Abstract
Under physiological conditions, guanine-rich sequences of DNA and RNA can adopt stable and atypical four-stranded helical structures called G-quadruplexes (G4). Such G4 structures have been shown to occur in vivo and to play a role in various processes such as transcription, translation and telomere maintenance. Owing to their high-thermodynamic stability, resolution of G4 structures in vivo requires specialized enzymes. RHAU is a human RNA helicase of the DEAH-box family that exhibits a unique ATP-dependent G4-resolvase activity with a high affinity and specificity for its substrate in vitro. How RHAU recognizes G4-RNAs has not yet been established. Here, we show that the amino-terminal region of RHAU is essential for RHAU to bind G4 structures and further identify within this region the evolutionary conserved RSM (RHAU-specific motif) domain as a major affinity and specificity determinant. G4-resolvase activity and strict RSM dependency are also observed with CG9323, the Drosophila orthologue of RHAU, in the amino terminal region of which the RSM is the only conserved motif. Thus, these results reveal a novel motif in RHAU protein that plays an important role in recognizing and resolving G4-RNA structures, properties unique to RHAU among many known RNA helicases.
INTRODUCTION
In cells, the strong inclination of RNA for mis-folding or adopting non-functional conformations is overcome by the presence of RNA chaperones that facilitate conformational transitions of RNA. Among these chaperones are the RNA helicases, which couple NTP hydrolysis with structural and functional rearrangement of the RNA. The DEAD/H-box proteins constitute a widely spread subgroup of RNA helicases that have been identified in all forms of life, including viruses. DEAD/H-box proteins have been shown to catalyse the disruption of RNA–RNA interactions (1,2), to remodel ribonucleoprotein (RNP) complexes (3,4) and to assist the correct folding of RNA (5,6). In this regard, DEAD/H-box proteins are essential cellular components that take part in many, if not all, aspects of RNA metabolism, ranging from transcription to RNA decay [for review see (7–9)].
Structurally, DEAD/H-box proteins consist of a highly conserved catalytic core composed of two RecA-like domains that couples NTP hydrolysis with the helicase activity. The helicase core domain is often flanked by ancillary N- and C-terminal regions of variable length and sequence. While the core domain of RNA helicases has been extensively studied, much less is known about the biological role of the N- and C-terminal regions. Because of the high degree of amino acid sequence conservation within the helicase core of DEAD/H-box proteins, this region may not contribute directly to the substrate specificity of the enzyme. In contrast to the helicase core, the N- and C-terminal flanking regions are usually unique, with the exception of certain identifiable sequence features. These regions have been shown to provide substantial substrate specificity through their interaction with RNAs or with protein partners that modulate the activity and/or the specificity of the helicase (1,10).
Genetic studies in yeast have demonstrated that DEAD/H-box proteins perform highly specific tasks in vivo. In most cases, they are required at a specific stage of RNA processing and most of them are highly specific for their substrates. As revealed by the lethality of null mutants in yeast, most DEAD/H-box proteins are essential, suggesting tight target specificity for each protein (10,11). However, with the exception of DbpA and related proteins (12,13) and to a lesser extent Prp5 (14,15), DEAD/H-box proteins show little or even no RNA specificity when analysed in vitro (8). This apparent non-discrimination of target RNA by in vitro analysis may be due to the absence of essential co-factors that would direct the helicase to its physiological RNA substrate or, more likely, due to the use of biologically non-relevant RNA substrates. It is, therefore, a prerequisite to identify the naturally occurring substrates of RNA helicases in order to characterize them in an in vitro context.
Unlike most of the RNA helicases that have been investigated biochemically, the human DEAH-box protein RHAU (alias DHX36 or G4R1) exhibits a unique ATP-dependent guanine-quadruplex (G4) resolvase activity with a high affinity and specificity for its substrate in vitro (16,17). G4–nucleic acid structures result from the propensity of guanine-rich sequences of DNA and RNA to form atypical and thermodynamically stable four-stranded helical structures under physiological conditions [for review see (18,19)]. Formation of G4 structures in vivo is related to impairment of cellular DNA replication, transcription or translation initiation (20). G4 structures have also been shown to play a role in immunoglobulin gene rearrangement, promoter activation and telomere maintenance (19). Owing to their high-thermodynamic stability, resolution of G4 structures in vivo requires specialized enzymes (21). RHAU binds G4-RNA with sub-nanomolar affinity (16) and unwinds G4 structures much more efficiently than double-stranded nucleic acid [(17) and Tran, H., unpublished data]. Consistent with these biochemical observations, RHAU was also identified as the major source of tetramolecular RNA-resolving activity in HeLa cell lysates (16). Despite these advances, we still lack a corresponding understanding of the mechanism by which RHAU recognizes its substrate.
Structurally, RHAU consists of a ∼400-amino acid helicase core comprising all signature motifs of the DEAH-box family of helicases (Figure 1A). The core region is flanked by N- and C-terminal regions of ∼200 and ∼400 amino acid, respectively. Previous work showed that RHAU associates with mRNAs and re-localizes to stress granules (SGs) upon translational arrest induced by various environmental stresses (22). Deletion analysis of the N-terminal region of RHAU revealed that a region of the first 105 amino acid was critical for RNA binding and re-localization of RHAU to SGs. Importantly, this 105 amino acid-long region alone had the ability in vivo to bind RNA and re-localize to SGs. The apparent significance of the first 105 amino acid for RHAU interaction with RNA prompted us to determine whether the N-terminal domain of RHAU also contributes to the recognition of G4 structures in vitro. To address this question, a series of N-terminal deletion mutants of RHAU was generated as shown in Figure 1A. Through biochemical analysis of these mutants, we have uncovered the functional importance of the first 105 amino acids of RHAU for interaction with G4 structures and further revealed among this region that the previously identified RSM (RHAU-specific motif, amino acids 54–66) domain (22) is essential to the high affinity for G4 structures shown by RHAU. Here, we show that the N-terminal region of RHAU alone can bind to G4-RNA structures, albeit with lower affinity than the full-length protein. We also show that the G4-resolving activity of RHAU is conserved in higher eukaryotes, insofar as CG9323, the Drosophila orthologue of RHAU, readily unwinds G4 structures in the presence of ATP. Finally, we show that a variant form of CG9323 harbouring mutations in the RSM domain manifests reduced G4-binding activity, suggesting that RSM functions similarly in Drosophila and human RHAU proteins.
Figure 1.
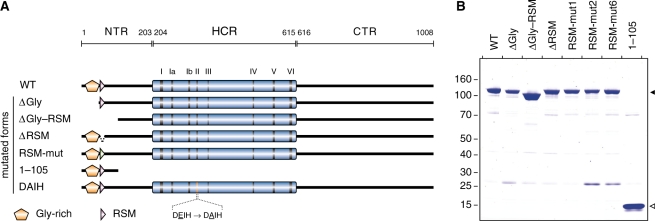
N-terminal deletion mutants of RHAU. (A) Schematic representation of the 1008 amino acid RHAU protein and its N-terminal truncated mutants. The conserved ATPase/helicase motifs I–VI of the DEAD/H-box family are indicated within the helicase core region (HCR) by vertical bars. The HCR is flanked by N-terminal (NTR) and C-terminal (CTR) regions of 203 and 393 amino acid, respectively. The N-terminal Gly-rich (amino acid 10–51) and RSM (RHAU-specific motif, amino acids 54–66) domains are indicated. WT, amino acids 1–1008; ΔGly, amino acids 50–1008; ΔGly–RSM, amino acids 105–1008; ΔRSM, RHAU harbouring a deletion of amino acids 54–66; RSM-mut, RHAU with mutagenized RSM (for details about amino acid substitutions, see Figure 6A); 1–105, amino acids 1–105; DAIH, D335A ATPase deficient mutant. (B) SDS–PAGE separation and Coomassie staining of purified FLAG-tagged recombinant wild-type (WT) and mutant RHAU proteins (2 μg protein per lane). The positions and sizes (kDa) of marker proteins are indicated at the left. The filled and open arrows on the right indicate the positions of the purified proteins.
MATERIALS AND METHODS
Plasmid constructs, cloning and mutagenesis
The plasmid used for the expression of GST-RHAU(1–200) protein in bacteria and the baculoviral expression vector used for the expression of GST-RHAU have been described previously (22,23). Plasmids for the expression of C-terminal FLAG-tagged recombinant RHAU proteins: RHAU-FLAG, RHAU(ΔGly)-FLAG, RHAU(ΔGly–RSM)-FLAG, RHAU(DAIH)-FLAG and RHAU(1–105)-FLAG have been described previously (22). CG9323 cDNA was obtained from the Drosophila Genomics Resource Center (Bloomington, IN, USA) and was sub-cloned by PCR amplification into pIRES.EGFP-FLAG-N1 (22). For the RHAU(ΔRSM)-FLAG plasmid, the RSM (amino acid 54–66) excision was performed using a standard overlapping PCR method (24,25) The resulting PCR product was sub-cloned into pIRES.EGFP-FLAG-N1. For RHAU(RSM-mutx)-FLAG and CG9323(RSM-mut), the RSM-coding sequence was mutagenized using a variation of the classical QuickChange (Stratagene) site-directed mutagenesis PCR method (26). Construction of all these plasmids was confirmed by sequencing. Sequences of oligonucleotides used in this work and detailed descriptions of the plasmid constructions are available upon request.
Cell culture
Human embryonic kidney HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS) and 2 mM l-glutamine at 37°C in a humidified 5% CO2 incubator. Spodoptera frugiperda Sf9 cells were maintained in Grace’s insect cells medium (Invitrogen) supplemented with 10% FCS, 4.11 mM l-glutamine, 3.33 g l−1 lactalbumin hydrolysate and 3.33 g l−1 yeastolate at 27°C.
Expression and purification of recombinant RHAU proteins
GST-RHAU(1–200) protein was expressed in Escherichia coli strain BL21-CodonPlus (DE3)-RIPL (Stratagene). Cultures were inoculated from a single colony of freshly transformed cells and maintained in logarithmic growth at 37°C in 2× YT medium supplemented with ampicillin (0.1 mg ml−1) to a final volume of 1 l. When the OD600 reached 0.3, the temperature was adjusted to 25°C and cells were cultured to an OD600 of 0.6. Isopropyl β-d-thiogalactopyranoside was added to 0.4 mM and the cultures were incubated for 12 h at 25°C with constant shaking. Cells were harvested by centrifugation and the pellets stored at −80°C. All subsequent operations were performed at 4°C. The cell pellets were resuspended in 50 ml of lysis buffer [1× PBS supplemented with NaCl to a final concentration of 300 mM, 1% Triton X-100, 5 mM EDTA, 5 mM DTT, 1× protease inhibitor cocktail (Complete EDTA-free, Roche)]. Lysozyme was added to 1 mg ml−1 and the suspensions were lysed for 30 min, followed by sonication (3× 10 s) to reduce viscosity. Insoluble material was removed by centrifugation (39 000g, 30 min, 4°C) in a Beckman JA-17 rotor and the resulting supernatant was filtered through a 0.22-μm Express PLUS Membrane (Millipore). The resulting filtrate was applied to an 1-ml GSTrap 4B column (GE Healthcare). The column was washed with 30 ml washing buffer (1× PBS supplemented with NaCl to a final concentration of 300 mM, 5 mM DTT) and the recombinant protein was recovered in elution buffer [50 mM Tris–HCl (pH 8.0 at 4°C), 10 mM reduced glutathione, 5 mM DTT)]. The fraction containing the recombinant protein were pooled and elution buffer was exchanged to storage buffer [20 mM HEPES-KOH (pH 7.7 at room temperature), 50 mM KCl, 0.01% Nonidet P-40, 0.5 mM EDTA, 5 mM DTT, 10% glycerol, 2 mM AEBSF] by repeated concentration and dilution steps in Amicon Ultra-15 centrifugal filters (Millipore). Recombinant proteins were stored at −80°C. Purity of protein preparations was assessed by SDS–PAGE and protein concentrations were determined photometrically at 280 nm using the calculated extinction coefficient ε = 71 905 M−1 cm−1.
GST-RHAU protein was expressed in Sf9 cells according to the supplier’s instructions (PharMingen). Three days post-baculoviral infection, cells were harvested by centrifugation and the pellets were stored at −80°C. All subsequent operations were performed at 4°C. The cell pellets were resuspended in insect cell lysis buffer [10 mM Tris–HCl (pH 7.5 at 4°C), 10 mM sodium phosphate, 300 mM NaCl, 1% Triton X-100, 10 mM sodium pyrophosphate, 10% glycerol, 5 mM EDTA, 5 mM DTT, 1× protease inhibitor cocktail (Complete EDTA-free, Roche)] and lysed for 30 min. All subsequent purification steps were carried out as described above for the purification of GST-RHAU(1–200) protein. Purity of protein preparations was assessed by SDS–PAGE and protein concentrations were determined photometrically at 280 nm using the calculated extinction coefficient ε = 166 615 M−1 cm−1.
All FLAG-tagged recombinant RHAU and CG9323 proteins [RHAU-FLAG, RHAU(ΔGly)-FLAG, RHAU(ΔGly–RSM)-FLAG, RHAU(ΔRSM)-FLAG, RHAU(RSM-mutx)-FLAG, RHAU(DAIH)-FLAG, RHAU(1–105)-FLAG, CG9323-FLAG and CG9323(RSM-mut2)-FLAG] were transiently expressed in HEK293T. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were harvested 24–36 h post-transfection, washed with ice-cold PBS and resuspended in lysis buffer [1× PBS supplemented with NaCl to a final concentration of 600 mM, 1% Nonidet P-40, 2 mM EDTA, 2 mM AEBSF (4-(2-aminoethyl)-benzenesulphonyl fluoride hydrochloride), 1× protease inhibitor cocktail (Complete EDTA-free, Roche) for 30 min. Cell lysates were sonicated (1× 10 s) to reduce the viscosity and insoluble material removed by centrifugation (39 000g, 20 min, 4°C) in a Beckman JA-17 rotor. The soluble lysates were mixed for 5 h with 200 μl of a 50% slurry of anti-FLAG M2-agarose affinity gel (Sigma) that had been equilibrated in lysis buffer. The resin was recovered by centrifugation, washed 3× with 1 ml of lysis buffer follow by three washes with 1 ml of IP-washing buffer [50 mM Tris–HCl (pH 7.5), 300 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA]. The bound proteins were eluted with 600 μl elution solution [0.1 mg ml−1 FLAG peptide, 10 mM Tris–HCl (pH 7.5), 150 mM NaCl] for 10 min at 37°C and stored at −20°C. Purity of protein preparations was assessed by SDS–PAGE and protein concentrations were determined by Bradford assay with BSA as the standard.
Tetraplex G4-RNA preparation
Unlabelled and 5′-TAMRA-labelled 35-mer oligoribonucleotides 5′-A15–G5–A15-3′ were used to form tetramolecular G4-RNA and are referred to hereafter as ‘rAGA’. rAGA were purchased from Dharmacon Research and were dissolved in RNase-free solution [100 mM KCl, 10 mM Tris–HCl (pH 7.5), 1 mM EDTA] to a final concentration of 500 μM. To form tetramolecular quadruplex by annealing of rAGA, the solution was aliquoted into PCR tubes and incubated in a PCR thermocycler at 98°C for 10 min and then held at 80°C. EDTA was added immediately to a final concentration of 25 mM and the solution was allowed to cool slowly to room temperature. rAGA aliquots were pooled together and stored at 4°C for 2–3 days. By this procedure, the conversion of monomeric rAGA to a stable tetramolecular quadruplex form was almost complete as judged by native PAGE (Supplementary Figure S1A and C). Circular dichroism (CD) analysis of the purified, annealed rAGA oligomers revealed a typical spectrum of a parallel G4 structure with positive and negative peaks at 263 and 245 nm, respectively (Supplementary Figure S1B). The above prepared tetramolecular G4-rAGA were stored at −20°C.
CD spectropolarimetry
CD experiments were performed with an AVIV Model 202 spectrophotometer equipped with a thermoelectrically controlled cell holder. G4-rAGA at a concentration of 1 μM were prepared in RNase-free solution [10 mM Tris–HCl (pH 7.5), 1 mM EDTA] supplemented with 50 mM KCl, NaCl or LiCl. Quartz cells with 1 cm path length were used for all experiments. CD spectra were recorded at 25°C in the UV region (200–350 nm) with 1 nm increments and an averaging time of 2 s.
Thermodynamic analysis of the stability of tetramolecular G4-rAGA structures
Preformed 5′-TAMRA-labelled tetramolecular G4-rAGA at a concentration of 100 nM were prepared in RNase-free solution [10 mM Tris–HCl (pH 7.5), 1 mM EDTA) supplemented with 50 mM KCl, NaCl or LiCl. Tetramolecular G4-rAGA structures were incubated for 5 min at various temperatures ranging from 20°C to 99°C and were immediately subjected to separation by electrophoresis for 3 h on a pre-electrophoresed 10% polyacrylamide native gel (19:1 acrylamide:bis ratio) in 0.5× TBE at 25°C. After electrophoresis, gels were scanned on a Typhoon 9210 Imager (GE Healthcare) and analysed with Multi Gauge software (Fuji). The fraction of undenatured G4-rAGA was quantitated as the ratio of the signal from the tetramolecular form to the sum of the tetramolecular and the denatured ssRNA. The apparent temperature of mid-transition (Tm) was determined by representing the fraction of undenatured G4-rAGA as a function of the temperature. Reported Tm values are representative of three independent experiments.
Electromobility shift assay and apparent Kd determination
Recombinant RHAU proteins at concentrations from 1 to 1000 nM were incubated with 100 pM 5′-32P-labelled G4-RNA in K-Res buffer [50 mM Tris–acetate (pH 7.8), 100 mM KCl, 10 mM NaCl, 3 mM MgCl2, 70 mM glycine, 10% glycerol], supplemented with 10 mM EDTA and 0.2 U μl−1 SUPERase-In (Ambion) in a 15-μl reaction. The reactions were incubated at 37°C for 30 min. RNA–protein complexes were resolved on a pre-electrophoresed 6% polyacrylamide native gel (37.5:1 acrylamide:bis ratio) in 0.5× TBE at 4°C for 90 min. After electrophoresis, gels were fixed for 1 h in 10% isopropanol/7% acetic acid. RNA–protein complexes were detected by Phosphor-Imaging, scanned on a Typhoon 9400 Imager (GE Healthcare) and analysed with ImageQuant TL software (Nonlinear Dynamics).
G4-RNA resolvase assay
Recombinant RHAU proteins at concentrations from 1 to 100 nM were incubated with 4 nM 5′-32P-labelled G4-RNA in K-Res buffer supplemented with 1 mM ATP and 0.2 U μl−1 SUPERase-In in a 15-μl reaction. Reactions were allowed to proceed at 30°C for 30 min, stopped by transfer to ice and addition of 1:10 volume of 10× loading buffer [33 mM Tris–HCl (pH 8.0), 25% (w/v) Ficoll-400, 110 mM EDTA, 0.17% SDS]. Reaction products were resolved on a pre-electrophoresed 10% polyacrylamide native gel (19:1 acrylamide:bis ratio) in 0.5× TBE at 4°C for 90 min. After electrophoresis, gels were fixed for 1 h in 10% isopropanol/7% acetic acid and exposed to a Phosphor-Imaging screen.
ATPase assay
Recombinant RHAU proteins at concentrations from 25 to 200 nM were incubated with 1 μl [γ-32P]ATP (3000 Ci mmol−1, 0.4 mCi ml−1) in ATPase assay buffer [50 mM Tris–HCl (pH 8.0), 100 mM KCl, 3 mM MgCl2, 1 mM ATP, 1 mM DTT] supplemented with 0.4 U μl−1 RNasin (Promega) and 1 μg μl−1 homopolymeric poly(U) RNA (Sigma) in a 20-μl reaction. Reactions were allowed to proceed at 37°C for 15 min. The reactions were stopped by addition of 1 ml of a 5% (w/v) suspension of activated charcoal (Sigma) in 20 mM phosphoric acid. The samples were incubated on ice for 10 min and the charcoal containing the adsorbed unhydrolysed ATP was pelleted by centrifugation (21 000g, 15 min, 4°C). The supernatants (containing free γ-32Pi) were transferred to new tubes and the radioactivity was quantified by liquid scintillation counting (Cerenkov counts).
RESULTS
The first 105 amino acids of RHAU are required for binding and resolving G4 structures
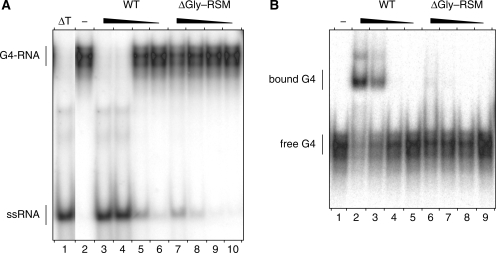
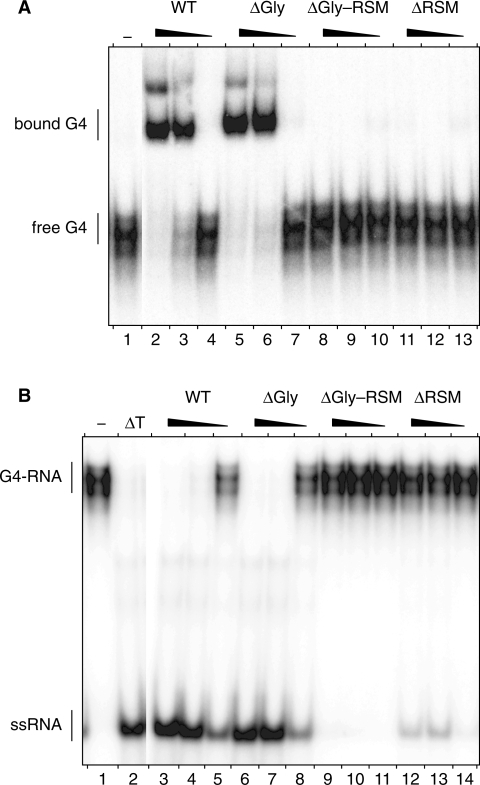
We demonstrated previously in vivo and in vitro that amino acids 1–105 of RHAU delineate a functional domain of prime importance for the interaction of RHAU with RNA (22). To assess the significance of this region for the G4-resolvase activity of RHAU, we constructed a series of FLAG-tagged N-terminal truncated mutants of RHAU (Figure 1A). These truncated forms were expressed in HEK293T cells and purified from soluble lysates by anti-FLAG immunoaffinity chromatography. The purity of these preparations as judged by Coomassie staining after SDS–PAGE was very similar (Figure 1B). To test whether the truncated form of RHAU lacking the first 105 amino acids [RHAU(ΔGly–RSM)] could resolve G4 structures, RHAU protein was incubated with 32P-labelled tetramolecular rAGA and ATP. The products were analysed by native PAGE after disruption of RNA–protein interactions by addition of SDS. The free single-stranded 32P-labelled oligos migrated faster than the quadruplex substrate. As shown previously, wild-type (WT) RHAU efficiently resolved G4 structures into single-stranded oligos (Figure 2A). The labelled product of the resolvase reaction co-migrated during electrophoresis with the single-stranded species released by thermal denaturation of the substrate and was proportional to the level of input protein. In contrast to RHAU(WT), RHAU(ΔGly–RSM) failed to resolve the G4 substrate, suggesting that the N-terminal region of RHAU is essential for its G4-resolvase activity.
Figure 2.
G4-RNA unwinding and binding by N-terminal truncated RHAU proteins. (A) G4-RNA unwinding assay: radio-labelled tetramolecular rAGA at a concentration of 4 nM was incubated in the presence of ATP without protein (−) or with increasing amounts (1, 3, 10 and 30 nM) of WT RHAU or ΔGly–RSM mutant. The reaction products were resolved by native PAGE after disrupting RNA–protein interactions with SDS. An autoradiogram of the gel is shown. The positions of the tetraplex substrate RNA (G4-RNA) and the unwound single-stranded product (ssRNA) are denoted on the left. An aliquot of the tetraplex substrate that was heat-denatured (95°C, 5 min) and then quenched (ΔT) serves as a marker for the position of ssRNA. (B) G4-RNA binding assay: radio-labelled tetramolecular rAGA at a concentration of 100 pM was incubated without protein (−) or with increasing amounts (1, 3, 10 and 30 nM) of WT RHAU or ΔGly–RSM mutant in the absence of ATP. The reaction mixtures were analysed by native PAGE. An autoradiogram of the gel is shown. The positions of the free tetramolecular RNA substrate and the protein–RNA complex are indicated on the left.
Being critical for the enzymatic activity and containing an atypical RNA-binding domain (22), the N-terminal region of RHAU is most likely to play a role in substrate recognition. Therefore, we examined whether this N-terminal region was also essential for the recognition of G4 structures. To address this question, we performed RNA electromobility shift assays (REMSA) using G4 substrate as the ligand. RHAU protein was incubated with 32P-labelled G4 in the absence of ATP (to prevent G4 resolution) and the mixtures were analysed by native PAGE. In the absence of protein, the 32P-labelled G4 structures migrated as a single species in the gel (Figure 2B). Addition of increasing amounts of RHAU(WT) protein resulted in the appearance of a protein–G4 complex with reduced mobility. The bound complexes appeared as two main band regions in the gel that might reflect multiple forms of the complexes. In contrast, the RHAU(ΔGly–RSM) mutant protein failed to form a stable complex with G4 structures. Taken together, these results demonstrate that the N-terminal region encompassing residues 1–105 of RHAU protein is indispensable for both binding and the resolving of G4 structures by RHAU. These results are in agreement with our previous report that the N-terminal region of RHAU is critical for its RNA binding in vivo and its relocalization to SGs (22).
The N-terminal region of RHAU binds but cannot alone resolve G4 structures
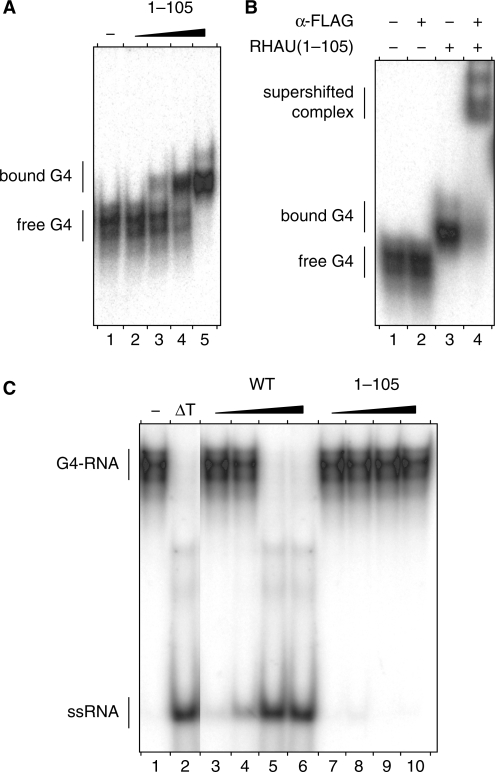
In view of their uniqueness, the N-terminal regions of DEAH-box proteins may have regulatory functions such as sub-cellular localization and/or interaction with RNAs or other proteins (1,10). With prima facie evidence that the N-terminal region of RHAU is important for both binding to and resolving G4 structures, we examined whether this region has G4-binding activity. As shown by REMSA, increasing amounts of RHAU(1–105) protein formed proportionately slow-migrating complexes with G4-RNA (Figure 3A). That the observed mobility shift was due to binding of RHAU(1–105) protein was confirmed by addition of anti-FLAG antibody to binding reactions. As shown in Figure 3B, anti-FLAG antibody caused a super shift of the protein–probe complex but not of the free probe.
Figure 3.
G4-RNA binding and resolving activities of the N-terminal domain of RHAU. (A) Gel mobility shift assay for G4-RNA binding: radio-labelled tetramolecular rAGA at a concentration of 100 pM was incubated without protein (−) or with increasing amounts (30, 100, 300 and 1000 nM) of RHAU(1–105) in the absence of ATP. The reaction mixtures were analysed by native PAGE. An autoradiogram of the gel is shown. (B) Super shift of G4-RNA-binding complex by a specific antibody: radio-labelled tetramolecular rAGA at a concentration of 100 pM was incubated with RHAU(1–105) at 300 nM in the presence or absence of anti-FLAG antibodies (2.5 μg per lane), as indicated. The reaction mixtures were electrophoresed on a native gel. An autoradiogram of the gel is shown. The positions of the free tetramolecular RNA substrate, the protein–RNA complex and the super-shifted complex are indicated on the left. (C) G4-RNA unwinding assay: radio-labelled tetramolecular rAGA at a concentration of 4 nM was incubated in the presence of ATP without protein (−) or with increasing amounts (1, 3, 10 and 30 nM) of WT RHAU or ΔGly–RSM mutant (30, 100, 300 and 1000 nM). The reaction products were resolved by native PAGE after disrupting of RNA–protein interactions with SDS. An autoradiogram of the gel is shown. An aliquot of the tetraplex substrate that was heat-denatured (95°C, 5 min) and then quenched (ΔT) serves as a marker for the position of ssRNA.
Given that some G4-binding proteins destabilize G4 structures in a non-catalytic fashion without requiring ATP hydrolysis (27), we examined whether the N-terminal region of RHAU may also destabilize G4 structures in a similar manner. However, as shown in Figure 3C, binding of the N-terminal region to G4 structures is not sufficient to destabilize it. In agreement with the observation that RHAU cannot resolve G4 structures in the absence of ATP (17), this result demonstrates that the N-terminal region alone lacks G4-destabilizing activity and that the G4-resolving activity is likely a property of the central catalytically active core domain.
The helicase core domain, together with the N-terminal region, contributes to tight G4 binding of RHAU
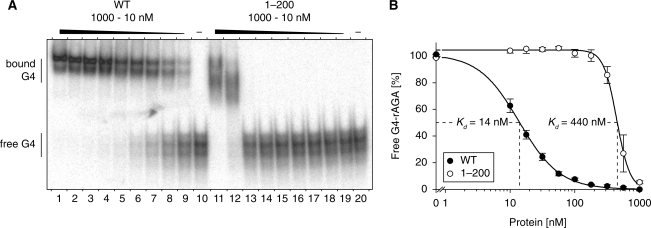
Considering that RHAU has a high affinity for G4 structures (16), we next examined whether the N-terminal region itself accounts for the high affinity for G4 structures of the whole protein. N-terminal region [RHAU(1–200)] and full-length RHAU(WT) were expressed as GST fusion proteins in bacteria and Sf9 insect cells, respectively, and purified to homogeneity as shown in Supplementary Figure S2. Their apparent dissociation constants (Kd) for G4 structures were determined by REMSA. Titration of RHAU(WT) and RHAU(1–200) proteins gave half-saturation points for G4 binding of 14 nM and 440 nM, respectively (Figure 4A and B). This 30-fold difference between the two proteins indicates that the N-terminal region does not constitute by itself an independent and high-affinity G4-RNA binding domain. We propose that the helicase core domain provides RHAU with substantial additional binding activity through interactions with the RNA phosphate backbone, as already shown for many DEAD/H-box proteins (28–34). It should be mentioned that the observed Kd value obtained here is higher than that previously reported (16), which may reflect the difference of the type and location of tags attached to RHAU (N-GST versus C-His6) and of the way how proteins were purified.
Figure 4.
G4-RNA-binding properties of RHAU and the N-terminal region. (A) Gel mobility shift assay for G4-RNA binding: radio-labelled tetramolecular rAGA at a concentration of 100 pM was incubated without protein (−) or with increasing amounts of either GST-tagged RHAU(WT) or RHAU(1–200) in the absence of ATP. The reaction mixtures were analysed by native PAGE. An autoradiogram of the gel is shown. At concentrations from 10 to 1000 nM, GST protein alone had no effect on G4-RNA mobility (data not shown). (B) Quantification of gel mobility shift assays for WT RHAU (WT, filled circle) and RHAU N-terminal region (1–200, open circle) binding to tetramolecular rAGA. The data represent means ± SEM from three independent experiments.
The RSM domain, but not the Gly-rich sequence, in the N-terminal region is crucial for the recognition and resolution of G4 structures by RHAU
The functional N-terminal region encompassing residues 1–105 of RHAU protein consists of two abutting domains (Figure 1A): a 41 amino acid-long low-complexity Gly-rich domain followed by the evolutionary conserved 13 amino acid-long RSM (amino acids 54–66). We demonstrated previously in vivo that deletion of the Gly-rich region [RHAU(ΔGly)] significantly impinged on the relocalization of the protein to SGs and on its association with RNA (22). To characterize the individual contributions of each of the two domains to the interaction of RHAU with G4 structures, we performed REMSA experiments using corresponding deletion mutants (Figure 5A). As already shown in Figure 2B, RHAU(ΔGly–RSM) lacking the first 105 amino acids failed to form a stable complex with G4 structures. In contrast, RHAU(ΔGly) still harbouring the RSM was able to bind to G4 structures with the same efficiency as RHAU(WT). Contrary to the deletion of the Gly-rich sequence, a RHAU mutant with an RSM deletion [RHAU(ΔRSM)] abrogated the interaction with G4 structures. We further assessed the consequence of loss of G4 recognition on the in vitro resolvase activity of RHAU (Figure 5B). As expected, RHAU(ΔRSM), like RHAU(ΔGly–RSM), failed to resolve G4 structures, while RHAU(ΔGly), like RHAU(WT), did. Similar results with strict RSM-dependency could also be reproduced with other G4-RNA structures (data not shown). Together, these results indicate that the presence of the RSM but not further sequences of the N-terminal region is a prerequisite for the recognition and the resolution of G4 structures by RHAU in vitro.
Figure 5.
G4-RNA binding and unwinding by Gly-rich and RSM-truncated RHAU proteins. (A) Gel mobility shift assay for G4-RNA binding: radio-labelled tetramolecular rAGA at a concentration of 100 pM was incubated without protein (−) or with increasing amounts (6, 20 and 60 nM) of either WT RHAU, or ΔGly, ΔGly–RSM or ΔRSM RHAU mutants in the absence of ATP. The reaction mixtures were analysed by native PAGE. An autoradiogram of the gel is shown. (B) G4-RNA unwinding assay: radio-labelled tetramolecular rAGA at a concentration of 4 nM was incubated in the presence of ATP without protein (−) or with increasing amounts (6, 20 and 60 nM) of either WT RHAU, or ΔGly, ΔGly–RSM or ΔRSM RHAU mutants. The reaction products were resolved by native PAGE after disrupting of RNA–protein interactions with SDS. An autoradiogram of the gel is shown. An aliquot of the tetraplex substrate that was heat denatured (95°C, 5 min) and then quenched (ΔT) serves as a marker for the position of ssRNA.
Conserved residues within the RSM domain are essential for the recognition of G4 structures by RHAU
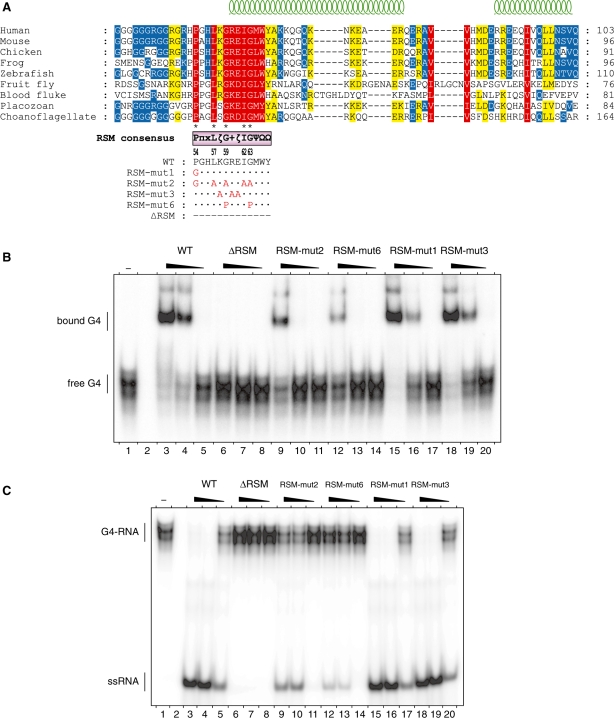
Multiple-sequence alignments of RHAU orthologues of various species from choanoflagelates to humans unveiled a highly conserved cluster of 13 amino acids presenting the RSM domain embedded in a moderately conserved region of about 50 amino acids (amino acids 54–100) (Figure 6A). In contrast, the rest of the N-terminus surrounding this region is poorly conserved. The RSM consensus sequence as determined from 40 different RHAU orthologue sequences is: P–π–x–L–ζ–G–[+]–ζ–I–G–Ψ–Ω–Ω [consensus RSM is listed according to the Seefeld convention (37), symbol nomenclature stands for: π (pi) = small side chain, ζ (zeta) = hydrophilic, [+] = basic, Ψ (Psi) = aliphatic, Ω (Omega) = aromatic] (Supplementary Figure S3). The motif consists of five invariant amino acids (Pro-54, Leu-57, Gly-59, Ile-62 and Gly-63) and seven highly conserved residues of similar biochemical properties (π-55, ζ-58, [+]-60, ζ-61, Ψ-64, Ω-65, Ω-66). In addition, computation-based secondary structure prediction of the sequences used for alignment suggested that RSM is partially structured, sitting astride an unstructured loop (amino acids 54–59) and an α-helix (amino acids 60–66) (Figure 6A; Supplementary Figure S4). We further examined by site-specific mutagenesis in vitro the contribution of conserved amino acids within the RSM domain to the recognition and resolution of G4 structures (Figure 6A). To this end, conserved residues of the RSM were substituted with alanines, prolines or glycines. We focused on Pro-54, Gly-59 and Gly-63 because they may provide the RSM with substantial structural rigidity (Pro-54) or flexibility (Gly-59 and Gly-63). As shown in Figure 6B, mutation of the five invariant residues of the RSM (RSM-mut2) considerably reduced the binding affinity of RHAU for G4-RNA, albeit to a lesser extent than the RSM-deleted form of RHAU (ΔRSM). Interestingly, substitution of the invariant Gly-59 and Gly-63 residues with prolines (RSM-mut6) caused a stronger reduction of G4-binding activity than the RHAU(RSM-mut2) in which these two amino acids are mutated to alanines. Since prolines may induce a structural constraint on the RSM, this result suggests that recognition of G4 structures may depend on the conformational organization of the RSM. Finally, as observed with RHAU(RSM-mut1) and RHAU(RSM-mut3) mutants, Pro-54 as well as the polar residues Lys-58, Arg-60 and Glu-61 appear to be dispensable.
Figure 6.
G4-RNA binding and unwinding by RSM-mutated forms of RHAU. (A) Conservation of the RSM among RHAU orthologues throughout evolution. Multiple sequence alignment was carried out with MAFFT (version 6) (35). Similarity analysis was made by GeneDoc (version 2.7) using the BLOSUM62 scoring matrix. Similarity is shown in red for 100%, yellow for 99–80% and blue for 79–60%. Amino acids that are identical between the nine sequences are indicated by asterisks below the alignment. Secondary structure prediction was performed by JPRED (36) and is indicated on the top. The RSM and its consensus sequence derived from 40 different RHAU orthologues are shown below the alignment [π (pi) = small side chain, ζ (zeta) = hydrophilic, [+] = basic, Ψ (Psi) = aliphatic, Ω (Omega) = aromatic]. The site-directed substitutions of the RSM mutants employed in this study are listed with the amino acid changes indicated underneath. Species and accession numbers of RHAU orthologues listed are: human (Homo sapiens, NP_065916), mouse (Mus musculus, NP_082412), chicken (Gallus gallus, XP_422834), frog (Xenopus tropicalis, ENSXETP00000016958), zebrafish (Danio rerio, NP_001122016), fruit fly (Drosophila melanogaster, NP_610056), blood fluke (Schistosoma mansoni, XP_002577014), placozoan (Trichoplax adhaerens, XP_002110272), choanoflagellate (Monosiga brevicollis, XP_001747335). (B) Gel mobility shift assay for G4-RNA binding: radio-labelled tetramolecular rAGA at a concentration of 100 pM was incubated without protein (−) or with increasing amounts (6, 20 and 60 nM) of either WT RHAU or the indicated RSM mutants in the absence of ATP. The reaction mixtures were analysed by native PAGE. An autoradiogram of the gel is shown. (C) G4-RNA unwinding assay: radio-labelled tetramolecular rAGA at a concentration of 4 nM was incubated in the presence of ATP without protein (−) or with increasing amounts (6, 20 and 60 nM) of either WT RHAU or the indicated RSM mutants. The reaction products were resolved by native PAGE after disrupting RNA–protein interactions with SDS. An autoradiogram of the gel is shown. An aliquot of the duplex substrate that was heat denatured (95°C, 5 min) and then quenched (ΔT) serves as a marker for the position of ssRNA.
As expected, RHAU proteins with mutated RSM and displaying strong G4-RNA binding deficiency (RSM-mut2 and 6) also had reduced G4-resolving activity (Figure 6C). In contrast, despite the slight reduction of G4-binding activity observed for RHAU(RSM-mut1) and RHAU(RSM-mut3), these two mutants unwound G4 structures with an efficiency comparable to WT RHAU. We concluded from these in vitro mutagenesis experiments that the highly conserved residues in the RSM domain are essential for RHAU G4-structure binding and resolving activities.
ATPase activity of RHAU N-terminal truncated mutants
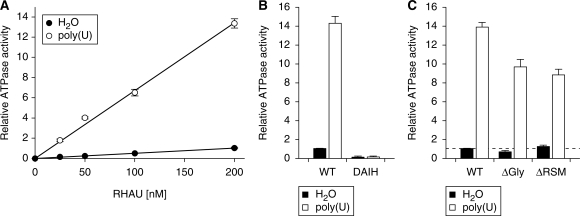
Hitherto, it could not be excluded that the introduced mutations caused significant conformational changes in the helicase core domain that resulted in enzymatically defective proteins. To define whether loss of G4-resolvase activity was due to loss of G4 binding by the N-terminal domain or to impairment of the basic activity of the helicase core domain, we compared the ATPase activities of RHAU mutants and WT RHAU. As shown previously (23), RHAU exhibited significant ATPase activity in the absence of nucleic acids, which was stimulated substantially by the presence of homopolymeric poly(U) (Figure 7A). Both the RNA-dependent and -independent ATPase activities were proportional to the amount of input RHAU protein. To exclude the possibility that the RNA-independent ATPase activity observed was due to contaminants derived from HEK293T cells, we substituted the Glu-335 residue with Ala within the Walker B site (DEIH → DAIH), which abolishes RHAU ATPase activity (23). RHAU(DAIH) protein was prepared according to the protocol employed for WT RHAU protein, with comparable yield and purity (data not shown). In contrast to WT RHAU, RHAU(DAIH) showed no ATPase activity, even in the presence of poly(U) (Figure 7B). Thus, we concluded that RHAU harbours intrinsic RNA-independent ATPase activity, which can be further stimulated by homopolymeric RNA.
Figure 7.
ATPase activity of WT RHAU and N-terminal mutants. (A) The extent of ATP hydrolysis by WT RHAU in the presence (open circle) or absence (filled circle) of poly(U) plotted as a function of input RHAU. The ATPase activity is expressed as a function of the activity obtained with WT RHAU at a concentration of 200 nM when poly(U) was omitted. (B) Influence of poly(U) on the ATPase activity of WT RHAU and DAIH ATPase-deficient mutant (200 nM). The ATPase activity was represented relatively to that of WT RHAU in the absence of poly(U) that was set to 1. (C) Influence of poly(U) on the ATPase activity of RHAU (WT) and ΔGly or ΔRSM RHAU mutants (200 nM). The basal ATPase activity (without nucleic acid co-factor) of WT RHAU is indicated (dashed line). The data represent the means ± SEM from three independent experiments.
To clarify the biochemical basis of the absence of G4-resolvase activity in the RHAU(ΔRSM) mutant, we measured the rate of ATP hydrolysis by this mutant and compared it to that of the WT RHAU and the RHAU(ΔGly) mutant that was still proficient in G4 unwinding. In the absence of RNA co-factor, the ATPase activity of RHAU(ΔRSM) protein was the same as RHAU(WT) and RHAU(ΔGly) proteins (Figure 7C), suggesting that the deletion of the Gly-rich (ΔGly) or the RSM (ΔRSM) domains does not affect the basal ATPase activity of RHAU. However, the extent of poly(U)-dependent stimulation of ATPase activity of RHAU(ΔRSM) was ∼60% of that of RHAU(WT) and similar to that of RHAU(ΔGly). Taken together, these results indicate that deletion of the N-terminal region of RHAU does not cause any significant conformational changes to the helicase core, but renders the RHAU protein less responsive to RNA in the stimulation of its ATPase activity. Given that RHAU(ΔRSM) retained significant RNA-dependent ATPase activity, equivalent to the G4-resolvase proficient RHAU(ΔGly) mutant, we concluded that the lack of G4-resolving activity in the RHAU(ΔRSM) mutant resulted from the loss of G4 binding by the N-terminal domain.
CG9323, the Drosophila ortholog of RHAU efficiently unwinds G4-RNA
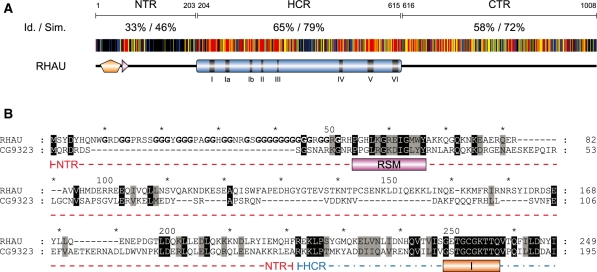
Based on sequence analysis, RHAU has clear orthologs in almost all species of the animal kingdom ranging from choanoflagellates to humans. A multiple sequence alignment between eight of these orthologs showed the helicase core (amino acids 204–615) together with the C-terminal region (amino acids 616–1008) of RHAU to be evolutionary conserved (Figure 8A). In contrast, sequences of the N-terminal region (amino acid 1–203) show little similarity with the exception of the highly conserved RSM (amino acids 54–66) and its surrounding region (amino acids 54–100). Considering the apparent importance of the RSM in the recognition of G4 structures and the high conservation of its sequence together with the rest of RHAU protein, we surmised that the G4-binding and resolving activity of the RHAU protein are conserved among higher eukaryotes. To validate this hypothesis, we cloned and characterized CG9323, the Drosophila ortholog of RHAU.
Figure 8.
Amino acid conservation of the N-terminal region of RHAU. (A) Schematic representation of amino acid conservation of human RHAU throughout evolution. The human RHAU sequence was aligned with eight RHAU orthologues (the sequences shown in Figure 6) by MAFFT (version 6) (35). Each residue of RHAU is represented with a colour code that indicates its level of conservation amongst the eight orthologous sequences. Similarity is shown in red for 100%, yellow for 99–80% and blue for 79–60%. Similarity analysis was made by GeneDoc (version 2.7) using the BLOSUM62 scoring matrix. Average values of identity (Id) and similarity (Sim) for N-terminal (NTR), helicase core (HCR) and C-terminal (CTR) regions are indicated. (B) Sequence alignment of the N-terminal region of RHAU with its Drosophila ortholog CG9323. Amino acids that are identical or similar between the two sequences are shaded in black and grey, respectively. The RSM domain as well as helicase motif I are indicated below the sequences. Gly residues of the Gly-rich domain (amino acid 10–51) of RHAU are bolded. N-terminal region and a part of the helicase core domain are delineated with a coloured dashed line in red and blue, respectively. For complete alignment between RHAU and CG9323, see Supplementary Figure S5.
CG9323 is a 942 amino acid-long protein, which like RHAU comprises all the typical signature motifs of the DEAH-box family of RNA helicases (Figure 8B; Supplementary Figure S5). The overall identity and similarity between CG9323 and RHAU are 34% and 52%, respectively, but not evenly distributed along the entire sequence. Sequence homology with RHAU is particularly high (41% identity and 60% similarity) for the helicase core and the C-terminal regions of CG9323. Nevertheless, apart from the conserved RSM, the CG9323 N-terminal region lacks evident sequence homology with RHAU. In addition, the CG9323 N-terminal region is 25% shorter than that of RHAU, mainly due to the absence of the Gly-rich sequence upstream of the RSM.
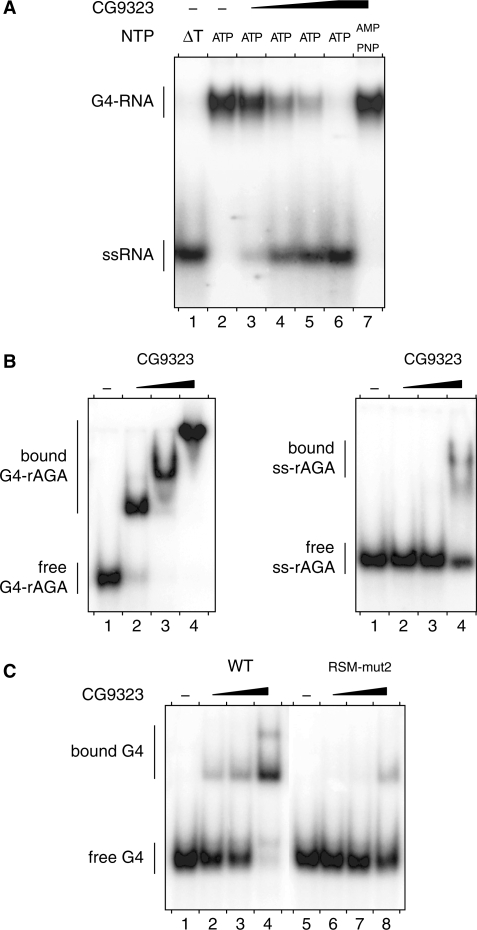
To check whether the Drosophila ortholog of RHAU retains G4 binding and resolving activities, CG9323 was expressed as a FLAG-tagged recombinant protein and immunopurified to homogeneity as shown in Supplementary Figure S6. The ability of purified recombinant CG9323 protein to unwind and to bind G4 structures was assessed under the conditions previously employed for RHAU. As shown in Figure 9A, CG9323 efficiently unwound the G4-RNA substrate. As for RHAU, the extent of products resolved by CG9323 was proportional to the input protein. Furthermore, CG9323 failed to resolve G4 substrates in the presence of the non-hydrolysable ATP analogue AMP-PNP, indicating that G4 unwinding by CG9323 requires the hydrolysis of nucleosides triphosphate.
Figure 9.
G4-RNA unwinding and binding by CG9323. (A) G4-RNA unwinding assay: radio-labelled tetramolecular rAGA at a concentration of 4 nM was incubated in the presence of ATP or AMP-PNP (as indicated) without protein (−) or with increasing amounts (2, 6, 20 and 60 nM) of purified recombinant CG9323 protein. The reaction products were resolved by native PAGE after disrupting RNA–protein interactions with SDS. An autoradiogram of the gel is shown. An aliquot of the duplex substrate that was heat-denatured (95°C, 5 min) and then quenched (ΔT) serves as a marker for the position of ssRNA. (B) G4- and ssRNA-binding assay: tetramolecular (left) or monomeric (right) radio-labelled rAGA at a concentration of 100 pM were incubated without protein (−) or with increasing amounts (10, 30 and 100 nM) of CG9323 in the absence of ATP. The reaction mixtures were analysed by native PAGE. An autoradiogram of the gel is shown. (C) G4-RNA-binding assay: radio-labelled tetramolecular rAGA at a concentration of 100 pM was incubated without protein (−) or with increasing amounts (3, 10 and 30 nM) of WT or RSM-mut2 CG9323 proteins in the absence of ATP. The reaction mixture were analysed by native PAGE. An autoradiogram of the gel is shown.
Similar to RHAU, CG9323 also formed a stable complex with G4 substrates in the absence of hydrolysable rNTPs (Figure 9B). In binding experiments, CG9323 turned out to be more specific for G4-RNA relative to ssRNAs, since CG9323 bound tetramolecular rAGA with a 10-fold higher affinity than monomeric single-stranded rAGA oligoribonucleotides of the same sequence. This observation, by analogy, is consistent with earlier observations that RHAU has poor sequence-specific recognition of ssRNAs (16,23). Finally, a variant form of CG9323, in which highly conserved residues of the RSM were mutagenized, displayed reduced G4-binding activity, suggesting that the RSM is essential for the recognition of G4-structures by CG9323 (Figure 9C). In conclusion, these results provide compelling evidence that the G4 binding and resolving activities of RHAU have been conserved from Drosophila to human, with the RSM playing a pivotal role.
DISCUSSION
Owing to their bulky and thermodynamically stable features, G4 structures have been shown in many respects to impede normal nucleic acid metabolism (38–45). To cope with this problem, proteins are produced that mitigate effects of these atypical stable structures. Four human helicases, including RHAU, have been shown so far to harbour G4-resolving activity in vitro. These include the RecQ family proteins BLM (46,47) and WRN (48), as well as FANCJ (41), a Rad3-like helicase. The latter are all DNA helicases that have been clearly implicated in the maintenance of genome integrity (49–51). RHAU, on the other hand, belongs to the DEAH-box family of RNA helicases and has very little sequence similarity with the above mentioned helicases. Curiously, RHAU was initially identified as a G4-DNA resolvase enzyme (17). The possibility that G4-RNA structures are targets of RHAU emerged from in vivo UV cross-linking results showing RHAU binding mainly to RNA (22). Subsequent characterization of RHAU demonstrated its aptitude to resolve G4-RNA better than G4-DNA (16). This finding was a remarkable breakthrough, since RHAU was the first and still the only known helicase to possess G4-RNA resolvase activity. Furthermore, RHAU is one of the rare DEAD/H-box proteins that exhibit high affinity and specificity for its substrate in vitro independent of accessory proteins. Since this finding, efforts have been made to understand the mechanism underlying recognition of G4-structures by RHAU. The present study shows that the N-terminal region of RHAU is essential and responsible for binding of RHAU to G4-structures. Further investigations dissecting the N-terminal region, coupled with site-directed mutagenesis, have demonstrated that the RSM makes a decisive contribution to the high affinity of RHAU for G4-structures. Sequence comparisons of RHAU orthologs from various species showed the RSM to be the unique highly conserved part of the N-terminal region. Hence, we predicted that all orthologs forms should possess G4-resolving activity based on the functional significance and sequence conservation of the RSM domain and the catalytic region. This hypothesis was supported by the robust ATP-dependent G4-RNA resolvase activity found for CG9323, the Drosophila form of RHAU. As expected, and also shown for RHAU, the binding of CG9323 to G4-RNAs depended on RSM integrity, which further indicated similar functions for this motif in both proteins.
Recognition of G4-RNA by RHAU depends on the N-terminal RSM
RHAU shares with most helicases a global scheme of modular architecture that combines a conserved central helicase core domain with peripheral regions of various lengths and sequences (52). Together with previous findings (22,53–55), our results indicate that the helicase core alone cannot account for the high specificity of function usually attributed to DEAD/H-box proteins. In this regard, these data also agree with numerous structural observations that the HCR of DEAD-box proteins interacts essentially in a non-sequence-specific manner with the phosphoribose backbone of single-stranded nucleic acid (28–32). Such contacts suffice to discriminate RNA from DNA by means of the 2′-hydroxyl groups of the ribose moieties, but not to distinguish between sequences of varying nucleotide composition. In the present work, we have shown the importance of the unique N-terminal flanking region in adapting the conserved catalytic core to a specific function. The present investigation has also shown clearly that, although necessary, ATPase activity by itself is not sufficient for RHAU to unwind G4 structures. Our data strongly suggest that the establishment of a stable complex between RHAU and its G4 substrate is a prerequisite for the subsequent ATPase-dependent unwinding of the G4 structure. We propose that the N-terminal RSM endows the enzyme with specificity by binding the G4 substrate, thereby positioning the helicase core in close proximity to the substrate. Likewise, critical roles for N- and C-terminal flanking regions have been reported for several DEAD/H-box proteins, exerting specific functions by interacting with particular RNA species/structures or with other regulatory proteins (53,56–58). For example, the prototypical yeast DEAH-box proteins Prp16 and Prp22 and the human ortholog HRH1 (alias DHX8 or hPrp22) have been shown to associate with the spliceosome via their non-conserved N-terminal regions (59–61). The Drosophila maleless (MLE) protein, more closely related to RHAU, harbours two copies of dsRNA-binding motifs in its N-terminal region (62). Their deletion, as with the N-terminal truncation of RHAU, caused the loss of RNA binding and unwinding activities in vitro and subcellular mis-localization of the protein in vivo (63). Together, these examples emphasize the role of the peripheral domains of DEAD/H-box proteins in adapting a common catalytic core to a broad spectrum of specific functions.
Potential role of the RSM in RHAU re-localization to SGs
The present work underscores the essential nature of the conserved RSM, which endows RHAU with a high affinity for G4 structures in vitro. Determining the mechanisms whereby RHAU resolve G4 substrates in vivo is an important issue to be addressed in future. It is tempting to draw a parallel between the present observation and a previous report that the stress-induced recruitment of RHAU to SGs is mediated by interactions of RHAU with RNA (22). Similar to our observations, the region identified as essential for this activity included the RSM together with the upstream Gly-rich domain, underscoring the functional relevance of the RSM and its surrounding region for the biological activity of RHAU. The N-terminal region of RHAU alone was also found to be sufficient to drive RNA binding in vivo as well to re-localize to SGs (22). This observation, however, contrasts with the data presented here in that, although required for functional specificity, the N-terminal region alone does not constitute an independent and high-affinity G4-RNA-binding domain. In this regard, our data rather suggest that both the N-terminal and HCRs are required for the productive interaction of RHAU with G4 structures. Since the nucleic acid binding motifs Ia, Ib, IV and V of DEAH-box proteins have been proposed to contact RNA (33,34), we surmise that, for RHAU, the helicase core may also provide the protein with substantial binding activity by interacting in a non-specific manner with the phosphoribose backbone of the single-stranded tail flanking the tetramolecular G4 structure. Further investigations into the functional contribution of the RNA-binding site of the helicase core are needed to further challenge this hypothesis. In addition, our previous finding that the ATPase-deficient form of RHAU [RHAU(DAIH)] stalls in SGs (22) agrees with the observation that, once bound to G4-RNA, RHAU(DAIH) cannot dissociate itself from its substrate, even in the presence of ATP (data not shown). This indicates that the G4-unwinding reaction requires ATP hydrolysis, rather than ATP binding per se, and further suggests that the release of the RNA substrate occurs only after G4 unwinding. SGs may constitute a favourable environment for the formation of intermolecular G4-RNA structures, as they are temporary sites of accumulation of stress-induced stalled translation initiation RNP complexes (64,65). However, an important question that we have not yet addressed is whether RHAU relocalizes to SGs upon binding to G4-RNA structures. Interestingly, however, we noticed that RSM-mutated forms of RHAU that are deficient for G4-structure recognition in vitro, manifest reduced association with RNA in vivo concomitantly with reduced relocalization to SGs (Chalupnikova, K., unpublished data). Thus, this finding raises the possibility that at least a fraction of RHAU is recruited to SGs via interactions with G4 structures.
The mechanism by which RHAU recognizes G4 structures is an important issue that needed to be addressed to improve our understanding of RHAU. So far, however, little is known about its biological function as a potential G4-resolvase enzyme. Recently, G4 structures in RNA have attracted considerable attention as a plausible means of regulating gene expression (42). Formation of G4 structures in the 5′-untranslated region has been shown to affect mRNA translation (43–45) and bioinformatics studies have identified more than 50 000 potential G4 structures near splicing and polyadenylation sites of various human and mouse genes. This raises the possibility that G4 formation impedes RNA metabolism at many different stages (66). At the moment, we lack a corresponding understanding of how cells negotiate with G4 structures in various RNA molecules. However, from the findings presented here, RHAU emerges as a promising regulator of G4 structure-based RNA metabolism and merits future investigation of its potential roles in different biological contexts.
ACCESSION NUMBER
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novartis Research Foundation. Funding for open access charge: Novartis Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Stéphane Thiry for excellent technical assistance and Patrick King, Janice Ching Lai and Sandra Pauli for critical comments on the manuscript
REFERENCES
- 1.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol. Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 2.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 3.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linder P. DEAD-box proteins: a family affair–active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- 6.Russell R. RNA misfolding and the action of chaperones. Front. Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 8.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol. Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 11.de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 12.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kossen K, Uhlenbeck OC. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 1999;27:3811–3820. doi: 10.1093/nar/27.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Day CL, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J. Biol. Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 15.Perriman R, Barta I, Voeltz GK, Abelson J, Ares M., Jr ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl Acad. Sci. USA. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, Nagamine Y, Akman SA. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005;280:38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 18.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 20.Wong HM, Payet L, Huppert JL. Function and targeting of G-quadruplexes. Curr. Opin. Mol. Ther. 2009;11:146–155. [PubMed] [Google Scholar]
- 21.Fry M. Tetraplex DNA and its interacting proteins. Front. Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 22.Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 24.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 25.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 26.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khateb S, Weisman-Shomer P, Hershco I, Loeb LA, Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 29.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 30.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 31.Collins R, Karlberg T, Lehtio L, Schutz P, van den Berg S, Dahlgren LG, Hammarstrom M, Weigelt J, Schuler H. The DEXD/H-box RNA helicase DDX19 is regulated by an {alpha}-helical switch. J. Biol. Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol. Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider S, Campodonico E, Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka N, Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6510–6521. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 36.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aasland R, Abrams C, Ampe C, Ball LJ, Bedford MT, Cesareni G, Gimona M, Hurley JH, Jarchau T, Lehto VP, et al. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 2002;513:141–144. doi: 10.1016/s0014-5793(01)03295-1. [DOI] [PubMed] [Google Scholar]
- 38.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, Boulton SJ, Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Shin-ya K, Brosh R.M., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris MJ, Basu S. An unusually stable G-quadruplex within the 5′-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 46.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 47.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 49.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Suhasini AN, Brosh R.M., Jr Welcome the family of FANCJ-like helicases to the block of genome stability maintenance proteins. Cell Mol. Life Sci. 2009;66:1209–1222. doi: 10.1007/s00018-008-8580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 52.Singleton MR, Wigley DB. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J. Biol. Chem. 2005;280:35499–35505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 54.Grohman JK, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, Lambowitz AM. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J. Mol. Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider S, Schwer B. Functional domains of the yeast splicing factor Prp22p. J. Biol. Chem. 2001;276:21184–21191. doi: 10.1074/jbc.M101964200. [DOI] [PubMed] [Google Scholar]
- 61.Ohno M, Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 62.Gibson TJ, Thompson JD. Detection of dsRNA-binding domains in RNA helicase A and Drosophila maleless: implications for monomeric RNA helicases. Nucleic Acids Res. 1994;22:2552–2556. doi: 10.1093/nar/22.13.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Izzo A, Regnard C, Morales V, Kremmer E, Becker PB. Structure-function analysis of the RNA helicase maleless. Nucleic Acids Res. 2008;36:950–962. doi: 10.1093/nar/gkm1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 66.Kostadinov R, Malhotra N, Viotti M, Shine R, D’Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.