Abstract
Non-homologous end-joining (NHEJ), the major repair pathway for DNA double-strand breaks (DSB) in mammalian cells, employs a repertoire of core proteins, the recruitment of which to DSB-ends is Ku-dependent. Lack of either of the core components invariably leads to a repair deficiency. There has been evidence that an alternative end-joining operates in the absence of the core components. We used chromosomal reporter substrates to specifically monitor NHEJ of single I-SceI-induced-DSB for detailed comparison of classical and alternative end-joining. We show that rapid repair of both compatible and non-compatible ends require Ku-protein. In the absence of Ku, cells use a slow but efficient repair mode which experiences increasing sequence-loss with time after DSB induction. Chemical inhibition and PARP1-depletion demonstrated that the alternative end-joining in vivo is completely dependent upon functional PARP1. Furthermore, we show that the requirement for PARP1 depends on the absence of Ku but not on DNA-dependent protein kinase (DNA-PKcs). Extensive sequencing of repair junctions revealed that the alternative rejoining does not require long microhomologies. Together, we show that mammalian cells need Ku for rapid and conservative NHEJ. PARP1-dependent alternative route may partially rescue the deficient repair phenotype presumably at the expense of an enhanced mutation rate.
INTRODUCTION
Non-homologous end-joining (NHEJ) is the prevailing pathway for repair of DNA double-strand breaks (DSB) in mammalian cells. NHEJ is executed by two core protein complexes: the DNA-dependent protein kinase (DNA-PKcs) complex composed of the Ku70/Ku80 heterodimer together with the catalytic subunit of the DNA-PKcs and a second complex of ligase IV and its co-factors XRCC4 and XLF (also known as Cernunnos) (1). If the DSB ends are not simply ligatable, terminal nucleotides are modified or removed by phosphokinases or nucleases such as PNK and Artemis and sequence gaps are replenished by polymerase λ or µ (2–4). Lack of any of the core components invariably creates a severe DSB repair defect and substantial sensitivity to ionizing radiation and various DNA damaging drugs like topoisomerase II inhibitors (5–9). Although most of the core components proved biochemically to be essential for the end-joining process, it was acknowledged for many years that cells completely lacking these proteins still rejoin the majority of radiation-induced DSBs (6,10–13). Using chromosomal reporter substrates that specifically monitor end-joining processes, we and others found that Ku-deficient and wild-type yeast and mammalian cells were equally capable of rejoining single I-SceI- and HO-induced breaks (14–18). This efficient end-joining has been ascribed to an alternative end-joining pathway, also named backup end-joining (B-NHEJ), which not only executes DSB repair but also class switch and to some extent V(D)J recombination (19–24). Genetic and biochemical in vitro analyses revealed that the alternative end-joining mode employs ligase III together with XRCC1, PNK and PARP1 (4,20,25). Accordingly, chemical inhibition of PARP1 significantly compromised the rejoining of episomal substrates in ligase IV- and Ku80-deficient cells (26). However, it is hitherto not clear whether the alternative end-joining route physically requires PARP1 for DSB repair in the chromosomal context.
Notably, the exquisite efficiency of Ku-independent end-joining was only observed for breaks with non-compatible ends (14–16). In contrast, rejoining of compatible ends was Ku-dependent. This raised the possibility that the alternative repair process prefers non-compatible ends which require further processing before ligation. Enforced recruitment of additional proteins into the repair complex including nucleases and polymerases (27,28) may increase the local stability of the junction and hence the probability of successful end-connection.
In this study, we used recently developed chromosomal reporter substrates to decipher genetics and structural requirements for Ku-dependent and independent end-joining. We describe that complementary and non-complementary ends similarly need Ku for rapid repair. In the absence of Ku, the cells employ a slow but error-prone alternative end-joining mode which is completely PARP1-dependent but does not need extensive microhomologies.
MATERIALS AND METHODS
Cell lines
The hamster cell lines CHOK1 (wild-type) and xrs5 (Ku80-deficient) were grown in α-medium (Gibco-Invitrogen) supplemented with 5% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C with 5% CO2. AA8 and V3 hamster cells were kindly provided by M. Löbrich, CHO9 and DNA-PK-deficient XR-C1 cells were a generous gift of M. Z. Zdzienicka.
Chemicals
The inhibitors 1,5-dihydroxyisoquinolinediol (DIQ), NU7026 and NU1025 were all purchased from Sigma-Aldrich.
Repair substrates
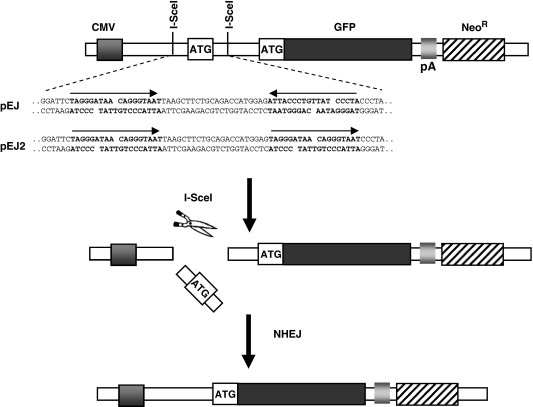
Two GFP-based repair substrates (pEJ and pEJ2) were used to investigate NHEJ in the chromosomal context as described previously (17). pEJ contains two repeat I-SceI recognition sites in inverted orientation which create double-stranded (ds)-ends that are non-cohesive. pEJ2 was newly cloned in order to create fully complementary DSB overhangs (Figure 1). Briefly, the proximal I-SceI recognition site was removed as EcoRI-HindIII fragment from the pEJ plasmid. A new ds-oligonucleotide (EcoRI-ATTACCCTGTTATCCCTAGTGCAC-HindIII) was reinserted that carried the I-SceI-site in the same direction as the distal copy. The advantage of this reporter system is that it is independent of how the ends are rejoined as long as any repair-associated deletions are no longer than 160 bp. The critical distance for base loss is reached at the transcription initiation site and the GFP open reading frame (ORF) (86-bp upstream of the first and 76-bp downstream of the second I-SceI site, respectively). Although in most cases both I-SceI sites are cut in vivo, the system allows to monitor single cleavage leading to in-frame shift of both ATGs, as described (9,29) (e.g. Supplementary Figure S1).
Figure 1.
Structure of the NHEJ reporter constructs pEJ and pEJ2. GFP translation is prevented by insertion of an artificial ATG (start codon) into the 5′-untranslated region between CMV promoter and the ORF. The artificial ATG is not in frame with the original ATG (17). Two repeat I-SceI recognition sequences (bold characters) flank the artificial ATG either in direct (pEJ2) or in inverted orientation (pEJ). Simultaneous cleavage of both I-SceI sites leads to pop out of the artificial ATG (middle) leaving either non-compatible ends (pEJ) or fully compatible ends (pEJ2). Repair of the I-SceI-induced DSB by NHEJ restores GFP translation leading to green fluorescence.
For stable chromosomal integration 0.5 µg of pEJ or pEJ2 linearized by AflIII were electroporated into the hamster cells as described (9). The cells were then grown in 14 µg/ml of G418 for 2 weeks. For each cell line and repair substrate, 30–60 individual colonies were picked, expanded and tested for green fluorescence after transient transfection of I-SceI expression vector as described earlier (17).
DSB-repair reporter assay
The cells containing single stably integrated copies of either repair substrate were electroporated with 50 µg of the I-Sce-I expression vector pCMV3xnls-I-SceI (17) to induce DSBs or with pCMV-Neo as a control. After 0–96 h of transfection, cells were assessed for green fluorescence by flow cytometry (FACScan, BD Bioscience). For further analysis of the individual NHEJ events, GFP-positive cells were sorted (FACS Aria, BD Bioscience) and proceeded in either of two ways: (i) cells were reseeded, raised to individual colonies and further expanded for DNA isolation (DNeasy tissue Kit, Qiagen). DNA of individual clones was amplified by PCR using the primers P1 and P2 that flank the I-Sce-I cleavage sites (forward 5′-GCAAATGGGCGGTAGGCGTGTA-3′, reverse 5′-TCGGGCATGGCGGACTTGAA-3′). PCR conditions were 35 cycles at (96°C for 20 s, 68°C for 20 s, and 72°C for 80 s) and an extension for 7 min at 72°C. Alternatively, (ii) total genomic DNA of the entire sorted cell population was isolated and subjected to PCR. The mixtures of those PCR products were ligated into the TOPO-cloning vector (TOPO TA cloning kit, Invitrogen), according to the manufacturer’s protocol and transformed into bacteria (one shot TOP10 competent Escherichia coli, Invitrogen). Single bacterial clones were scraped off the plate, directly subjected to PCR using the primers P1 and P2. PCR conditions are the same as above except for a preceding 10-min incubation for bacterial lysis at 96°C. PCR products from either of two procedures were subjected to sequencing using an automated ABI 3100 sequencer, (Applied Biosystems-Hitachi).
To assess the transfection efficiency, 30 µg of pEGFP-N1 (Clontech) were electroporated into both strains and analyzed by FACS 24 h later. CHOK1 and xrs5 showed 70.8 ± 3.9 and 64.2 ± 8.4% of green fluorescent cells, respectively. All subsequently given repair results are corrected for the 1.1-fold lower transfection efficiency of xrs5 cells.
PARP1 down-regulation by siRNA and western blot
Cells were transfected with the previously described siRNA sequence (5′-GGGCAAGCACAGUGUCAA-3′) to target the hamster PARP1 transcript (30). 5′-UAGGCAUUGCGCGUGUGUC-3′ (non-targeting RNA) was used as control. Cells were transfected using Trans-IT-TKO (Mirus, USA) according to the manufacturer’s protocol. The protein expression was monitored by western blotting according to standard procedures (9) using anti-PARP1 mouse monoclonal antibody (Enzo life sciences, Germany) and anti-actin antibodies (Sigma). Protein signals were visualized using the ECL detection kit (Amersham).
Colony formation assay
The effect of PARP1 inhibition on the radiosensitivity was assessed by colony formation. Cells were seeded and incubated for about 2–3 h to allow adhesion. Thereafter, specific PARP competitive inhibitor (75 µM DIQ or 300 µM NU1025) was added and the cells were incubated at 37°C for 2 h before irradiation. After X-irradiation (200 keV, 15 mA, additional 0.5 mm Cu filter at a dose rate of 2 Gy/min), the cells were incubated for 2 weeks with/without PARP inhibitor for colony formation. The surviving fraction of irradiated cells was normalized to the plating efficiency of non-irradiated controls.
Immunofluorescence staining of PAR moieties
Cells grown on slides were treated with or without PARP inhibitors and subsequently irradiated with 20 Gy. After 20 min, cells were fixed using 4% paraformaldehyde in PBS for 30 min at room temperature. The slides were then blocked with 1% BSA in PBS for 30 min at RT and incubated with anti-PAR monoclonal IgG (clone C2-10, Trevigen Inc. MD, USA) for 1 h (1 : 100 dilution in 0.5% BSA/0.1% Tween 20 in PBS). After washing, Alexa fluor 594-conjugated anti-mouse monoclonal IgG (1 : 600) was used as secondary antibody. Slides were counterstained with DAPI, examined and photographed under a fluorescent microscope (Axioplan2, Zeiss, Germany, Excitation at 590/617 nm).
RESULTS
Alternative end-joining is slow and error-prone
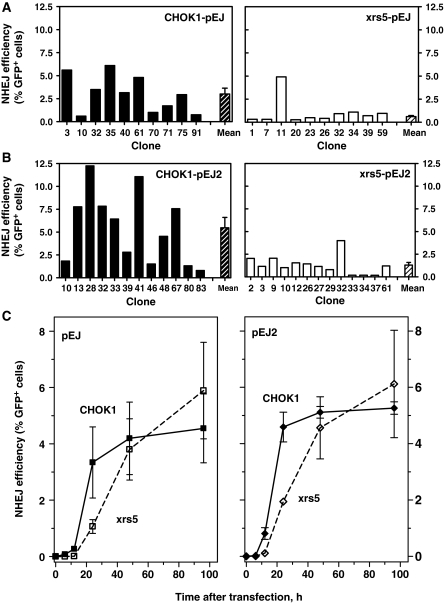
The Ku protein is a well-established essential enjoining factor. However, we and others have recently found that DSB with non-complementary ends were equally well rejoined by clones of wild-type and Ku-deficient rodent cells (14–18). In a series of experiments, using reporter constructs previously designed to specifically monitor chromosomal NHEJ (9,17), we sought to elucidate this apparent contradiction. The system makes use of the green fluorescent protein (GFP), the translation of which is only activated after successful rejoining of two nearby I-SceI-induced DSBs (Figure 1), which is then monitored by flow cytometry. Single copies of the reporter pEJ (Figure 1) (17) were stably integrated into CHOK1 and xrs5 (Ku80-deficient) hamster cells. NHEJ efficiency 24 h after transient expression of I-SceI was assessed in 10–13 independent clones of either cell line to appreciate inter-clonal variation that possibly impacts on those reporter assays (Figure 2). Despite substantial variations, CHOK1-pEJ cells showed on average 4-fold higher frequency of GFP-positive cells compared to xrs5-pEJ (Figure 2A) (3.0 ± 0.6 versus 0.71 ± 0.1%, Mann–Whitney test P = 0.0025).
Figure 2.
Alternative end-joining pathway is efficient but slow for both compatible and non-compatible ends. Ten to thirteen independent clones of CHOK1 and xrs5 cells harboring either pEJ (A) or pEJ2 (B) were transfected with the I-SceI-expressing vector. After a 24-h repair interval the fraction of GFP-positive/total cells (%GFP+) was measured by flow cytometry. Hatched columns represent mean values (±SE). xrs5-pEJ clone #11 was not included as deviation from others was larger than 2σ. NHEJ efficiency for both ends and was significantly lower in xrs5 than in CHOK1 cells (Mann–Whitney, P = 0.001). (C) NHEJ kinetics for non-cohesive (left panel) and cohesive (right panel) ends. Cells were harvested at different time intervals after transfection and analyzed for GFP+. The mean (±SE) values of three to four representative clones of each variant (CHOK1-pEJ #3, 40, 75; CHOK1-pEJ #33, 39, 48; xrs5pEJ #20, 23, 26, 59; and xrs5-pEJ2 #9, 10, 12) are shown. Differences between the two strains were significant only after 24 h (Mann–Whitney, P = 0.0025).
The structure of the double-stranded ends might affect the requirement for the Ku-protein (14,15). In order to test this possibility, we employed a second substrate pEJ2 which, in contrast to pEJ, provided full complementary ends (Figure 1). However, similar differences between both strains were observed for rejoining of these compatible ends (Figure 2B, 5.47 ± 1.1% GFP-positive CHOK1-pEJ2 cells versus 1.5 ± 0.3% GFP-positive xrs5-pEJ2 cells, P = 0.0012). Here, we investigated relatively short repair intervals compared to the preceding studies where repair efficiency was monitored after 48 h or later (15–18). In order to further understand the impact of the repair time, we analyzed the kinetics of GFP-positive cells between 6 and 96 h post-transfection (Figure 2C). In wild-type cells, the earliest repair events were found after only 6 h. The number of GFP-positive cells then rapidly increased with similar kinetics for non-cohesive and cohesive ends and reached a plateau after 48 h. In contrast, repair in Ku80-deficient cells was delayed by more than 6 h and GFP-positive cells accumulated more slowly than in CHOK1 cells. However, rejoining in xrs5 cells continued significantly beyond 24 h and the number of GFP-positive events almost reached wild-type levels after 48 h, confirming our previous observations (17,18). After 96 h, repair in xrs5 cells appeared to be even more efficient than in wild-type cells suggesting that Ku-independent end-joining includes the possibility to rejoin open breaks very lately after damage induction. However, the differences between wild-type and mutants at 96 h were statistically not significant. As expected from in vitro results (31), repair of fully compatible ends was slightly (n.s.) more efficient than of non-complementary ends.
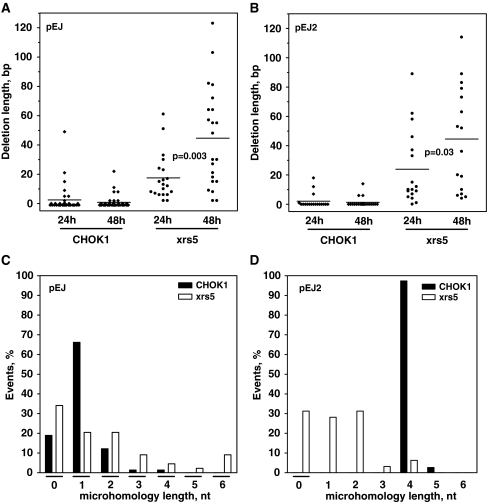
The slower kinetics of the end-joining process in xrs5 cells means that DSBs stayed open for long which might affect end stability. Given the time between DSB induction and closure matters, we hypothesized that in Ku-deficient cells repair fidelity decreases with increasing time. To address this, GFP-positive cells were sorted 24 and 48 h after transfection and 186 individual repair junctions were sequenced (Supplementary Figure S1). These two time points, in particular, appeared suitable to pick up differences between Ku-dependent and independent end-joining. Most CHOK1 repair events were highly conservative (Figure 3A and B). In many cases processing of the non-compatible overhangs led to fill-in of one additional base (Figure 3A, deletion of −1 bp). After 24 and 48 h, only few of the individual cells harvested showed deletions of up to 22 bp (one exception of Δ49 bp in CHOK1-pEJ after 24 h). Notably, high repair fidelity was maintained independently of the time of cell harvest. In contrast, nearly all xrs5 junctions showed substantial loss of flanking sequences, the amount of which drastically increased between 24 and 48 h reaching up to 123 bp (Figure 3A and B). Accordingly, in the absence of Ku the mean deletion length almost doubled between 24 and 48 h (for xrs5-pEJ from 17 ± 3 to 45 ± 7 bp and for xrs5-pEJ2 from 22.6 ± 6 to 45.5 ± 7.9 bp, respectively), while it remained constant in Ku-proficient cells (Figure 3A and B). Interestingly, neither in wild-type nor in Ku-deficient cells the structure of the DSBs affected the extent of base loss. Even full complementary overhangs (pEJ2) did not provide sufficient stability to prevent end degradation in xrs5 cells.
Figure 3.
Fidelity of the alternative end-joining. After 24 or 48 h of transfection, GFP+ cells were sorted and subjected to sequencing (Supplementary Figure S1). Scatter plots show deletion length of all individual repair junctions of (A) non-cohesive (pEJ) or (B) cohesive ends (pEJ2). Deletions are defined as the sum of base pairs lost at both external DSB ends. The 34-bp internal pop-out between both I-SceI sites was not defined as a deletion. P-values indicate the significance level of differences between 24 and 48 h in xrs5 clones (Mann–Whitney test). (C) and (D) Distribution of length of microhomologies used for formation of the repair junctions at both substrates. The 4-nt microhomologies employed by CHO-K1-pEJ2 cells correspond to the unmodified 4-bp complementary overhangs created directly through I-SceI cleavage. Sequence results were pooled from three independent sorting experiments for each cell line.
Sequence analyses (Supplementary Figure S1) demonstrate that 85% of pEJ-repair junctions in CHOK1 cells were formed upon using either no or one overlapping base and only 15% by longer microhomologies (Figure 3C). Xrs5 cells (Figure 3C and D, pEJ and pEJ2) used significantly more often two or more overlapping bases compared to the wild-type (Fisher’s exact test, P < 0.001). However, the average length of microhomologies was not significantly different (1.03 ± 0.09 nt for CHOK1-pEJ versus 1.8 ± 0.3 nt for xrs5-pEJ and 1.25 ± 0.2 nt xrs5-pEJ2) in contrast to previous reports (7,16,32–34). CHOK-pEJ2 employed directly the 4-nt complementary overhang resulting from double I-SceI cleavage without further modifying the ends (Figure 3D, see 4-nt column).
Together, our results revealed a slow but efficient alternative end-joining mode in the absence of Ku. This Ku-independent process is highly error-prone, as DSB termini are subjects of continuous end degradation before cells succeed to join the ends. Extended microhomologies are not required for efficient end ligation in xrs5 cells.
PARP1 inhibition sensitizes Ku80-deficient cells to ionizing radiation
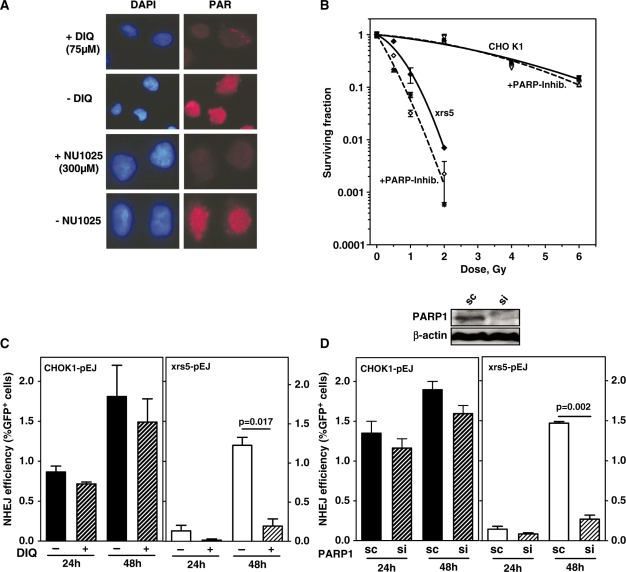
The slow repair in xrs5 cells might reflect either the insufficient classical end-joining in the absence of the critical Ku component or alternatively another genetically distinct end-joining pathway which operates slowly and inaccurately as reported recently (20,21,26,35,36). Biochemical studies and rejoining of episomal plasmids suggested that PARP1 is involved together with ligase III, XRCC1 and PNK (4,19,20,25). However, it is hitherto not known whether PARP1 is physically required for the alternative rejoining of chromosomal breaks in vivo. To address this question, we used chemical PARP inhibition as well as PARP1 depletion strategies. Two PARP inhibitors were used at non-toxic concentrations (Supplementary Figure S2A and B). Both 75 µM DIQ and 300 µM NU1025 completely abrogated IR-induced nuclear accumulation of poly(ADP)ribose (PAR) moieties (Figure 4A). However, PARP-inhibition did not modulate survival of irradiated CHOK1 cells (Figure 4B), while xrs5 cells were significantly radio-sensitized (1.9-fold). This suggests that inhibition of PARP1 compromises the residual repair capacity in Ku-deficient cells.
Figure 4.
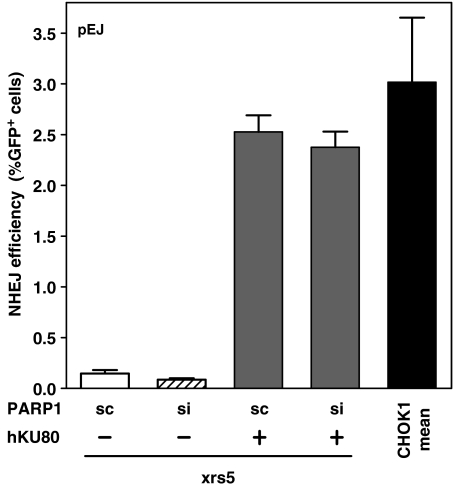
Alternative end-joining mechanism is PARP1-dependent. (A) xrs5 cells were X-irradiated (20 Gy) and immunostained after 20 min for obtaining PAR. Pretreatment with the PARP-inhibitors (DIQ or NU1025) completely abrogated nuclear PAR accumulation. (B) Cell survival of CHOK1 and xrs5 cells after X-irradiation (closed symbols, solid lines) alone or in the presence of either PARP inhibitor (open symbols, dashed lines) was carried out. An amount of 75 µM DIQ (asterisk) and 300 µM NU1025 (diamonds) led to equal radiosensitization in xrs5 cells (dashed line), but had almost no effect on CHOK1 (open triangles). The mean values of three independent experiments are shown. Data were fitted to the linear quadratic equation S/S0 = exp(–αD – βD2). The parameters obtained (α, β) were used to calculate the mean inactivation dose, Dbar (87) as a measure for the radiosensitivity. (C) NHEJ efficiency in the presence or absence of DIQ was determined as shown before (Figure 2) in CHOK1-pEJ and xrs5-pEJ clones (CHOK1-pEJ #70 and 71, and xrs5-pEJ #20 and 26). DIQ, given immediately after I-SceI transfection, significantly inhibited end-joining efficiency only in xrs5 cells. Data show the mean (±SE) of two experiments performed with two clones and three replicates each. (D) PARP1 was depleted from cells carrying pEJ by transient siRNA transfection 24 h before end-joining experiments were started as described before. (Immunoblot shows PARP1 expression 24 h after siRNA treatment.) The significant difference is indicated. sc indicates transfection with unspecific ‘scrambled’ siRNA. si indicates siRNA specifically targeting PARP1. Data show the mean (±SE) of two experiments performed with three replicates using a single clone of each strain.
PARP1 is physically required for alternative end-joining
In order to more specifically determine the impact upon NHEJ, repair of I-SceI induced DSB was followed in the presence of PARP inhibitors. DIQ only slightly reduced the end-joining efficiency in CHOK1 cells (Figure 4C). In contrast, successful end-joining was almost completely abolished in xrs5 cells at 24 and 48 h post-transfection, suggesting that the slow alternative end-joining mode requires functional PARP1. Similar results were obtained for complementary and non-complementary DSB ends (Figure 4C and Supplementary Figure S2C), indicating that the end-structure does not modulate the need for PARP1 when Ku80 is lacking.
PARP1 function not only includes rapid recruitment to damaged sites but also its subsequent release from DNA which is achieved through auto-modification by loading of (ADP)ribose-polymers (37). Exactly the latter function is inhibited by DIQ (or NU1025), raising the possibility that the inhibitors ‘freeze’ inactive PARP1 molecules in situ. Un-rejoined DNA termini may then be blocked for any further repair. In order to exclude this unspecific repressive effect, PARP1 was depleted in vivo by transient knock-down and the repair efficiencies were again measured 24 and 48 h after DSB induction. siRNA treatment 24 h before I-SceI transfection reduced cellular PARP1 content by >90% (Figure 4D and Supplementary Figure S3), which, however, did not significantly affect end-joining in wild-type cells at both time points (Figure 4D and Supplementary Figure S2C). Similar to PARP1 inhibition, its depletion in xrs5 cells virtually precluded successful end-joining as measured by lack of GFP-positive cells. Complementation of xrs5 cells with hKU80 reverted the end-joining efficiency to a level previously measured in CHOK1 (Figure 5, compare columns 3 and 5). Besides, PARP1 depletion did no longer exhibit a strong inhibitory effect. The slightly reduced end-joining efficiency (Figure 5, column 4) was found to be of the same magnitude as in wild-type cells (Figure 4D). These in vivo observations prove for the first time that PARP1 is physically needed to execute the alternative end-joining pathway in the absence of the Ku-protein.
Figure 5.
Complementing the Ku defect abrogates the PARP1-dependency of the end-joining process. Experiments were performed as described earlier. Xrs5-pEJ cells (clone #20) were co-transfected with I-SceI- and hKU80-expressing vectors (17) and analyzed for GFP+ after 24 h. Reversion of the NHEJ capability in xrs5 cells from three independent experiments was compared with the mean of CHOK1-pEJ taken from Figure 2A.
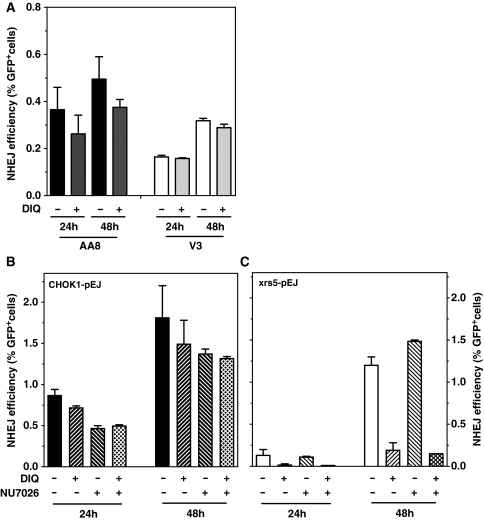
We next tested whether the switch to this alternative end-joining mode is a general consequence of lacking any of NHEJ core proteins or an exclusive response when Ku is absent. To this end we transiently transfected DNA-PK-deficient V3 hamster cells and their parentals (AA8) with I-SceI-linearized pEJ. Repair was then monitored after 24 and 48 h in the presence or absence of DIQ. Figure 6A shows that inhibition of PARP did not affect the end-joining in V3 cells suggesting that repair in this case was not PARP-mediated. Wild-type cells experience the same minor response to DIQ as seen before (Figures 6A and 4C, see ‘Discussion’ section). It could be speculated that the repair observed in mutant cells (DNA-PKcs- and PARP-independent) is brought about by XRCC4/LigIV. In line with this notion Ku-dependent recruitment of XRCC4/LigIV was observed in DNA-PKcs-deficient cells (38). Virtually, identical results were obtained with a second DNA-PKcs proficient and deficient cell pair (CHO9 and XR-C1) (data not shown), confirming that not the end-joining defect per se (i.e. lack of DNA-PKcs), but in particular the absence of Ku guides the choice towards PARP1-dependent NHEJ.
Figure 6.
PARP1-dependent end-joining is hindered by the presence of Ku but not modulated by DNA-PKcs. Cells were treated with either DIQ (75 µM) or the DNA-PK-inhibitor NU7026 (15 µM) or with both inhibitors simultaneously. (A) I-SceI was used to linearize pEJ plasmid in vitro of which 1.6 µg were then transfected using lipofectamine 2000 (Invitrogen, Germany) into DNA-PK-deficient (V3) and proficient (AA8) hamster cells. The % GFP+ cells were measured as an indicator of the repair after 24 and 48 h with or without DIQ. (B) Experiments were performed as described in Figure 4C. Inhibition of DNA-PKcs but not of PARP compromised the repair in CHOK1. (C) Inhibition of PARP but not of DNA-PK almost completely inhibited the slow repair in xrs5 cells.
It was previously shown that PARP1 may stimulate DNA-PKcs kinase activity even in the absence of Ku proteins (39). Hence, PARP1 might support DSB repair by DNA-PKcs and perhaps other downstream core components. In order to verify this possibility in xrs5 cells, we inhibited DNA-PKcs by a potent inhibitor (NU7026) which proved to inhibit classical end-joining as it efficiently sensitized CHOK1 but not xrs5 cells to IR [Supplementary Figure S4 and (40,41)]. The presence of NU7026 did not change the end-joining efficiency in xrs5 cells which fully rely on PARP1-mediated end-joining (Figures 6C and 4C). Adding both PARP and DNA-PKcs inhibitors simultaneously did not further reduce the residual end-joining activity below the effect of the PARP inhibitor alone (Figure 6C). Thus, xrs5 cells do not use PARP1 to direct the repair into ‘classical’ DNA-PK-dependent end-joining. The wild-type cells showed moderately reduced repair upon DNA-PK inhibition [Figure 6B; (29)], which could not be further reduced when combined with PARP inactivation (Figure 6B). Altogether these experiments demonstrate that PARP1 is not significantly involved in the DSB repair as long as Ku is present. In the absence of Ku, the repair is directed to a PARP1-dependent end-joining mode which, however, does not require functional DNA-PKcs.
DISCUSSION
Alternative end-joining is slow and error-prone but not dependent upon micro-homologies
Here we characterize an alternative mode of DSB end-joining which received increasing attention during the last years as a backup pathway in case classical NHEJ is not operative (19,24). Chromosomal DSB were rejoined independent of Ku and DNA-PKcs at markedly slow kinetics. However, alternative end-joining finally reaches efficiencies (fraction of GFP-positive cells in our assay) comparable to those in Ku-proficient wild-type cells (Figure 2). A slow but widely complete repair of radiation induced DSB was similarly found for hypomorphic ligase IV and DNA-PKcs deletion mutants (42,43), suggesting that the backup pathway is not restricted to enzymatic breaks. However, this delayed repair option in Ku80-deficient cells does not confer radioresistance (Figure 4) perhaps due to its error-proneness. We observed extensive sequence loss at the junctions (Figure 3), other studies described in addition frequent chromosomal rearrangements resulting from alternative end-joining (21–24,44).
Interestingly, we found that neither the efficiency nor the fidelity of NHEJ significantly depends upon the end structure. Wild-type cells rejoined cohesive and non-cohesive ends equally well. Necessary modifications were restricted to the 4-nt protrusions of the I-SceI-induced DSB ends, representing the minimum effort to render overhangs ligatable. In Ku80-deficient cells, the ends were invariably degraded, fully compatible overhangs to the same extent as non-compatible ends (Figure 3). The deletion length increased with time, confirming the protective function of the Ku-protein against nucleolytic degradation (18,31,45,46). Again, degree of end modification and overall efficiency of Ku-independent end-joining was not determined by the end structure. This result contrasts to the recent observations in yeast and hamster cells which suggested that only non-compatible ends are efficiently repaired in the absence of Ku while complementary ends absolutely require Ku (14–16,47). This discrepancy may be partly due to the different reporter constructs used or to the small number (one to three) of clones investigated that were not truly representative. The variation observed here among the 10–13 independent clones of each kind (Figure 1A and B) illustrates the potential bias inherent in those transgenic systems.
Alternative end-joining was also named microhomology-mediated end-joining (MMEJ) (14,21,33,34,48,49), suggesting that the requirement of stretches of homology sequences (up to 8 bp) is a hallmark of this pathway and distinct from classical end-joining. In contrast, we demonstrate here that Ku-independent repair does not necessarily need extensive microhomologies for efficient ligation (Figure 3C). In agreement with preceding studies including our own (7,16–18,32–34,48), wild-type cells were found to use mostly no or one overlapping base. Although 0/1-nt overlap was used efficiently by Ku-deficient cells (Figure 3C), they significantly more often employed microhomologies of ≥2 nt. However, the mean length of microhomologies was not strikingly different between CHOK1 and xrs5 cells (1.03 versus 1.8 nt, n.s.), although our system included many options for longer microhomologies of up to 7 bp (Supplementary Figure S5). Xrs5 cells simply did not use them for synapsis. On the contrary, the best assessable homologous stretch, the 4-nt complementary overhang resulting directly from I-SceI cleavage, was used only in wild-type (95% of pEJ2 junctions, see Supplementary Figure S1), but not in Ku80-deficient cells. Similar observations were also reported previously (13–16,31,34,48,50,51). We thus conclude that usage of longer microhomologies is not a distinct feature of the alternative end-joining (34,47,49). Without Ku (or other core components), however, the end degradation process may eventually expose longer microhomologies which transiently enhance the stability of the junction and hence facilitate the ligation (52–55). It could well be that a favorable position of microhomologies within reporter constructs bias repair towards MMEJ but efficient ligation is not at all dependent upon longer stretches of homologies.
Alternative end-joining requires functional PARP1
Recently, the base excision repair protein PARP1 was appreciated as a potential component of alternative end-joining (20,25) and class switch recombination (56). Importantly, chemical PARP inhibitors compromised the rejoining of episomal plasmids and chromosomal DSB in Ku- and ligase-IV-deficient but not in wild-type cells [(26); Figure 4C]. To exclude an unspecific suppressive effect upon DSB rejoining we depleted PARP1 specifically by siRNA and proved for the first time that PARP1 is absolutely required for the alternative Ku-independent end-joining in vivo (Figure 4D). The fact that other PARP isoforms were not co-depleted underscores the unique role of PARP1 for this pathway. In contrast, knock-down of ligase III, known to be involved in the alternative end-joining (20,25), but also of ligase I or IV and even combined depletion had a surprisingly moderate effect upon end-joining (57) either because low levels of those proteins are sufficient for effective NHEJ or due to the functional overlap of the three ligases.
Recruitment of PARP1 to sites of DNA damage and subsequent poly-(ADP)ribosylation of target proteins, including itself, is a rapid process starting within seconds and implicates PARP1 in the ATM-mediated DNA damage response (37,58,59). PARP-automodification has a dual role: it amplifies the DNA damage signal and at the same time leads to dissociating PARP1 from the DNA and hence to self-termination of the signal within 30 min (37). Here we find PARP1 involved in a delayed and slow repair process and propose a ‘second wave’ of PARP1 activity completely separated from initial signaling and base excision repair.
A current model would predict that the majority of DSB ends are occupied by Ku due to its high binding affinity (60) but presumably in competition with PARP1 (26). Ku virtually ‘blocks’ the entry to the PARP1-dependent end-joining pathway which could well explain that: (i) PARP-inhibition is not effective when DNA-PK is lacking but Ku is present [Figure 6C; (26)], (ii) alternative end-joining in XRCC4 mutants is much less efficient (16,18,61) than in mutants lacking Ku and (iii) in ligase IV−/− cells end-joining efficiency and radioresistance increase upon knockdown of Ku70 (26,62). In the latter case PARP1 efficiently gets access to the DSB and directs repair towards alternative end-joining (26). This competition model would on the other hand also predict that even in the presence of Ku a small fraction of DSBs is repaired by alternative end-joining, probably those DSBs that lead to longer deletions (14% in CHOK1 cells, Figure 3A and B). Consistent with this notion, PARP1 inactivation in CHOK1 cells slightly reduces end-joining efficiency (on average by 16% in Figures 4C, D and 6A). In addition DNA-PK is a target of PARP1, poly(ADP)ribosylation increases its kinase function and vice versa catalytically dead PARP suppresses DNA-PK activity (39,63). However, the significance of these mechanisms is not clear yet since neither general inhibition of DSB repair nor radiosensitization were uniformly observed in wild-type cells upon PARP inactivation [Figure 4B; (26,63–68)].
Impact of Ku and PARP1 upon regulation of DSB repair pathways
In a preceding paper we have shown that in the absence of Ku DSB repair is not only channeled into alternative end-joining but also towards single-strand annealing (SSA) and homologous recombination (17). A common theme of all three modes is the resection of primarily 5′-DNA ends though to different extents. Long-distance resection of remote homologous repeats which is required for SSA appears to be driven by ExoI, DNA2 and RecQ family helicases (69,70). Limited removal up to 100 nt is achieved through Mre11 nuclease activity in line with CtIP (69,71–73). Resulting stretches of 3′ss-DNA constitute intermediates for subsequent long-track resection but may also directly serve as substrates for alternative end-joining. Accordingly, both CtIP and Mre11 have been shown to be essential components of the alternative end-joining (14,34,61,74–76). Interestingly, PARP1 is a necessary recruitment factor of Mre11 and NBS1 and physically interacts with both proteins (59). It is thus tempting to speculate that PARP1 may promote the resection step during alternative end-joining beside its interaction with XRCC1, PNK and ligase III (4,20,25). Regulation of resection may also contribute to the impact of PARP1 upon homologous recombination. The HR promoting role (77–80) was hitherto explained by inactivating poly(ADP)ribosylation of Ku which then dissociates from DNA ends leaving space for HR and replication factors (79–81). However, it is not known whether PARP1 could also stimulate HR in the absence of Ku possibly by supporting end resection.
CONCLUSION
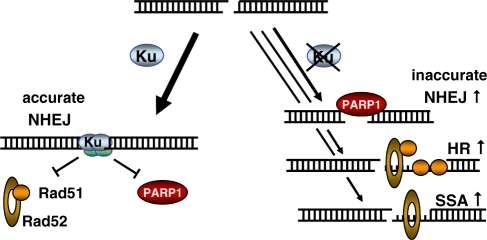
Extending our previous observations (17,18) we conclude that the majority of chromosomal DSB are repaired by classical NHEJ in a rapid and highly accurate manner (see model in Figure 7). In the absence of Ku, rather than of other core components, repair is mainly channeled to an alternative end-joining route which is slow, error-prone and completely dependent upon PARP1. The end synapsis appears to be facilitated by overlapping bases but does not require longer microhomologies. Besides alternative end-joining, Ku-deficient cells used SSA and intra-chromosomal gene conversion more frequently than wild-type cells (17,34,82). All three repair modes may compensate for the classical end-joining defect but do not effectively complement the radiosensitivity probably due to the reduced repair fidelity.
Figure 7.
Model of Ku-dependent and independent DSB repair. In the presence of Ku, DSB repair is mainly executed by accurate NHEJ. A minor fraction is normally also repaired by HR or SSA (17). In the absence of Ku, repair is shuttled to all three other pathways which likely share a common initial end resection step. The lack of Ku binding leaves space for key proteins to bind such as PARP1, Rad51 or Rad52, respectively. The majority of DSBs are still repaired by end-joining, however by an alternative PARP1-dependent mode.
Given its robustness and its error-prone character, however, PARP1-dependent end-joining can be considered to augment genetic instability and hence carcinogenesis (24). Support to this notion comes from the fact that NHEJ-deficient mice are characterized by high rate of large deletions, translocations including particularly those between the Ig heavy chain (IgH) locus and c-Myc which frequently induce lymphomas (23,56,83–86).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Federal Ministry of Education and Research (BMBF), Projekttraeger Karlsruhe (Grant No 02S8427 to JDD). Funding for open access charge: Institutional.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Löbrich for providing us with AA8 and V3 cells and M. Z. Zdzienicka, for gifting CHO9 and XR-C1 cells.
REFERENCES
- 1.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 2.Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J. 2002;21:2827–2832. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y, Schwarz K, Lieber MR. The Artemis: DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair. 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Audebert M, Salles B, Weinfeld M, Calsou P. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J. Mol. Biol. 2006;356:257–265. doi: 10.1016/j.jmb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 5.Jeggo PA, Kemp LM. X-ray-sensitive mutants of Chinese hamster ovary cell line. Isolation and cross-sensitivity to other DNA-damaging agents. Mutat. Res. 1983;112:313–327. doi: 10.1016/0167-8817(83)90026-3. [DOI] [PubMed] [Google Scholar]
- 6.Dikomey E, Dahm-Daphi J, Brammer I, Martensen R, Kaina B. Correlation between cellular radiosensitivity and non-repaired double-strand breaks studied in nine mammalian cell lines. Int. J. Radiat. Biol. 1998;73:269–278. doi: 10.1080/095530098142365. [DOI] [PubMed] [Google Scholar]
- 7.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeggo PA. DNA breakage and repair. Adv. Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 9.Dahm-Daphi J, Hubbe P, Horvath F, El-Awady RA, Bouffard KE, Powell SN, Willers H. Nonhomologous end-joining of site-specific but not of radiation-induced DNA double-strand breaks is reduced in the presence of wild-type p53. Oncogene. 2005;24:1663–1672. doi: 10.1038/sj.onc.1208396. [DOI] [PubMed] [Google Scholar]
- 10.Caldecott K, Banks G, Jeggo P. DNA double-strand break repair pathways and cellular tolerance to inhibitors of topoisomerase II. Cancer Res. 1990;50:5778–5783. [PubMed] [Google Scholar]
- 11.Jeggo PA. Studies on mammalian mutants defective in rejoining double-strand breaks in DNA. Mutat. Res. 1990;239:1–16. doi: 10.1016/0165-1110(90)90028-a. [DOI] [PubMed] [Google Scholar]
- 12.Kruger I, Rothkamm K, Lobrich M. Enhanced fidelity for rejoining radiation-induced DNA double-strand breaks in the G2 phase of Chinese hamster ovary cells. Nucleic Acids Res. 2004;32:2677–2684. doi: 10.1093/nar/gkh586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhfittig-Kulle S, Feldmann E, Odersky A, Kuliczkowska A, Goedecke W, Eggert A, Pfeiffer P. The mutagenic potential of non-homologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis. 2007;22:217–233. doi: 10.1093/mutage/gem007. [DOI] [PubMed] [Google Scholar]
- 14.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl Acad. Sci. USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Perrault AR, Takeda Y, Qin W, Wang H, Iliakis G. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 21.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 22.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 24.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 26.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brissett NC, Doherty AJ. Repairing DNA double-strand breaks by the prokaryotic non-homologous end-joining pathway. Biochem. Soc. Trans. 2009;37:539–545. doi: 10.1042/BST0370539. [DOI] [PubMed] [Google Scholar]
- 28.Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 29.Willers H, Husson J, Lee LW, Hubbe P, Gazemeier F, Powell SN, Dahm-Daphi J. Distinct mechanisms of nonhomologous end joining in the repair of site-directed chromosomal breaks with noncomplementary and complementary ends. Radiat. Res. 2006;166:567–574. doi: 10.1667/RR0524.1. [DOI] [PubMed] [Google Scholar]
- 30.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verkaik NS, Esveldt-van Lange RE, van Heemst D, Bruggenwirth HT, Hoeijmakers JH, Zdzienicka MZ, van Gent DC. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.van Heemst D, Brugmans L, Verkaik NS, van Gent DC. End-joining of blunt DNA double-strand breaks in mammalian fibroblasts is precise and requires DNA-PK and XRCC4. DNA Repair. 2004;3:43–50. doi: 10.1016/j.dnarep.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thode S, Schafer A, Pfeiffer P, Vielmetter W. A novel pathway of DNA end-to-end joining. Cell. 1990;60:921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- 36.Woodhouse BC, Dianova II, Parsons JL, Dianov GL. Poly(ADP-ribose) polymerase-1 modulates DNA repair capacity and prevents formation of DNA double strand breaks. DNA Repair. 2008;7:932–940. doi: 10.1016/j.dnarep.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 40.Losada R, Rivero MT, Slijepcevic P, Goyanes V, Fernandez JL. Effect of Wortmannin on the repair profiles of DNA double-strand breaks in the whole genome and in interstitial telomeric sequences of Chinese hamster cells. Mutat. Res. 2005;570:119–128. doi: 10.1016/j.mrfmmm.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Iwabuchi K, Hashimoto M, Matsui T, Kurihara T, Shimizu H, Adachi N, Ishiai M, Yamamoto K, Tauchi H, Takata M, et al. 53BP1 contributes to survival of cells irradiated with X-ray during G1 without Ku70 or Artemis. Genes Cells. 2006;11:935–948. doi: 10.1111/j.1365-2443.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 42.DiBiase SJ, Zeng ZC, Chen R, Hyslop T, Curran WJ, Jr, Iliakis G. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000;60:1245–1253. [PubMed] [Google Scholar]
- 43.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 44.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 46.Mimori T, Ohosone Y, Hama N, Suwa A, Akizuki M, Homma M, Griffith AJ, Hardin JA. Isolation and characterization of cDNA encoding the 80-kDa subunit protein of the human autoantigen Ku (p70/p80) recognized by autoantibodies from patients with scleroderma-polymyositis overlap syndrome. Proc. Natl Acad. Sci. USA. 1990;87:1777–1781. doi: 10.1073/pnas.87.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haber JE. Alternative endings. Proc. Natl Acad. Sci. USA. 2008;105:405–406. doi: 10.1073/pnas.0711334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katsura Y, Sasaki S, Sato M, Yamaoka K, Suzukawa K, Nagasawa T, Yokota J, Kohno T. Involvement of Ku80 in microhomology-mediated end joining for DNA double-strand breaks in vivo. DNA Repair. 2007;6:639–648. doi: 10.1016/j.dnarep.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 49.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl Acad. Sci. USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Secretan MB, Scuric Z, Oshima J, Bishop AJ, Howlett NG, Yau D, Schiestl RH. Effect of Ku86 and DNA-PKcs deficiency on non-homologous end-joining and homologous recombination using a transient transfection assay. Mutat Res. 2004;554:351–364. doi: 10.1016/j.mrfmmm.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Daza P, Reichenberger S, Gottlich B, Hagmann M, Feldmann E, Pfeiffer P. Mechanisms of nonhomologous DNA end-joining in frogs, mice and men. Biol. Chem. 1996;377:775–786. doi: 10.1515/bchm3.1996.377.12.775. [DOI] [PubMed] [Google Scholar]
- 53.Labhart P. Nonhomologous DNA end joining in cell-free systems. Eur J Biochem. 1999;265:849–861. doi: 10.1046/j.1432-1327.1999.00805.x. [DOI] [PubMed] [Google Scholar]
- 54.Roth DB, Porter TN, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol. Cell. Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 56.Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J. Exp. Med. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Windhofer F, Wu W, Iliakis G. Low levels of DNA ligases III and IV sufficient for effective NHEJ. J. Cell. Physiol. 2007;213:475–483. doi: 10.1002/jcp.21120. [DOI] [PubMed] [Google Scholar]
- 58.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 59.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 60.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 61.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adachi N, Iiizumi S, So S, Koyama H. Genetic evidence for involvement of two distinct nonhomologous end-joining pathways in repair of topoisomerase II-mediated DNA damage. Biochem. Biophys. Res. Commun. 2004;318:856–861. doi: 10.1016/j.bbrc.2004.04.099. [DOI] [PubMed] [Google Scholar]
- 63.Veuger SJ, Curtin NJ, Smith GC, Durkacz BW. Effects of novel inhibitors of poly(ADP-ribose) polymerase-1 and the DNA-dependent protein kinase on enzyme activities and DNA repair. Oncogene. 2004;23:7322–7329. doi: 10.1038/sj.onc.1207984. [DOI] [PubMed] [Google Scholar]
- 64.Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–1691. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rudat V, Bachmann N, Kupper JH, Weber KJ. Overexpression of the DNA-binding domain of poly(ADP-ribose) polymerase inhibits rejoining of ionizing radiation-induced DNA double-strand breaks. Int. J. Radiat. Biol. 2001;77:303–307. doi: 10.1080/09553000010009026. [DOI] [PubMed] [Google Scholar]
- 66.Russo AL, Kwon HC, Burgan WE, Carter D, Beam K, Weizheng X, Zhang J, Slusher BS, Chakravarti A, Tofilon PJ, et al. In vitro and in vivo radiosensitization of glioblastoma cells by the poly (ADP-ribose) polymerase inhibitor E7016. Clin. Cancer Res. 2009;15:607–612. doi: 10.1158/1078-0432.CCR-08-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–3882. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- 68.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 69.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhuang J, Jiang G, Willers H, Xia F. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J. Biol. Chem. 2009;284:30565–30573. doi: 10.1074/jbc.M109.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 76.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25:1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saberi A, Hochegger H, Szuts D, Lan L, Yasui A, Sale JE, Taniguchi Y, Murakawa Y, Zeng W, Yokomori K, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol. Cell. Biol. 2007;27:2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell. Biol. 2008;183:1203–1212. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J. Biol. Chem. 2004;279:13659–13667. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- 82.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 84.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 85.Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 86.Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett CF, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 87.Fertil B, Malaise EP. Intrinsic radiosensitivity of human cell lines is correlated with radioresponsiveness of human tumors: analysis of 101 published survival curves. Int. J. Radiat. Oncol. Biol. Phys. 1985;11:1699–1707. doi: 10.1016/0360-3016(85)90223-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.