Abstract
Cyclic-di-GMP is a bacterial second messenger that controls the switch between motile and sessile states. It is synthesized by proteins containing the enzymatic GGDEF domain and degraded by the EAL domain. Many bacterial genomes encode several copies of proteins containing these domains, raising questions on how the activities of parallel c-di-GMP signalling systems are segregated to avoid potentially deleterious cross-talk. Moreover, many ‘hybrid’ proteins contain both GGDEF and EAL domains; the relationship between the two apparently opposing enzymatic activities has been termed a ‘biochemical conundrum’. Here, we present a computational analysis of 11 248 GGDEF- and EAL-containing proteins in 867 prokaryotic genomes to address these two outstanding questions. Over half of these proteins contain a signal for cell-surface localization, and a majority accommodate a signal-sensing partner domain; these indicate widespread prevalence of post-translational regulation that may segregate the activities of proteins that are co-expressed. By examining the conservation of amino acid residues in the GGDEF and EAL catalytic sites, we show that there are predominantly two types of hybrid proteins. In the first, both sites are intact; an additional regulatory partner domain, present in most of these proteins, might determine the balance between the two enzymatic activities. In the second type, only the EAL catalytic site is intact; these—unlike EAL-only proteins—generally contain a signal-sensing partner domain, suggesting distinct modes of regulation for EAL activity under different sequence contexts. Finally, we discuss the role of proteins that have lost GGDEF and EAL catalytic sites as potential c-di-GMP-binding effectors. Our findings will serve as a genomic framework for interpreting ongoing molecular investigations of these proteins.
INTRODUCTION
Signal transduction pathways often use small molecule second-messengers to integrate, amplify and transmit information to intracellular sensors and effectors (1). Among the most important are cyclic nucleotides such as cyclic adenosine monophosphate and cyclic guanosine monophosphate which regulate a variety of functions ranging from sugar metabolism to ion channel conductance in prokaryotes and eukaryotes.
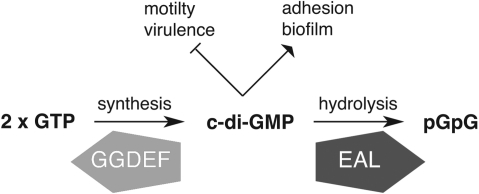
In contrast to the well-established roles of cyclic mono-nucleotides, cyclic di-nucleotides have gained prominence only recently as major prokaryotic signalling molecules. The bacterial second messenger bis-(3′–5′)-cyclic-dimeric-guanosine monophosphate (c-di-GMP), the focus of this study, was identified in 1987 as an allosteric activator of cellulose synthase in Gluconacetobacter xylinus and Agrobacterium tumefaciens (2,3). Since then, the molecule has become recognized as a key regulator for complex cellular functions (Figure 1). Most notably, c-di-GMP controls the switch between motile and sessile lifestyles: high cellular levels of c-di-GMP promote exopolysaccharide production and surface adhesion, eventually leading to biofilm formation; conversely, low c-di-GMP levels result in flagellar gene- expression and increased cellular motility (4,5). Further, there is now substantial evidence that c-di-GMP has a role beyond these functions; for example, in regulating virulence in pathogens such as Vibrio cholerae and Pseudomonas aeruginosa (6) and responding to nutrient starvation in Mycobacterium smegmatis (7).
Figure 1.
Turnover of c-di-GMP. A schematic representation of the synthesis (from GTP) and hydrolysis (to pGpG) of c-di-GMP by the GGDEF and EAL domains, respectively. Also shown are general cellular functions that are activated (arrow-shaped head) and repressed (flat head) by c-di-GMP. Figure concept adapted from Ref. (4).
C-di-GMP levels are modulated through the activity of di-guanylate cyclases (DGCs) that convert two molecules of GTP to c-di-GMP, and phosphodiesterases (PDEs) that linearize c-di-GMP to pGpG, which is subsequently hydrolyzed to GMP (Figure 1). DGCs are characterized by the active site GG[D/E]EF amino acid motif in the enzyme catalytic site, whereas PDEs contain either the EAL domain or the more recently described HD-GYP domain (4). The simplest proteins involved in c-di-GMP turnover have only one of these domains: these we call GGDEF-only, EAL-only or HD-GYP-only proteins. In what has been termed a ‘biochemical conundrum’, there are also hybrid proteins containing both GGDEF and EAL domains (8). Early biochemical studies identified such proteins in which only one of the domains was catalytically active, leading to suggestions that the other domain had acquired a regulatory function (4). However, there are now several examples of hybrid proteins that retain both DGC and PDE activities (7,9,10), raising questions on how the two activities are co-ordinately or reciprocally regulated.
An early computational survey of signalling proteins in 30 prokaryotic genomes showed that GGDEF and EAL proteins are ubiquitous in bacteria, but absent from archaea (8,11). In general, genomes were found to encode several GGDEF and EAL proteins with a particularly striking expansion in Gamma Proteobacteria. Given that most of these proteins are likely to be mainly involved in c-di-GMP signalling, it is intriguing that organisms should encode multiple proteins containing the same enzymatic domain. This has led to questions on how the activities of different GGDEF and EAL proteins are separated in order to minimize cross-talk among distinct outputs.
The main method for managing parallel c-di-GMP signalling systems is through tight regulation of the DGC and PDE activities. The first point of regulation is at the level of transcription; however, most GGDEF and EAL proteins in Escherichia coli K12 are expressed during the stationary phase of growth, with only a few being produced during exponential growth state (12). Thus, given that many proteins are expressed under the same condition, their spatial control and post-translational regulation are also probably critical. The genomic study above identified a number of small molecule-sensing and phosphorylation-receiving domains that frequently occur in GGDEF and EAL-containing proteins (11) thus illustrating the importance of signal-dependent post-translational regulation. However, the prevalence of these partner domains has not been systematically investigated. Further, apart from individual examples (10,13,14), the implications of partner domains for GGDEF-only, EAL-only and hybrid protein activity have not been discussed.
In this computational study, we investigate how cells might manage potentially detrimental cross-talk between multiple c-di-GMP signalling pathways through spatial localization and post-translational control of GGDEF- and EAL-containing proteins. In addition, we interrogate the ‘biochemical conundrum’ of hybrid proteins by investigating the prevalence of DGC and PDE activity and associated regulatory sequence motifs in such proteins. Our results complement the detailed findings from numerous molecular studies and provide a genome-scale framework for understanding c-di-GMP signalling and control.
MATERIALS AND METHODS
Data sources
Proteomic sequences for 867 prokaryotic genomes were downloaded from the KEGG database (15) (results are based on data downloaded in May 2009; Supplementary Material 1). Phylogenetic and habitat-based classification of organisms were obtained from the NCBI Microbial Genomes database, and cross-database mapping of organism names was performed using the NCBI taxonomy IDs. Protein family models were obtained from the PFAM database (16) (downloaded in October 2008). Distance-based operon predictions were downloaded from http://popolvuh.wlu.ca/gmh/TUpredictions (recently updated to http://popolvuh.wlu.ca/public/TUpredictions/) (17), which includes information for 97.5% (10 964) of GGDEF and EAL proteins (May 2009).
Annotation of domains
All protein sequences were searched against the PFAM database using the hmmpfam tool available in HMMER 2.3.2 (http://hmmer.janelia.org). For all searches (PFAM entries: ‘GGDEF’, ‘EAL’ and ‘HD’ for HD-GYP), a score threshold equal to the ‘trusted cutoff’ for each PFAM sequence alignment model was used; this is a stringent cutoff that gives high confidence in family annotations (see HMMER manual). In total, 9443 proteins contained the GGDEF domain and 5574 the EAL domain. We identified a further set of 5951 proteins containing the HD domain (18). Since this represents a broad set of protein families, we selected 1034 proteins with HHExxDGxxGYP motif [described in (19)] as the most likely to contain an HD-GYP domain.
For validation, we performed reverse psi-blast (rpsblast from BLAST version 2.2.19), at a E-value cutoff of 10−6, using position-specific scoring matrices for the GGDEF and EAL domains (20).
PFAM models matching outside the GGDEF, EAL and HD-GYP domains were assigned as partner domains. We also flagged protein sequences containing an unannotated stretch of more than 100 amino acids as potentially containing a partner domain.
Annotation of spatial localization signals
Spatial localization signals were identified using TMHMM 2.0 (21), SignalP 3.0 (22), LipoP 1.0A (23) and TatP 1.0 (24), adopting default parameters. For SignalP, the Gram-positive prediction was used for Firmicutes and Actinobacteria, and the Gram-negative prediction was applied to all other organism groups (25). Here, a sequence was flagged as containing the secretion signal if it gave a positive result in either the Hidden Markov Model- or the Neural Network-based algorithms.
Divergence of pairs of sequences
In order to assess divergence between two instances of a given PFAM domain, the amino acid sequences concerned were aligned using the Needleman–Wunsch global alignment algorithm (26) implemented in EMBOSS (http://emboss.sourceforge.net/). The BLOSUM-62 matrix (27), a gap-opening penalty of 10 and a gap extension penalty of 0.5 were used, and the scores obtained were divided by the length of the alignment and used to assess sequence divergence (higher scores signify smaller divergence and vice versa).
Identification of catalytic and allosteric site motifs
Multiple sequence alignments of the GGDEF and EAL domains were produced using MUSCLE (28) in order to identify positions corresponding to catalytic site residues.
We used the protein sequence of PleD (GGDEF-only protein in Caulobacter crescentus) as the reference for GGDEF domains. The catalytic AGGDEF site was considered intact domains retained in the GG[D/E]EF signature motif (29). The c-di-GMP-binding allosteric IGGDEF site in the GGDEF domain was considered intact if it contained an RxxD motif five residues upstream of catalytic site (30). The large-scale nature of the multiple alignment means that domains containing degenerate catalytic sites were occasionally mis-aligned, leading to an underestimation of the number of intact allosteric sites among these proteins. To correct for this, we also used a more relaxed 20-residue window for the position of the allosteric site relative to the catalytic site. The general conclusions of this article remain the same for both definitions (Supplementary Material 2), and we present the results of the more stringent search criterion in the main text.
RocR (EAL-only protein in P. aeruginosa) (31) was used as reference for EAL domains. The catalytic AEAL site was considered intact if seven non-contiguous positions comprising the Mg2+-chelating site (E175, N233, E265, E268, D295, K316 and E352, where the numbers correspond to amino acid positions in RocR) were all retained. An additional set of four c-di-GMP recognition residues (Q161, R179, D296 and D318) was considered when appropriate.
Statistical methods
Pearson and Spearman correlation coefficients were calculated using the R statistical package (http://www.r-project.org). Pearson correlation was calculated between (i) numbers of transcription factor (TFs) encoded per genome and the total number of genes per genome and (ii) numbers of GGDEF and EAL genes per genomes and the total number of genes per genome. Spearman correlation coefficients were calculated between the fraction of proteins of a given type (GGDEF-only, EAL-only or hybrid) that contained a spatial localization signal or a partner domain and the total number of proteins of that type encoded in a genome.
RESULTS AND DISCUSSION
GGDEF and EAL proteins in prokaryotic genomes
We assembled a set of 867 completely sequenced prokaryotic genomes of which 813 were bacterial and the remainder archaeal [downloaded on 15 May 2009 from the KEGG database (15); Supplementary Material 1]. Using the HMMER suite of programs, we searched the coding regions of these genomes for sequences containing the GGDEF and EAL domains, both of which show comparable levels of sequence divergence (average pairwise identity of 28% for both domain sequence models). We identified a total of 11 248 GGDEF and EAL proteins, across 618 genomes (Table 1); thus, over 75% of all bacterial genomes in the data set code for at least one GGDEF or EAL protein. Compared to the significantly more sensitive HMMER3 suite of programs, which was in Beta version at the time this work was performed, our analysis has a sensitivity of 97.6 and 98.8% for GGDEF and EAL domains, respectively. Almost all GGDEF (99.8%; 9422 proteins) and EAL (99.9%; 5572 proteins) domains identified in our study are also detected by the position-specific scoring matrices-based reverse psi-blast, thus demonstrating the consistency of our domain assignments. The less well-characterized HD-GYP domain is discussed later.
Table 1.
Spatial localization signals and partner domain occurrence for GGDEF and EAL proteins
| GGDEF-Only | EAL-Only | Hybrid | |
| Total proteins, N | 5674 | 1805 | 3769 |
| Localization signalsa | |||
| With localisation signal, n(%) | 3193 (56) | 643 (36) | 2142 (56) |
| With transmembrane helices, n | 3090 | 605 | 2030 |
| Sec signal peptide, n | 1438 | 417 | 1076 |
| Tat signal peptide, n | 112 | 14 | 100 |
| Lipoprotein signal, n | 22 | 21 | 54 |
| Partner domains | |||
| With partner domains | 2550 (45) | 473 (26) | 2636 (70) |
| With PAS domain | 801 | 21 | 1740 |
| With GAF domain | 445 | 41 | 357 |
| With REC domain | 552 | 123 | 237 |
| With HAMP domain | 327 | 6 | 394 |
| With HDOD domain | 28 | 128 | 0 |
| With unannotated sequenceb | 1195 | 491 | 507 |
aThe sets of proteins corresponding to each of the four localization signals are not mutually exclusive. These signals are dominated by TMHs, present in 96% of all proteins containing at least one of the four signals (Supplementary Material 8).
bProteins with unannotated sequence stretches (>100 amino acids) are not included in the total number of proteins with partner domains.
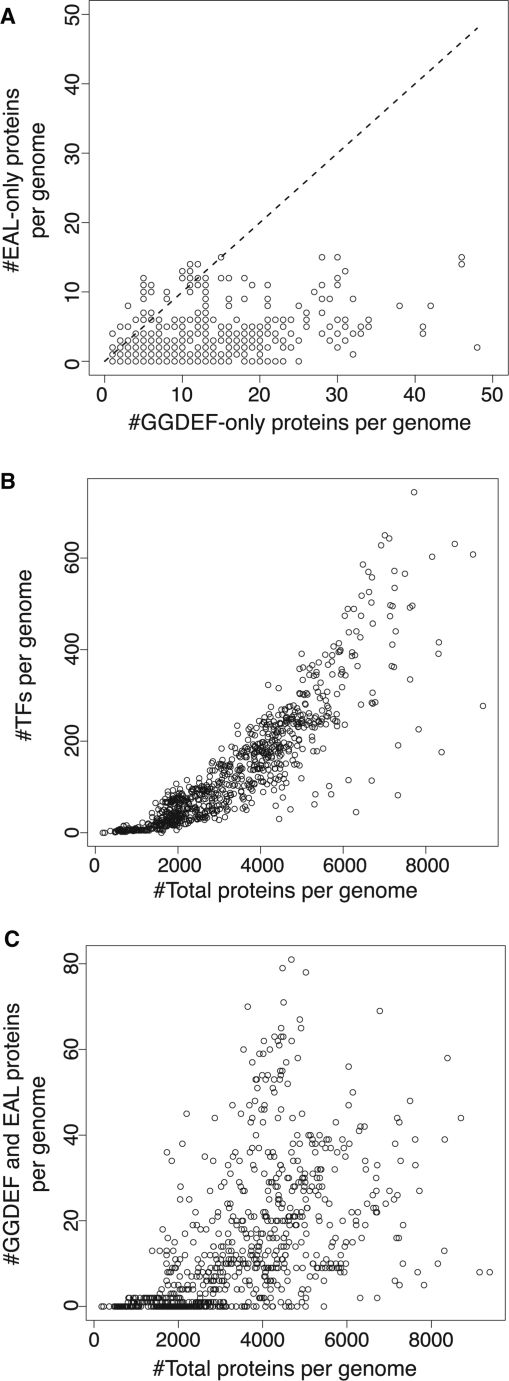
Of the proteins identified here, 5674 are GGDEF-only (i.e. do not contain the EAL domain), 1805 are EAL-only and 3769 are hybrid (i.e. contain both GGDEF and EAL domains; Table 1). It is notable that most organisms code for more GGDEF-only than EAL-only proteins (Figure 2A), although there are exceptions like E. coli K12 MG1655 that contain nearly equal numbers of GGDEF-only and EAL-only proteins.
Figure 2.
Occurrence of GGDEF and EAL proteins. (A) Plot of the number of EAL-only proteins versus the number of GGDEF-only proteins encoded in prokaryotic genomes. (B) Plot of the number of transcription factors versus the total number of genes per prokaryotic genome. (C) Plot of the number of GGDEF and EAL-containing proteins versus the total number of genes per prokaryotic genome.
To test the accuracy of our PFAM-based search, we compared our results with experimentally characterized c-di-GMP systems. In E. coli K12, we recovered all 29 GGDEF- and EAL-containing proteins (32). One of the EAL-only proteins (b2503; YfgF) in our data set was previously classed as a hybrid protein; however, the GGDEF domain has a degenerate catalytic site, and was not identified under the stringent thresholds used here (see ‘Materials and Methods’ section). Similarly, in P. aeruginosa PA01 we identified all 38 known proteins (17 GGDEF-only, 5 EAL-only and 16 hybrid proteins) (33) as well as an additional EAL-only protein (PA0707; exotoxin regulatory protein ToxR, which has a degenerate catalytic site). We obtained identical results when we scanned the above two genomes for GGDEF and EAL domains using (i) the HMMER3 programs and (ii) reverse psi-blast. This agreement with detailed molecular characterizations and with other software for domain identification demonstrate the reliability of the PFAM models and the stringent thresholds used in this study.
The phylogenetic distribution of GGDEF and EAL protein families across the prokaryotic kingdom follows known trends (Supplementary Material 3); they are expanded in Gamma Proteobacteria (median = 22.0 genes per genome), whereas Firmicutes (median = 1.0) and Actinobacteria (median = 3.5) generally have only a few per genome. GGDEF and EAL proteins are not known in Archaea; however, we detected one GGDEF-containing protein with an intact catalytic site in the genome of an uncultured methanogenic archaeon (Supplementary Material 4). In terms of habitat distributions, host-associated organisms tend to have only a few (median = 1.0) or no GGDEF or EAL protein, whereas organisms belonging to other habitat groups have substantially more (median = 10.0).
In our data set, 72 genomes code for a single GGDEF or EAL protein (Supplementary Material 5). Since c-di-GMP turnover requires both DGC and PDE activities, one might expect these proteins to be hybrid. However, we find that 57 are GGDEF-only proteins; since these organisms appear to lack proteins associated with PDE activity, this presents the intriguing question of how these organisms might maintain c-di-GMP homeostasis. Such organisms include Proteus mirabilis—a leading model system for studies of motility and adhesion (34,35)—in which the GGDEF protein is uncharacterized. Another example is the major pathogen Staphylococcus epidermidis (36): the GGDEF protein regulates biofilm formation in this organism, although it is not involved in c-di-GMP synthesis. A recently studied example is the cattle pathogen Anaplasma phagocytophilum (37), in which the catalytically active GGDEF-only protein is a homologue of PleD, a well-studied protein in C. crescentus (13).
GGDEF and EAL gene occurrence correlate weakly with genome size
Previous studies have shown that the number of regulatory proteins, such as transcription factors, encoded in the genome correlates strongly with genome size (R2 = 0.77; Figure 2B) (38). In contrast, the number of GGDEF and EAL proteins correlates only weakly with genome size (R2 = 0.18 for c-di-GMP genes in genomes with more than 2000 genes; Figure 2C). A majority of genomes (64%) coding for less than ∼2000 genes in total contain no GGDEF or EAL genes, whereas most larger genomes (85%) contain at least one. A possible explanation is the unusual expansion of these genes in Gamma Proteobacteria, which have significantly more GGDEF and EAL proteins than comparably sized genomes from other phylogenetic groups (Supplementary Material 6). But, even within Gamma Proteobacterial genomes coding for more than 2000 genes, the correlation with genome size is weak (R2 = 0.19; Supplementary Material 7).
A more likely explanation is the range of cellular functions targeted by different regulatory systems. In general, transcription factors control the expression of most genes in an organism. Barring a few global regulators, each factor responds to a single or a few types of signals and regulates a small set of functionally related target genes. Therefore, the number of transcription factor genes is expected to correlate with the total number of genes encoded in the genome. In contrast, GGDEF and EAL numbers do not scale with total gene numbers because c-di-GMP regulates a limited set of complex, possibly accessory, functions such as the switch between motility and adhesion. Instead, the number of genes involved in c-di-GMP turnover probably depends on the number of signals that they respond to, and the number of distinct spatial foci of c-di-GMP required, rather than genome size.
Spatial localization and signal-dependent post-translational control of GGDEF and EAL proteins: spatio-temporal segregation of different GGDEF and EAL systems
Genomic organisation and transcriptional regulation
As with most other prokaryotic enzymes, the concentration and activities of GGDEF and EAL proteins are regulated at the transcriptional and post-translational levels. We are unable to assess systematically the extent of transcriptional regulation because of the paucity of high-throughput gene-expression data for most bacteria. However, specific examples illustrate the intricate control of GGDEF and EAL gene expression. In Salmonella enterica Typhimurium, the GGDEF-only protein AdrA is required for biofilm formation when grown in rich media, whereas another GGDEF protein GcpA regulates the same process under nutrient-deficient conditions (39). In E. coli K12, only a few GGDEF and EAL proteins are expressed and active during the exponential phase of growth (12); for example, the protein YhjH is regulated by the flagella master regulator FlhDC (40). Numerous other proteins, notably those involved in curli biosynthesis, are expressed during stationary phase (12,32). Therefore, the expression of multiple GGDEF and EAL proteins under the same condition, at least in E. coli, indicates the need for spatial and temporal post-translational regulation in separating their activities.
To gain further insights into possible transcriptional co-regulation of different GGDEF and EAL proteins on a genomic scale, we analysed their gene neighbourhoods. We find that only over a quarter of these proteins (27%) are encoded in multi-gene operons, which is considerably less than our observation that over 55% of all genes are so encoded. Further, less than 6% of multi-gene operons contain more than one GGDEF or EAL protein, suggesting that there is little pressure for co-evolution or co-expression of sets of such proteins. Therefore, different GGDEF and EAL proteins could come under the control of distinct, specific transcription factors. These, together with the observation that there are many more GGDEF than EAL proteins, may indicate that the c-di-GMP synthesizing activity of a GGDEF protein may not be linked to the hydrolyzing function of a cognate EAL protein in a one-to-one fashion.
Spatial localization
One mechanism for ensuring separation of different GGDEF and EAL proteins in a cell is to sequester them at specific locations. For instance, the GGDEF protein WspR in P. aeruginosa clusters into cytoplasmic foci in response to a surface growth-associated signal (41) and cell-pole localization of PleD in C. crescentus is critical to regulation of the cell cycle (14).
Given the lack of comprehensive measurements of protein localization in bacteria, we used protein sequence information to predict the following localization signals: (i) transmembrane helices (TMHs) identified by the TMHMM software, which is reported to have false positive and false negative rates of 2.5% each (21); (ii) signal peptides for membrane transport through the Sec translocon predicted using SignalP (22); (iii) signals for transport of folded proteins through the Tat pathway using TatP (24); and (iv) lipid-mediated anchoring of proteins to the outer membrane predicted by LipoP (23). We classified a protein as spatially localized if it contained at least one of the four signatures (see Supplementary Material 8 for details). Note that these predictions are likely to underestimate spatial localization since some signals–such as those for cell pole localization of PleD in C. crescentus—are independent of the above sequence motifs, and may be impossible to predict from sequence data alone (42).
In total, over half of all GGDEF and EAL proteins contain localization signals (Table 1). This represents a substantial enrichment compared with expectations, since 15–20% of proteins encoded in bacterial genomes typically contain TMHs (43). Thus in a majority of GGDEF and EAL proteins, signal sensing and enzymatic activity occur at the membrane. Different proteins may be localized to distinct segments of the membrane thus further separating their activities in the cell.
Of note is the difference in the proportions of GGDEF-containing (i.e. GGDEF-only and hybrid) and EAL-only proteins containing localization signals (Table 1): whereas a majority of GGDEF-only and hybrid proteins are spatially localized, only a third of EAL-only proteins are.
Signal-dependent post-translational regulation
Previous studies on smaller data sets reported the prevalence of signal-sensing partner domains in GGDEF and EAL proteins, notably the small molecule-binding Per-Arnt-Sim (PAS) (44) and the two-component receiver domains (REC) (11). The roles of such signal-sensing partner domains in determining protein activity are illustrated by the REC domain in the classical GGDEF protein PleD (13). More recently, a regulatory role for a flavin-nucleotide-binding PAS domain has been demonstrated in the hybrid protein AxDGC2 in G. xylinus (45). We extended the above observations to a larger set of GGDEF and EAL proteins. In particular, we tested whether the extent of such post-translational control differs between GGDEF-containing and EAL-only proteins.
Using PFAM, we found that over half of GGDEF and EAL proteins contain an additional domain (Table 1). We also have an additional 20% of proteins with an unannotated stretch of more than 100 amino acids (the approximate length of the PAS domain) as potentially containing a partner domain. Thus, between 50% and 70% of GGDEF and EAL proteins comprise multiple domains.
Most proteins contain only a single partner domain (4419 out of 5659 proteins with annotated partner domains; 78%). Among these, the most common partners are the PAS and REC domains. Also common are the HAMP domain (46), which is generally found in signal-sensing histidine kinases and the GAF domain, which may bind nucleotides (47).
Again, there is a striking difference between GGDEF-containing and EAL-only proteins. Similar to our observations on spatial localization, the former tend to associate more with partner domains (Table 1): nearly half of GGDEF-only and over two-thirds of hybrid proteins contain at least one identifiable partner domain. However, only about a quarter of EAL-only proteins do so. The proteins also differ in the types of partner domains they contain. Whereas GGDEF-containing proteins favour signal-sensing domains (4650 proteins; 90%), the partner domain in nearly half of EAL-only proteins (191 proteins; 40%) is more likely to perform a second output function, rather than sense a signal. The most prominent example is the HDOD domain, a dinucleotide phosphohydrolase which might convert the pGpG product of the EAL activity to GMP (see Supplementary Material 9 for other such domains). Thus, only a small minority of EAL-only proteins (16%) contain an identifiable signal-sensing partner domain, of which the phosphorylation-receiving REC domain is the most common. Unfortunately, there is little information on the specifics of the signals to which these proteins might respond to.
Sequence divergence of signal-sensing and enzymatic domains
Since a few signal-sensing partner domains dominate, we measured the degree of sequence divergence within each domain using pairwise alignments of sequences from the same genome, to see whether they are likely to respond to distinct signals (Supplementary Material 10). We restricted this analysis to GGDEF-only proteins containing one of PAS, GAF or REC domains, ensuring that only proteins containing the signature motif for catalytic activity were included. Further, only proteins with one instance of the partner domain were considered.
In general, we find that the catalytic domain is much more conserved than the signal-sensing partner domain (median alignment scores are GGDEF = 1.45; REC = 1.13; PAS = 0.48; GAF = 0.47). Moreover, the PAS and GAF domains, which bind to small molecules, show greater pairwise variability than the phosphorylation-receiving REC domain where a few selected mutations are sufficient to change the identity of the cognate kinase (48).
Thus, significant variability in the most common small molecule-sensing domains is likely to allow different c-di-GMP systems to respond to distinct environmental and cellular signals.
Association of transmembrane localization and signal-dependent regulation with family expansion
Since spatial and post-translational control help separate the activities of distinct DGCs and PDEs, we tested whether such forms of regulation are more prevalent in organisms encoding more enzymes (Supplementary Material 11). We find that EAL-only proteins are more likely to contain localization signals if they occur in relatively large numbers in a given genome (Spearman correlation between total number of EAL-only proteins and the fraction with localization signals = 0.48, P-value = 2 × 10−28). However, there is no equivalent trend in GGDEF-only and hybrid proteins (Spearman correlations of 0.00 and −0.06 for GGDEF and hybrid proteins, respectively).
In contrast to localization, there is little association between family expansion and the fraction of proteins containing partner domains, across GGDEF-only, EAL-only or hybrid proteins (Spearman correlations of −0.17, 0.06 and −0.03, respectively). The slight negative correlation for GGDEF-only proteins is explained by organisms coding for a single GGDEF-only protein, most of which have a partner domain [53 of 72 proteins, such as the PleD homolog in A. phagocytophilum which is controlled by its REC domain (37)]; removal of such organisms gives an insignificant association between GGDEF-only family expansion and its propensity to contain partner domains (Spearman correlation = 0.02). Therefore, signal-dependent post-translational control is not necessitated by family expansions and may depend instead on the nature of the signal and the required kinetics of response.
Catalytic activity of GGDEF and EAL proteins: implications for the ‘biochemical conundrum’ of hybrid proteins
Most GGDEF and EAL proteins retain intact catalytic site motifs
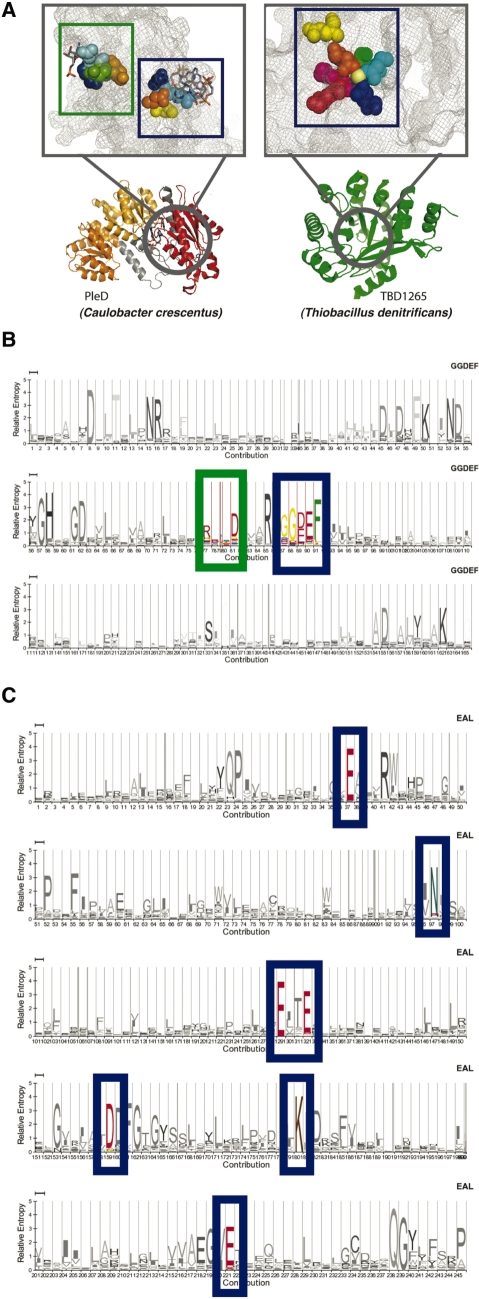
Biochemical and structural studies of GGDEF and EAL domains have identified the amino acid residues that are essential for catalytic activity (Figure 3). The GGDEF domain contains the GG[D/E]EF signature motif (termed the AGGDEF site) (29) that is necessary for substrate binding and catalysis, and an additional RxxD motif (IGGDEF site) that provides c-di-GMP-dependent allosteric control of DGC activity (30). In the EAL domain, the catalytic site comprises seven discontinuous residue positions (AEAL site) that chelate Mg2+, and an additional four residue positions determine c-di-GMP binding (not included in our definition of the AEAL site since mutations at these sites do not eliminate activity) (31). In both domains, amino acid substitutions at the AGGDEF and AEAL positions abolish enzymatic function.
Figure 3.
Catalytic and allosteric sites in GGDEF and EAL domains. (A) Representative structures of GGDEF- (left; PleD from C. crescentus) and EAL-domain-containing (right; TBD1265 from Thiobacillus denitrificans) proteins. The grey circles indicate the protein regions which contain the catalytic and allosteric motifs. The insets are enlarged versions of the circled regions. The catalytic and allosteric residues are shown in the form of space-fill diagrams. The catalytic sites AGGDEF and AEAL are highlighted in blue boxes and the allosteric IGGDEF site is enclosed within a green box. Multi-line HMM logos (http://logos.molgen.mpg.de), which graphically represent a HMM sequence model of (B) GGDEF and (C) EAL PFAMs. Larger the size of the residue, the more information it provides about that position in the sequence family. The AGGDEF and IGGDEF motifs and the residues forming the AEAL site are highlighted in (B) and (C), respectively (the catalytic sites in blue and the allosteric site in green); all other residues are in grey. Note that the seven residues forming the EAL catalytic site, though separated in sequence (C) are brought together in a three-dimensional space (A, right).
Overall, 9764 GGDEF and EAL domains retain intact catalytic site residues (87%; Table 2 ); however, distinct patterns of conservation emerge once we examine the context in which the domains occur. The AGGDEF site is conserved in most GGDEF-only proteins (Table 2; Supplementary Material 12), compared with only a small majority among hybrid proteins. In contrast, EAL domains tend to retain the AEAL site regardless of their occurrence in EAL-only and hybrid proteins.
Table 2.
Occurrence of catalytic sites in GGDEF and EAL proteins
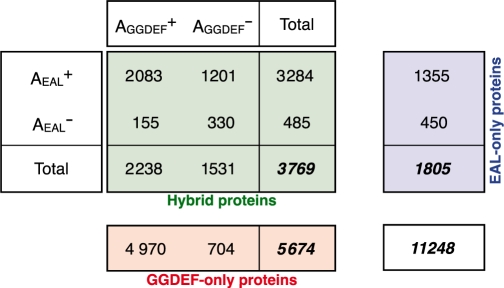 |
The numbers of GGDEF-only (red), EAL-only (blue) and hybrid proteins (green) with different combinations of intact (AGGDEF+ and AEAL+) and degenerate (AGGDEF– and AEAL–) catalytic sites.
Among GGDEF domains with intact AGGDEF sites, the IGGDEF site is more prevalent among GGDEF-only proteins than in hybrid proteins (Table 3 ). It has been hypothesized that an IGGDEF site might be essential for controlling the rate of c-di-GMP synthesis by GGDEF domains with high activity, but probably not in those with low activity (4). Under this model, one might expect GGDEF-only proteins to be more enriched for high activity than hybrid proteins. We observe that GGDEF-only proteins tend to make use of the GGEEF motif (3299 proteins; 66%), whereas intact sites in hybrid proteins are almost always GGDEF (2203; 98%). Although the biochemical consequence of such a contrast—also observed earlier in P. aeruginosa (8)—is not known, we speculate that this might lead to differences between the kinetics of GGDEF-only and hybrid DGC activity.
Table 3.
Occurrence of catalytic and allosteric sites in GGDEF domains
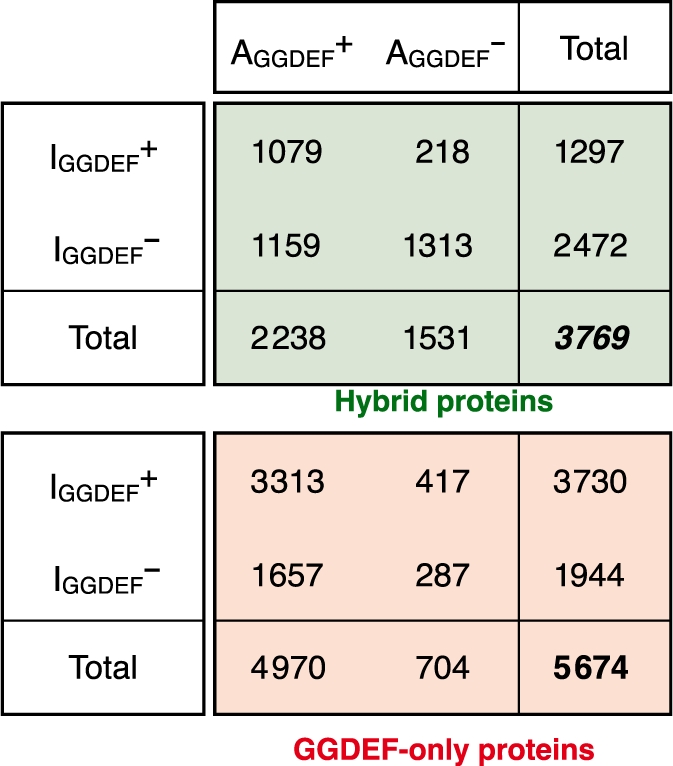 |
The numbers of GGDEF-only (red) and hybrid proteins (green) with different combinations of intact (AGGDEF+ and IGGDEF+) and degenerate (AGGDEF– and IGGDEF–) catalytic (A) and allosteric (I) sites.
Catalytic site motifs in GGDEF and EAL proteins
Early biochemical studies of hybrid proteins suggested that catalytic activity is generally retained only in one of the two domains (e.g. dgc and pdeA gene products in G. xylinus). However, more recent work has identified hybrid proteins that possess both DGC and PDE enzymatic functions [MSDGC1 in M. smegmatis (7), BphG1 in Rhodobacter sphaeroides (10) and ScrC in Vibrio parahaemolyticus (9)]. Based on patterns of amino acid conservation, our analysis above indicates that out of the four possible combinations of AGGDEF and AEAL sites in hybrid proteins, the most common are (i) the retention of both sites and (ii) retention of the AEAL site only (Table 2).
Most hybrid proteins retaining both sites probably operate through preferential activation of DGC or PDE function. For example, the proteins BphG1 and ScrC display switching behaviour between the two catalytic functions. Although the protein MSDGC1 shows both activities simultaneously in vitro, it is clear that DGC function is prioritized in vivo during nutrient starvation as intracellular c-di-GMP levels rise during this condition. Since the GGDEF domain is known to function as a dimer whereas the EAL domain activity is independent of dimerization (4), the switching of catalytic activities may be mediated by the transition between dimerization states. For BphG1, activity switching is controlled by the partner domain, and in ScrC this is achieved through accessory proteins. As a large proportion of this class of hybrid proteins contain signal-sensing partner domains (1575 proteins; 76%), we speculate that their catalytic activities may be regulated post-translationally. However, as illustrated by the study on BphG1, the mechanism by which activity switching is regulated may be complex, involving a series of steps including proteolysis and signal sensing by the partner domain (10).
The other major class of hybrid proteins are those that retain only the AEAL site, which probably function primarily as PDEs. We noted earlier that very few EAL-only proteins contain a partner domain; in contrast, a majority of hybrid proteins retaining only the AEAL site possess a partner domain (787 proteins; 66%), which could provide signal-dependent control of PDE activity. In fact, the PAS-GGDEF and GGDEF-EAL combinations are the two most common domain pairs among c-di-GMP proteins (Supplementary Material 13), and can be considered as ‘supra-domains’ that co-occur under many sequence contexts (49). Therefore, we speculate that the EAL domain in hybrid proteins, but not in EAL-only proteins, gains a signal-sensing partner domain due to the sequence context of GGDEF domains. Additionally, since degenerate GGDEF domains may retain substrate-binding capabilities, they might provide GTP-dependent control of PDE activity. This has been experimentally confirmed in a protein in C. crescentus (CC3396) (50), and proposed to occur in a GGDEF protein in P. aeruginosa (FimX) (51,52). Therefore, PDE activity is post-translationally regulated in a signal-dependent manner in hybrid proteins, but rarely in EAL-only proteins.
Alternative functions of GGDEF and EAL proteins not retaining any catalytic motif
A significant minority of GGDEF-only, EAL-only and hybrid proteins appear to lack catalytic function, and may have acquired alternative functions including roles as c-di-GMP effectors.
We first investigated degenerate GGDEF domains. Among such GGDEF-only proteins, a majority retain the IGGDEF site, whereas only a small minority of in hybrid proteins do so (Table 3). This indicates that a majority of degenerate GGDEF-only proteins could act as effectors by binding c-di-GMP at the IGGDEF site. This has been experimentally verified in a protein called PopA in C. crescentus, in which c-di-GMP binding to the IGGDEF site triggers a change in protein localization which is essential to cell division control (53). Nonetheless, the most common partner domains in degenerate GGDEF-only proteins are still the signal-sensing PAS, GAF and REC instead of output domains that might respond to c-di-GMP binding to the IGGDEF site (Supplementary Material 14). Therefore, one might expect many of these degenerate GGDEF-only proteins to integrate multiple signals or possess novel activity. In hybrid proteins, degenerate GGDEF domains can bind GTP and control the activity of the associated EAL domain (50). However, in the small minority of hybrid proteins lacking both AGGDEF and AEAL sites alternative functions are possible; for example, YhdA (CsrD) in E. coli can bind two small RNAs controlling motility and sugar metabolism (54).
Among EAL-only proteins, a quarter do not retain signatures of catalytic activity (Supplementary Material 15). Since most EAL-only proteins without the AEAL site lose the c-di-GMP binding site (411 proteins; 91%), these are probably not c-di-GMP effectors and could have acquired novel functions. This has been experimentally verified in YcgF in E. coli which, acting independently of c-di-GMP, binds to and controls the activity of a transcription factor YcgE (55). It is striking that functions downstream of the actions of YcgF include biofilm formation, as is typical of canonical c-di-GMP-associated proteins (5).
The HD-GYP domain
A study of proteins involved in c-di-GMP turnover is complete only with an analysis of the HD-GYP domain (56), which is a relatively less characterized domain that hydrolyzes c-di-GMP to GMP (PDE activity like EAL, but leading to a different end product). We identified 1034 proteins with this domain in 295 organisms, all of which contain at least one GGDEF or EAL-containing protein. We did not discuss proteins with this domain earlier since the identification of this domain involves selecting a subset of the larger HD superfamily containing a conserved motif (19), which might decrease the accuracy of a purely PFAM-based search.
Spatial localization signals are present in 200 HD-GYP proteins (19%), which is around the average for bacterial proteins (43). We are able to identify partner domains in 452 HD-GYP proteins (44%; excluding the HDOD domain, part of which generally overlaps with the HD-GYP domain), of which 347 (77%) contain a single type of partner. The most common is the REC domain (Supplementary Material 16). Thus, HD-GYP domains show a greater tendency to associate with signal-sensing partner domains than EAL-only proteins.
The GGDEF domain is present in 74 HD-GYP proteins; these could be considered as hybrid proteins, though such a definition is not used traditionally. In contrast to HD-GYP-only proteins, 62% (46 of 74) of these hybrid HD-GYP proteins contain partner domains—of which 32 contain the PAS domain—making them similar to EAL-GGDEF hybrid proteins. All but two hybrid HD-GYP proteins (GAU1030 in Gemmatimonas aurantiaca T-27 and Rxyl1018 in Rubrobacter xylanophilus DSM 9941) contain an intact AGGDEF site, and might therefore retain both enzymatic activities, with the balance between the two activities being determined by the partner domain.
In summary, the HD-GYP domain rarely associates with the GGDEF domain to form hybrid proteins (only 7% of HD-GYP proteins are hybrid), compared with the EAL domain (68% of EAL proteins are hybrid). Moreover, nearly all HD-GYP-GGDEF hybrid proteins are likely to retain both catalytic activities. The proportion of HD-GYP proteins with signal-sensing partner domains (378 of 960 HD-GYP-only proteins; 39%) is similar to that for proteins with DGC-only (2324 of 5125 proteins retaining the AGGDEF site but not the AEAL site; 45%) and EAL-based PDE-only activities (1069 of 2556 proteins retaining only the AEAL site but not the AGGDEF site; 42%). This probably precludes the need for a degenerate GGDEF domain to allow signal-dependent control of HD-GYP activity.
CONCLUSION
Our study is the first example of a comprehensive genome-scale computational assessment of post-translational control of c-di-GMP turnover at multiple levels in bacteria. Many bacterial genomes code for several proteins with GGDEF and EAL domains, which has led to questions on how the activities of distinct c-di-GMP signalling systems are segregated in order to avoid potentially deleterious cross-talk. Moreover, the observation that a large number of hybrid proteins contain both GGDEF and EAL domains raised questions about the relationship between the two opposing activities and how the two are coordinately or reciprocally regulated. Our study contributes to answering these two questions by systematically determining the extent of post-translational regulation and analysing the presence of catalytic motifs across a large set of GGDEF and EAL proteins.
We began by demonstrating remarkable differences between GGDEF-containing and EAL-only proteins in the occurrence of spatial localization signals and signal-sensing partner domains, and therefore the extent of post-translational control. The relative absence of signal-sensing partner domains in EAL-only proteins, together with the observation that EAL-only proteins are few in number in most genomes, might indicate that many EAL-only proteins are regulated largely at the transcriptional level, and may contribute towards maintaining c-di-GMP homeostasis rather than respond directly to environmental or cellular cues. Among GGDEF-containing proteins, hybrid proteins tend to show a much greater tendency to contain signal-sensing partner domains than GGDEF-only proteins.
We then show that hybrid proteins, which have been termed a biochemical conundrum (48), would retain both DGC and PDE activities or only the PDE activity. In hybrid proteins retaining both activities, partner domains could specify the nature of enzymatic activity. The two activities could be mutually exclusive (9,10). Alternatively, both domains might be simultaneously active with the EAL domain hydrolyzing excess c-di-GMP, thus acting as an enzymatic substitute for the IGGDEF site, which is absent in many hybrid proteins. PDE activity, determined by the EAL domain, has little scope for signal-dependent post-translational control in EAL-only proteins, but gains such regulation in the context of hybrid proteins that have lost DGC activity. Overall, a higher proportion of GGDEF domains (2235 proteins; 23.7%) have lost their catalytic motif than EAL domains (935 proteins; 16.8%). This might imply that GGDEF domains have a greater capacity to assume different functions, which include (i) the ability to act as a c-di-GMP effector through the IGGDEF site primarily in GGDEF-only proteins; and (ii) the potential to allosterically regulate EAL activity in hybrid proteins by binding to GTP. An important caveat of our study is that predictions of catalytic activity are based on occurrences of certain sequence motifs and may not be comprehensive; for example, roles of conserved residues outside the GG[D/E]EF motif in DGC catalytic activity are not considered here. Though the identity of partner domain might point to a broad class of molecules that can be sensed, the exact nature of these signals remains a major question in the field. Further, the mechanisms of signal transduction, spatial sequestration and activity switching in hybrid proteins remain largely unresolved.
We identified a number of genomes (>10% of genomes with GGDEF or EAL proteins) coding for only a single GGDEF or EAL protein. Our observation that most of these proteins are GGDEF-only and lack known PDE proteins raises the question of how c-di-GMP homeostasis may be maintained in these organisms. They might harbour novel proteins with PDE activity or transport c-di-GMP out of the cell potentially as a form of intercellular signalling (57). As suggested recently, these systems could be used as models for studying c-di-GMP signalling at its lowest complexity (58).
A major area of interest in the field is understanding the mechanisms by which c-di-GMP exerts its effects. Current knowledge points to the presence of both protein and RNA-based effector molecules (5). We did not study outputs of c-di-GMP signalling here, except as determined by degenerate catalytic and intact allosteric sites. A majority of GGDEF-only proteins—but not hybrid proteins—with a degenerate AGGDEF site retain an intact c-di-GMP allosteric site and could act as c-di-GMP effectors. It appears unlikely that degenerate EAL-only proteins could retain c-di-GMP-binding. Even GGDEF and EAL proteins which do not have a role in c-di-GMP signalling, where characterized, control aspects of motility and adhesion (5). Combination of computational and experimental approaches have led to the identification and characterization of c-di-GMP effectors such as the PilZ protein domain (58–61) and the GEMM riboswitch RNA motif (62). However, characterization of novel classes of c-di-GMP-binding molecules is necessary to broaden our understanding of this signalling mechanism.
Finally, the data sets used in this study and the results of our analysis (Supplementary Data sets) will serve as a genomic framework for interpreting results of ongoing molecular investigation of c-di-GMP signalling.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cambridge Commonwealth Trust; St John’s College, University of Cambridge; Girton College, University of Cambridge to A.S.N.S.; Biotechnology and Biological Sciences Research Council (BBSRC) grant ‘Genomic Analysis of Regulatory Networks for Bacterial Differentiation and Multicellular Behaviour’ to G.M.F. and N.M.L.; European Molecular Biology Laboratory (EMBL) to N.M.L. Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 2009;12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Amikam D, Benziman M. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 1989;171:6649–6655. doi: 10.1128/jb.171.12.6649-6655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, et al. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 5.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 6.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar M, Chatterji D. Cyclic di-GMP: a second messenger required for long-term survival, but not for biofilm formation, in Mycobacterium smegmatis. Microbiology. 2008;154:2942–2955. doi: 10.1099/mic.0.2008/017806-0. [DOI] [PubMed] [Google Scholar]
- 8.Ryan RP, Fouhy Y, Lucey JF, Dow JM. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 2008;190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarutina M, Ryjenkov DA, Gomelsky M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 2006;281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 11.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 12.Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N, Hengge R. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology. 2009;155:1318–1331. doi: 10.1099/mic.0.024257-0. [DOI] [PubMed] [Google Scholar]
- 13.Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 2003;47:1695–708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 14.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer E.LL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno-Hagelsieb G, Collado-Vides J. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18:S329–S336. doi: 10.1093/bioinformatics/18.suppl_1.s329. [DOI] [PubMed] [Google Scholar]
- 18.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 19.Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1999;1:303–305. [PMC free article] [PubMed] [Google Scholar]
- 20.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32(W):327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 22.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 23.Juncker AS, Willenbrock H, Heijne GV, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protocol. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 26.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 27.Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malone JG, Williams R, Christen M, Jenal U, Spiers AJ, Rainey PB. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology. 2007;153:980–994. doi: 10.1099/mic.0.2006/002824-0. [DOI] [PubMed] [Google Scholar]
- 30.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 31.Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 2008;190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 2006;62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 33.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl Acad. Sci. USA. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, et al. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 2008;190:4027–4037. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rather PN. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 2005;7:1065–1073. doi: 10.1111/j.1462-2920.2005.00806.x. [DOI] [PubMed] [Google Scholar]
- 36.Holland LM, O'Donnell ST, Ryjenkov DA, Gomelsky L, Slater SR, Fey PD, Gomelsky M, O'Gara JP. A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J. Bacteriol. 2008;190:5178–5189. doi: 10.1128/JB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai TH, Kumagai Y, Hyodo M, Hayakawa Y, Rikihisa Y. The Anaplasma phagocytophilum PleC histidine kinase and PleD diguanylate cyclase two-component system and role of cyclic Di-GMP in host cell infection. J. Bacteriol. 2009;191:693–700. doi: 10.1128/JB.01218-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- 40.Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 41.Güvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bendtsen JD, Binnewies TT, Hallin PF, Ussery DW. Genome update: prediction of membrane proteins in prokaryotic genomes. Microbiology. 2005;151:2119–2121. doi: 10.1099/mic.0.28181-0. [DOI] [PubMed] [Google Scholar]
- 44.Ponting CP, Aravind L. PAS: a multifunctional domain family comes to light. Curr. Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 45.Qui Y, Rao F, Luo Z, Liang ZX. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry. 2009;48:10275–10285. doi: 10.1021/bi901121w. [DOI] [PubMed] [Google Scholar]
- 46.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 47.Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 48.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 51.Huang B, Whitchurch CB, Mattick JS. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 2003;185:7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kazmierczak BI, Lebron MB, Murray TS. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 2006;60:1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jonas K, Tomenius H, Romling U, Georgellis D, Melefors O. Identification of YhdA as a regulator of the Escherichia coli carbon storage regulation system. Microbiol. Lett. 2006;264:232–237. doi: 10.1111/j.1574-6968.2006.00457.x. [DOI] [PubMed] [Google Scholar]
- 55.Tschowri N, Busse S, Hengge R. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 2009;23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl Acad. Sci. USA. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Römling U. Cyclic Di-GMP (c-Di-GMP) goes into host cells-c-Di-GMP signaling in the obligate intracellular pathogen Anaplasma phagocytophilum. J. Bacteriol. 2009;191:683–686. doi: 10.1128/JB.01593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 60.Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 2007;282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 62.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.