5.
Protein sequence alignment of B. subtilis MTTases YqeV (NP_390421) and YmcB (NP_389583). Boundaries of the three domains as defined by Pfam are boxed in pink (UPF0004; PF00919), cyan (radical-SAM; PF04055) and brown (TRAM; PF01938). Arrows indicate the breakpoints 1 (red) and 2 (green) used to construct chimeric proteins (see text).
We attempted to rescue the modification defect in both BSF2608 and YQEVd with each of the four chimeric constructs as was done for ymcB, yqeV and mj0867. tRNA was purified and the modified nucleosides examined for each of these eight strains (four chimeras in two backgrounds), and we observed successful rescue in two of them: in strains B(B1V) and B(B2V), chimeras B1V and B2V rescued the loss of ms2i6A modification (Table 3 and Supplementary Figure S2). These results are consistent with differential recognition of the i6 or t6 substituent residing in either the N-terminal or radical-SAM domain of these MTTases rather than in the TRAM domain. The inability of V1B and V2B to rescue the ms2t6A defect (or the ms2i6A defect) is likely due to misfolding of the proteins. Indeed, all four chimeric proteins appeared largely insoluble in the cell extracts, which was not the case with the two native proteins (data not shown). However, the B1V and B2V proteins were significantly more highly expressed than V1B and V2B, and enough protein may have folded correctly in these cases to observe enzymatic activity.
In order to pinpoint residues within the N-terminal and/or radical-SAM domains that may be responsible for substrate recognition, we performed a computational analysis that is described more fully in the Supplementary Data. Briefly, we examined a collection of sequences from the three characterized MTTase subfamilies, MiaB, YqeV and RimO, to identify residues that are conserved within each subfamily but differ between them. Such residues are candidates for discriminating the three types of substrates specific for each subfamily. Of the 13 highest scoring residues in our analysis, shown in Supplementary Figure S3, 6 are closely proximal to the 6 invariant cysteines involved with coordinating the FeS clusters, suggesting they may be of structural importance, and 2 more are within the TRAM domain. The remaining five, all within the radical-SAM domain yet distal in the primary sequence to the FeS cluster motif, represent strong candidates for substrate interaction; these are residues 195, 231, 232, 297 and 328, using numbering from B. subtilis YqeV. Note that the highest scoring of these, residue 328, is technically outside of the Pfam-defined radical-SAM domain, but is just upstream of breakpoint 1 in our chimera construction, and therefore segregated with the radical-SAM domain in all of the chimeras we constructed.
Relative modification levels
A semi-quantitative investigation of the LC/UV data revealed apparent differences in the relative amounts of the modifications noted in Table 3 for the 16 strains listed there. We first calculated the peak area ratios of t6A, ms2t6A, i6A and ms2i6A to m5U, a modified nucleoside found at position U54 of all sequenced B. subtilis tRNA species (http://modomics.genesilico.pl), and whose modification is independent of those studied in this work; these ratios can therefore be interpreted as the fractions of all tRNA molecules bearing t6A, ms2t6A, i6A or ms2i6A, respectively. We then calculated the ratios of ms2i6A and ms2t6A to (ms2i6A + i6A) and (ms2t6A + t6A), respectively, that is, the fractions of i6A- and t6A-containing residues that are methylthiolated. This information is summarized in Supplementary Table S2, and the original peak areas from which the ratios were calculated can be found in Supplementary Table S1.
The fraction of t6A residues that are methylthiolated is roughly similar to the wild-type across ms2t6A+ strains at ∼10%, regardless of whether YqeV is expressed from the chromosome or the plasmid (Supplementary Table S2). The wild-type strain 168 exhibits roughly equal amounts of i6A and ms2i6A (50% methylthiolation), but in other ms2i6A+ strains this fraction ranges as high as 70% in strain B(B), where wild-type YmcB is expressed from the plasmid, to as low as 8% for strain B(B1V) (Supplementary Table S2). These more extreme values are likely caused by promiscuous modification of non-cognate tRNA by overexpressed YmcB in the case of B(B), and misfolding of a substantial fraction of chimeric protein in the case of B(B1V), as described earlier.
A final observation from the LC/UV and LC/MS data indicates possible crosstalk between the YqeV and YmcB modification pathways. In three of the four strains where no ms2i6A peak could be detected by LC/UV, trace amounts of the diagnostic MH+ and BH2+ ions could still be detected by LC/MS (Table 3 and Supplementary Figure S2). One of these three was the original insertion mutant strain BSF2608, suggesting that this trace modification could be due either to incomplete inactivation of ymcB by the insertion, or to trace promiscuous activity by the fully active YqeV in these strains. Levels of this nucleotide are low enough that stochastic variation between samples could explain the absence of LC/MS-detectable amounts in the fourth strain.
Other tRNA modifications present
In addition to enabling identification of t6A, ms2t6A, i6A and ms2i6A, Figure 2 represents one of the most comprehensive censuses of tRNA modification in B. subtilis to date, with a total of 23 modified nucleosides identified as present in physiologically relevant amounts. The spectrum of modified nucleosides in Figure 2 is largely consistent with previous studies, including those of Vold and coworkers (36,37), and the collection of sequenced tRNA species in the MODOMICS database (2). All nucleosides positively identified in at least one of those studies can be identified in Figure 2 with the exception of s4U and cmnm5s2U. In addition, we identified k2C, which was previously identified only in a more specialized analysis (38). We detected three nucleosides (m2G, m2A and m26A) that are typical of rRNA and therefore may be indicative of trace rRNA contamination in our tRNA preparation. One other nucleoside, ms2A, is present in trace amounts in all samples and is assumed to be a breakdown product created during tRNA purification. We could identify no modified nucleosides besides those observed in previous studies.
DISCUSSION
Regulation of MTTase activity
Early studies of ms2i6A in bacteria revealed that the methylthiol group, in modulating codon–anticodon interactions, acts as an environmental sensor, mediating cellular responses to changing growth conditions. In E. coli, most i6A residues are methylthiolated to ms2i6A at all stages of growth under normal conditions (31,39). However, i6A is undermodified under conditions of iron limitation in E. coli and other Proteobacteria (40–42). An effect of the loss of ms2 from tRNATrp and tRNAPhe is to specifically reduce the efficiency at which these tRNAs read the regulatory leader sequences preceding the tryptophan and phenylalanine biosynthetic operons, respectively. The slower rate at which these sequences are read by ribosomes disfavors the formation of attenuator structures that prematurely terminate transcription, with the end result being specific up-regulation of the trp and phe operons in response to iron depletion.
The same effect of iron-limitation has been observed in B. subtilis (43), but in addition, the degree of ms2 modification in this differentiating bacterium is growth-phase dependent. During exponential growth, ms2i6A levels are low relative to i6A, whereas the situation is reversed at stationary phase and sporulation (31,43). These two conditions are not independent, since cells grown in iron-deficient media sporulate much less efficiently than in iron-replete media. However, ms2i6A modification also covaries with other activities affecting sporulation such as catabolite repression, NADPH oxidase and isocitrate dehydrogenase, and as a result has been hypothesized to be itself required for sporulation (43). The effect of growth conditions and growth phase on ms2t6A modification has been less closely studied. However, in tRNALys, the sole ms2t6A-modified species identified in B. subtilis, the extent of ms2 modification appears to be somewhat greater in exponentially growing cells than in stationary phase cells (32,44), in direct contrast to the pattern of ms2i6A methylthiolation. Thus, although it is tempting to speculate that growth phase-related changes in methylthiolation are due to fluctuations in intracellular iron concentration, most likely acting through the FeS clusters of MTTases, the inconsistency of behavior of YmcB and YqeV suggests there are additional levels of regulation at work.
Confirmation of the functions encoded by the ymcB and yqeV MTTase genes should facilitate better understanding of the precise regulatory networks that coordinate their activities. The ymcB gene appears to be co-transcribed with ymcA, a master regulator of biofilm formation that is required for pellicle formation and colony differentiation (33). This gene pair is extensively conserved within the Bacilli, but the functional relationship between the two genes remains unclear. The fact that both genes are associated with cell differentiation is nonetheless intriguing. Based on published results, these genes do not appear to be part of any well-characterized regulon, particularly those associated with sporulation (σE, σH, Spo0A) or iron utilization (Fur) (45–49). Given the condition-dependent pattern of ms2i6A modification described earlier, this is somewhat surprising and suggests that some regulatory element remains to be discovered. Transcription of yqeV appears to be dual regulated: a background level of transcript (along with other modification-related genes and dnaJ) expressed from a vegetative promoter, supplemented with expression from a second, longer dnaK transcript governed by a CIRCE-regulated σA promoter. Again the functional connection between the hrcA-related genes and the translation-related modification genes is unclear, but this gene order is again well conserved among both Bacilli and Clostridia, so there appears to be some selective pressure to maintain this arrangement.
Substrate recognition
The substrate elements recognized by MTTases are not completely defined. However, the absence of observed ms2A in miaA mutants indicates that the i6A group is absolutely required for MiaB modification. Given the strong similarity among all MTTases, it seems reasonable to expect that YqeV should correspondingly require the t6A group for modification; however, no t6A− phenotype has been created in an organism harboring an YqeV-type MTTase to test this. Our results with YmcB/YqeV chimeric proteins, while incomplete, nonetheless suggest that recognition of the N6 substituent lies in either the N-terminal or radical-SAM domain, not in the C-terminal TRAM domain. The TRAM domain may function as a non-specific RNA clamp by which the MTTase holds its substrate, such that TRAM domains are completely interchangeable, or alternatively, TRAM may confer additional substrate specificity, perhaps at the RNA sequence level. We have shown that the B1V and B2V chimeras form ms2i6A, but without further characterization of individual tRNA species, it cannot be assumed that these chimeras modify the same set of tRNAs as YmcB.
Bacillus subtilis YmcB and YqeV both appear relatively restrictive in their additional substrate requirements. Of the 7 i6A-modified tRNA species, only tRNA1Phe and tRNA1Tyr are methylthiolated (2), and of the 9 t6A-modified species, only tRNA1Lys is methylthiolated (2,21). Pierrel and coworkers have shown that E. coli MiaB can modify a 17 base RNA oligonucleotide derived from the anticodon stem-loop of tRNAPhe in vitro when the residue corresponding to A37 is isopentenylated (18) so we examined the corresponding regions from the tRNA species above for elements common to those that are methylthiolated. (Alignments of these regions are provided as Supplementary Figure S4.) In the case of i6A-bearing tRNAs, the only sequence feature outside the anticodon that is unique to the two methylthiolated species is the A31:Ψ39 bp at the end of the stem-loop. Ψ:A base pairs at the corresponding position of a synthetic stem-loop oligonucleotide have been shown to have a stabilizing effect on the structure (50). However, tRNA1Trp, which has a Ψ31:A39 bp, is not methylthiolated, suggesting that if this base pair has any significance with respect to YmcB recognition, the important element may be Ψ39 itself (and/or A31) rather than the stem structure it confers. In the case of t6-bearing tRNAs, there is only a single methylthiolated example, tRNA1Lys. It again contains an A31:Ψ39 bp, but tRNA1Thr also contains this base pair yet is not methylthiolated. Indeed, the sequences of tRNA1Lys and tRNA1Thr are nearly identical in this region outside the anticodon, except for the U27:A39 bp, which is unique to tRNA1Lys.
Escherichia coli MiaB appears much less restrictive in its modification than B. subtilis YmcB: of 10 i6A-modified tRNA species, nine are methylthiolated. In addition, mutant tRNAs that have been altered to become MiaA substrates, such as tRNA2Gly C36A, tRNA1Lys U36A and tRNAfMet A35U U36A (51), acquire ms2i6A37, indicating they too are MiaB substrates when isopentenylated. It is unclear to what degree the difference in the suite of methylthiolated tRNA species between these two organisms is due to changes in the proteins and to what degree it is due to changes in tRNA, as both enzyme and substrate coevolve. In B. subtilis, ms2i6A modification has come to be associated with sporulation, a process that has no analog in E. coli. We speculate that this additional role may have influenced these changes, but alternative explanations are certainly possible. In any case, proper assessment of any differences in substrate specificity between E. coli MiaB and B. subtilis YmcB will require complementation of one with the other and full analysis of the modified tRNA species. This work provides a first step towards an understanding of the process of substrate recognition by MTTases, and further progress in this area may shed light on multiple key biological processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (CHE 0602413 cofunded by the MPS/CHE and BIO/IDBR Divisions and by the MPS Office of Multidisciplinary Activities, and CHE 0910751 cofunded by the MPS/CHE and BIO/MCB Divisions, to P.A.L.); National Institutes of Health (NIH RR019900 to P.A.L.); and National Institutes of Health (NIH 1RC2GM092602-01 to S.K., J.V. and R.J.R.). Funding for open access charge: New England Biolabs.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to Alessandro Galizzi, Tsutomu Sato and the National BioResource Project (NIG, Japan): B. subtilis for strains; to Jack Benner, Deana Martin, Marta Meda and Agnieszka Sekowska for plasmids; and to Antoine Danchin for additional help in obtaining materials. The authors also thank Marion Sibley for maintaining bacterial strains, and Irena Nikcevic and Balasubrahmanyam Addepalli for technical advice. Special thanks to Melanie Berkmen for protocols and advice related to B. subtilis. Finally, we thank Don Comb for support.
REFERENCES
- 1.Nishimura S. In: Transfer RNA: Structure, Properties, and Recognition. Schimmel PR, Soll D, Abelson JN, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1979. pp. 59–79. [Google Scholar]
- 2.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller JP, Hussain Z, Schweizer MP. The involvement of the anticodon adjacent modified nucleoside N-(9-(BETA-D-ribofuranosyl) purine-6-ylcarbamoyl)-threonine in the biological function of E. coli tRNAile. Nucleic Acids Res. 1976;3:1185–1201. doi: 10.1093/nar/3.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weissenbach J, Grosjean H. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions. A thermodynamic and kinetic evaluation. Eur. J. Biochem. 1981;116:207–213. doi: 10.1111/j.1432-1033.1981.tb05320.x. [DOI] [PubMed] [Google Scholar]
- 5.Caillet J, Droogmans L. Molecular cloning of the Escherichia coli miaA gene involved in the formation of delta 2-isopentenyl adenosine in tRNA. J. Bacteriol. 1988;170:4147–4152. doi: 10.1128/jb.170.9.4147-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol. Cell Biol. 1987;7:177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Najarian D, Dihanich ME, Martin NC, Hopper AK. DNA sequence and transcript mapping of MOD5: features of the 5′ region which suggest two translational starts. Mol. Cell Biol. 1987;7:185–191. doi: 10.1128/mcb.7.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore JA, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: a binding mechanism for recombinant enzyme. Biochemistry. 1997;36:604–614. doi: 10.1021/bi962225l. [DOI] [PubMed] [Google Scholar]
- 9.Vold BS, Lazar JM, Gray AM. Characterization of a deficiency of N6-(delta 2-isopentenyl)-2-methylthioadenosine in the Escherichia coli mutant trpX by use of antibodies to N6-(delta 2-isopentenyl)adenosine. J. Biol. Chem. 1979;254:7362–7367. [PubMed] [Google Scholar]
- 10.Elkins BN, Keller EB. The enzymatic synthesis of N-(purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry. 1974;13:4622–4628. doi: 10.1021/bi00719a024. [DOI] [PubMed] [Google Scholar]
- 11.El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierrel F, Hernandez HL, Johnson MK, Fontecave M, Atta M. MiaB protein from Thermotoga maritima. Characterization of an extremely thermophilic tRNA-methylthiotransferase. J. Biol. Chem. 2003;278:29515–29524. doi: 10.1074/jbc.M301518200. [DOI] [PubMed] [Google Scholar]
- 13.Pierrel F, Bjork GR, Fontecave M, Atta M. Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 2002;277:13367–13370. doi: 10.1074/jbc.C100609200. [DOI] [PubMed] [Google Scholar]
- 14.Esberg B, Leung HC, Tsui HC, Bjork GR, Winkler ME. Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1999;181:7256–7265. doi: 10.1128/jb.181.23.7256-7265.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agris PF, Armstrong DJ, Schafer KP, Soll D. Maturation of a hypermodified nucleoside in transfer RNA. Nucleic Acids Res. 1975;2:691–698. doi: 10.1093/nar/2.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connolly DM, Winkler ME. Structure of Escherichia coli K-12 miaA and characterization of the mutator phenotype caused by miaA insertion mutations. J. Bacteriol. 1991;173:1711–1721. doi: 10.1128/jb.173.5.1711-1721.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, Roberts RJ, Krebs C, Booker SJ. Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry. 2009;48:10162–10174. doi: 10.1021/bi900939w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierrel F, Douki T, Fontecave M, Atta M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J. Biol. Chem. 2004;279:47555–47563. doi: 10.1074/jbc.M408562200. [DOI] [PubMed] [Google Scholar]
- 19.Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2008;105:1826–1831. doi: 10.1073/pnas.0708608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaminska KH, Baraniak U, Boniecki M, Nowaczyk K, Czerwoniec A, Bujnicki JM. Structural bioinformatics analysis of enzymes involved in the biosynthesis pathway of the hypermodified nucleoside ms(2)io(6)A37 in tRNA. Proteins. 2008;70:1–18. doi: 10.1002/prot.21640. [DOI] [PubMed] [Google Scholar]
- 21.Yamada Y, Ishikura H. The presence of N-[(9-beta-D-ribofuranosyl-2-methylthiopurin-6-yl)carbamoyl]threonine in lysine tRNA1 from Bacillus subtilis. J. Biochem. 1981;89:1589–1591. doi: 10.1093/oxfordjournals.jbchem.a133353. [DOI] [PubMed] [Google Scholar]
- 22.Reddy DM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Stetter KO, Widdel F, McCloskey JA. Structure determination of two new amino acid-containing derivatives of adenosine from tRNA of thermophilic bacteria and archaea. Nucleic Acids Res. 1992;20:5607–5615. doi: 10.1093/nar/20.21.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez HL, Pierrel F, Elleingand E, Garcia-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. MiaB, a bifunctional radical-S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe-4S] clusters. Biochemistry. 2007;46:5140–5147. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
- 25.Booker SJ, Cicchillo RM, Grove TL. Self-sacrifice in radical S-adenosylmethionine proteins. Curr. Opin. Chem. Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee TT, Agarwalla S, Stroud RM. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 27.Arragain S, Garcia-Serres R, Blondin G, Douki T, Clemancey M, Latour JM, Forouhar F, Neely H, Montelione GT, Hunt JF, et al. Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase. J. Biol. Chem. 2010;285:5792–5801. doi: 10.1074/jbc.M109.065516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pomerantz SC, McCloskey JA. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 31.Vold BS. Post-transcriptional modifications of the anticodon loop region: alterations in isoaccepting species of tRNA's during development in Bacillus subtilis. J. Bacteriol. 1978;135:124–132. doi: 10.1128/jb.135.1.124-132.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vold BS, Keith D.E., Jr, Buck M, McCloskey JA, Pang H. Lysine tRNAs from Bacillus subtilis 168: structural analysis. Nucleic Acids Res. 1982;10:3125–3132. doi: 10.1093/nar/10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J. Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W. The DNAK operon of Bacillus subtilis is heptacistronic. J. Bacteriol. 1997;179:1153–1164. doi: 10.1128/jb.179.4.1153-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Homuth G, Mogk A, Schumann W. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol. Microbiol. 1999;32:1183–1197. doi: 10.1046/j.1365-2958.1999.01428.x. [DOI] [PubMed] [Google Scholar]
- 36.Vold B. Modified nucleosides of Bacillus subtilis transfer ribonucleic acids. J. Bacteriol. 1976;127:258–267. doi: 10.1128/jb.127.1.258-267.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vold BS, Longmire ME, Keith D.E., Jr Thiolation and 2-methylthio- modification of Bacillus subtilis transfer ribonucleic acids. J. Bacteriol. 1981;148:869–876. doi: 10.1128/jb.148.3.869-876.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsugi J, Murao K, Ishikura H. Characterization of a B. subtilis minor isoleucine tRNA deduced from tDNA having a methionine anticodon CAT. J. Biochem. 1996;119:811–816. doi: 10.1093/oxfordjournals.jbchem.a021312. [DOI] [PubMed] [Google Scholar]
- 39.Bartz J, Soll D, Burrows WJ, Skoog F. Identification of the cytokinin-active ribonucleosides in pure Escherichia coli tRNA species. Proc. Natl Acad. Sci. USA. 1970;67:1448–1453. doi: 10.1073/pnas.67.3.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLennan BD, Buck M, Humphreys J, Griffiths E. Iron-related modification of bacterial transfer RNA. Nucleic Acids Res. 1981;9:2629–2640. doi: 10.1093/nar/9.11.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg AH, Gefter ML. An iron-dependent modification of several transfer RNA species in Escherichia coli. J. Mol. Biol. 1969;46:581–584. doi: 10.1016/0022-2836(69)90197-1. [DOI] [PubMed] [Google Scholar]
- 42.Buck M, Griffiths E. Iron mediated methylthiolation of tRNA as a regulator of operon expression in Escherichia coli. Nucleic Acids Res. 1982;10:2609–2624. doi: 10.1093/nar/10.8.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menichi B, Heyman T. Study of tyrosine transfer ribonucleic acid modification in relation to sporulation in Bacillus subtilis. J. Bacteriol. 1976;127:268–280. doi: 10.1128/jb.127.1.268-280.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vold BS. Analysis of isoaccepting transfer ribonucleic acid species of Bacillus subtilis: changes in chromatography of transfer ribonucleic acids associated with stage of development. J. Bacteriol. 1973;114:178–182. doi: 10.1128/jb.114.1.178-182.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feucht A, Evans L, Errington J. Identification of sporulation genes by genome-wide analysis of the sigmaE regulon of Bacillus subtilis. Microbiology. 2003;149:3023–3034. doi: 10.1099/mic.0.26413-0. [DOI] [PubMed] [Google Scholar]
- 46.Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 48.Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl Acad. Sci. USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- 50.Meroueh M, Grohar PJ, Qiu J, SantaLucia J., Jr, Scaringe SA, Chow CS. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res. 2000;28:2075–2083. doi: 10.1093/nar/28.10.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mangroo D, Limbach PA, McCloskey JA, RajBhandary UL. An anticodon sequence mutant of Escherichia coli initiator tRNA: possible importance of a newly acquired base modification next to the anticodon on its activity in initiation. J. Bacteriol. 1995;177:2858–2862. doi: 10.1128/jb.177.10.2858-2862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.