Abstract
We used mouse hepatic chromatin enriched with an FXR antibody and chromatin immunoprecipitation-sequencing (ChIP-seq) to evaluate FXR binding on a genome-wide scale. This identified 1656 FXR-binding sites and 10% were located within 2 kb of a transcription start site which is much higher than predicted by random occurrence. A motif search uncovered a canonical nuclear receptor IR-1 site, consistent with in vitro DNA-binding studies reported previously. A separate nuclear receptor half-site for monomeric receptors such as LRH-1 was co-enriched and FXR activation of four newly identified promoters was significantly augmented by an LRH-1 expression vector in a co-transfection assay. There were 1038 genes located within 20 kb of a peak and a gene set enrichment analysis showed that genes identified by our ChIP-seq analysis are highly correlated with genes activated by an FXR-VP16 adenovirus in primary mouse hepatocytes providing functional relevance to the genome-wide binding study. Gene Ontology analysis showed FXR-binding sites close to many genes in lipid, fatty acid and steroid metabolism. Other broad gene clusters related to metabolism, transport, signaling and glycolysis were also significantly enriched. Thus, FXR may have a much wider role in cellular metabolism than previously appreciated.
INTRODUCTION
The farnesoid X nuclear receptor (FXR; NR1H4) is mainly expressed in the liver and distal small intestine and is a key regulator of enterohepatic bile acid metabolism (1). Bile acids are secreted by the liver and released into the small intestine during a meal where they aid the absorption of dietary fat and fat-soluble vitamins (2). Bile acids have been proposed as endogenous FXR agonists and are natural detergents that become toxic at high levels, which can occur when the normally tight regulation of their synthesis, transport and excretion is perturbed (3). Studies with mice fed synthetic FXR agonists have suggested that FXR plays a key role not only in cholesterol and bile acid metabolism but also in the regulation of glucose metabolism (3–5).
FXR interacts with retinoid X receptor (RXR; NR2B) as a requisite heterodimeric partner and binds to DNA elements called FXR response elements (FXREs) (6). All nuclear receptors have a highly conserved zinc finger DNA-binding domain that binds to a similar response element and individual nuclear receptors bind to either a single half-site if they bind as a monomer or to a dimeric response element composed of two half-sites with a variable orientation and spacing relative to one another (7). An in vitro DNA sites selection assay showed that FXR prefers binding to an inverted repeat of the ideal sequence 5′-AGGTCA-3′ where the monomers are separated by 1 nt (IR-1) (8). This specificity is also supported by the functional analysis of a limited number of FXR activated promoters (9).
To extend the limited information available from the relatively small set of individually characterized FXR target genes and to identify putative new targets and provide more insight into the mechanism for how FXR activates gene expression, we have evaluated FXR binding on a genome-wide scale in hepatic chromatin using a combination of chromatin immunoprecipitation (ChIP) coupled with high-throughput DNA sequencing (ChIP-seq) (10) We identified 1656 binding sites for FXR in the liver (with an estimated false discovery rate of <5%) and a motif search suggested that all contain an identifiable IR-1 site. Most of the sites are located primarily in intergenic and intronic regions but there is a significant enrichment of FXR-binding sites within 2 kb of transcription start sites (TSS) for known genes. Interestingly, an additional nuclear receptor half-site was significantly co-enriched along with the IR-1 element suggesting that FXR activates gene expression in combination with a co-binding monomeric nuclear receptor. Transient reporter studies analyzing four genes where the IR-1 and associated additional half-site are located in promoter regions shows that FXR activation was significantly augmented by including an expression construct for LRH-1 (11), a liver enriched monomeric nuclear receptor that is known to preferentially bind nuclear receptor half sites and has already been shown to augment promoter activation by LXR (12–14).
MATERIALS AND METHODS
Additional data
Additional information related to this study can be found at http://cbcl-1.ics.uci.edu/public_data/FXR/.
Chromatin immunoprecipitation followed by sequencing (ChIP-seq)
Twelve-week-old C57BL6 male mice were purchased from Jackson Laboratory and were fed a standard chow diet and allowed to adapt to a 12 h dark/12 h light cycle for 2 weeks (15). All animals were sacrificed at the end of the dark cycle and ChIP assays from liver were performed as previously described (15) with a minor modification. Chromatin was harvested and subjected to an immunoselection process, which required the use of antibodies against FXR (sc-13063; Santa Cruz Biotechnology, Santa Cruz, CA) or mouse IgG (Sigma) as a control. To prepare samples for the ChIP-seq, after isolating the ChIP-enriched DNA, gene-specific enrichment for known FXR target promoters from SHP in the FXR chromatin relative to IgG control chromatin was verified. The qPCR primers for the mouse SHP promoters were as follows: Forward, 5′gagagcctgagaccttggtg3′; Reverse 5′cgtggccttgctatcacttt3′. Approximately 20 ng of ChIP enriched DNA or control DNA was sent to Ambry Genetics (Aliso Viejo, CA) for high throughput DNA sequencing. The samples were blunt ended and adapters were ligated to the ends, according to the library preparation protocol from Illumina. Then DNA fragments with 200 ± 25 bp in length were selected for the construction of ChIP-seq DNA library. After size selection, all the resulting ChIPed DNA fragments were amplified and sequenced simultaneously using Solexa/Illumina Genome Analyzer (by Ambry Genetics).
Quantitative PCR, microarray analysis
Manual ChIP confirmation on the randomly selected putative FXR target genes from lipid metabolism category or negative control regions was by quantitative PCR (qPCR) (16). Final ChIPed and control DNA samples were analyzed in triplicate with L32 as internal control. For this assay, we used pre-designed and validated qPCR primer specific to the genomic region being interrogated.
Cell culture, transient transfection and reporter gene assays
Transfection assays were slightly modified from a previous report (14). 293T cells were maintained in high glucose Dulbecco’s Modified Eagle Medium (DMEM) (Gibco), 4.5 g/ml glucose, 0.1 mM non-essential amino acids, 100 units/ml penicillin, 100 mg/ml streptomycin and 10% fetal bovine serum (FBS). 293T cells (2 × 105 cells/well) were seeded in 24-well plates and transfected with luciferase reporter and expression (FXR and RXR) plasmids using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s protocol. A pCMV–β-gal expression construct was included in every transfection as a normalization control. After 6 h cells were treated with either DMSO or GW4064. After 24 h of treatment, cells were harvested and assayed for luciferase and β-gal activities.
Adenovirus preparation and infection
Adenovirus containing either the transactivation domain of herpes simplex virus, VP16, or mouse FXRα2 fused to VP16 were generated as previously described (17). Primary mouse hepatocytes were isolated from wild-type C57BL/6 mice and maintained in DMEM (25 mM glucose/10% fetal bovine serum) as described (18). Hepatocytes were cultured for three days before infection with adenovirus expressing VP16 or FXRα2-VP16 at a m.o.i. of 10. Infected cells were then incubated with vehicle (0.01% DMSO) or 2 µM GW4064 in DMSO for 48 h. RNA was isolated using Trizol reagent (Invitrogen, CA) and microarrays were performed as described (19).
Plasmid constructs for promoter activation assay
The promoter regions of Adfp (–826 to +287) and Pcx (–1246 to +110) were cloned by PCR amplification using mouse genomic DNA as template, followed by recombination with pDONR221™ vector (Invitrogen, Carlsbad, CA) according to Gateway Cloning technology (Invitrogen, Carlsbad, CA). The entry clone constructs were then transferred into the luciferase reporter vector pLUC-GW kindly provided by J. Imbert (Institute Paoli-Calmettes, Marseille, France). All constructs were verified by DNA sequencing. Oligo primers for the Adfp and Pcx promoter region used in PCR amplification are as follows:
Adfp 5′, tccctgaacccttatgactcc; Adfp 3′, cagaaggacgtgcaaacaga; Pcx 5′, caccctaggtgctctgcttc; Pcx 3′, gagccatacctgctctgga, Adfp 5′ with ATT sites, ggggacaagtttgtacaaaaaagcaggcttccctgaacccttatgactcc; Adfp with ATT site 3′, ggggaccactttgtacaagaaagctgggtcagaaggacgtgcaaacaga; Pcx with ATT site 5′, ggggacaagtttgtacaaaaaagcaggctcaccctaggtgctctgcttc; Pcx with ATT site 3′, ggggaccactttgtacaagaaagctgggtagagccatacctgctctgga.
Plasmid constructs containing FXR-binding sites for LRH-1 co-activation assay
The DNA fragments containing FXR-binding sites from SHP, Pcx, Rdh9 and Pemt and which also contain an additional half site near them were cloned by PCR amplification using mouse genomic DNA as template, and using primers containing sites for restriction enzymes KpnI and NheI on either side. The DNA fragments and pGL3 Luciferase Reporter Vector (Promega) were then digested with KpnI and NheI, which generated compatible ends for cloning. All constructs were verified by DNA sequencing. The luciferase activity was observed during the reporter assay after transfection of 293T cells. Primers for PCR amplifications: SHP 5′, actttgaggtccgacacacc; SHP 3′, agtggctgtgagatgcaggt; Pemt 5′, agaaagtccaggtggcttga; Pemt 3′, gccagtgtcagatggtcctt; Rdh9 5′, gcaggagccgtatgtaaagc; Rdh9 3′, gaagccaagcagagagagaga; Pcx 5′, caccctaggtgctctgcttc; Pcx 3′, gagccatacctgctctgga. Restriction enzyme sites were then added to these primer oligos by PCR amplification: KpnI, atggacggtacc; NheI, atggacgctagc.
ChIP-Seq data analysis
Preprocessing sequence data
The ChIP-seq dataset was analyzed to identify peaks which contain binding sites of FXR binding. Short reads of 39 bp were produced from Solexa/Illumina Genome Analyzer, and mapped to a reference genome by Ambry Genetics using ELAND, allowing one mismatch. Short sequence reads that mapped to simple and complex repeats or that were not unique by chance were removed from the analysis. We converted these files to BED files using the ChIP-Seq mini 2.0.1 suite (10). The BED files were used as input to downstream processing, as well as visualization in the UCSC Genome Browser (http://genome.ucsc.edu/index.html). The wiggle (WIG) files for display of our data as custom annotation tracks in UCSC Genome Browser can be downloaded from http://cbcl-1.ics.uci.edu/public_data/FXR/.
Finding peaks
To determine where the FXR bound to the genome, we looked for areas where there were significantly more enriched reads mapped in the ChIP sample than in the IgG. This was accomplished using MACS (20) with the parameters of mfold 10, bandwidth 300 bp, P-value 1 × 10−5 and false discovery rate (FDR) 5%.
Distance to IR-1 sites from the summit of each peak
MACS provides a summit for every peak, which can be regarded as the center of the peak. It is where there is the maximum number of overlapping reads, and is the most likely location of the true binding site. For each peak with an IR-1 site, we determined the distance from the best IR-1 site to this summit. If they overlapped, we score the distance as zero. To give a sense of the enrichment, we placed an arbitrarily located site of the same length in each peak, determined the distance to the summit, and plotted the results on the same histogram.
Distance from peak to transcription start sites (TSSs)
For each peak, the distance from the peak to the nearest transcription start site was determined, and plotted. The TSSs were taken from a RefSeq file obtained from NCBI. The background was determined by placing peaks at random locations on the genome and by determining distances to TSS.
Motif analysis
DNA sequences were retrieved using Galaxy (http://main.g2.bx.psu.edu) and used for motif search using MEME (21). MEME represents motifs as position-dependent letter-probability matrices (PWM). The PWM was used to find a score for any 13-bp sequence; each letter in the sequence has a likelihood given in the PWM, these are summed to find a score for the sequence, with a higher score meaning it is more likely to be the motif in question. We used the PWM to find scores for every position along an entire chromosome (excepting coding and repeat regions), and found the average score and standard deviation. Then when a new sequence was tested, we obtained its score from the PWM, subtracted the average, and divided by the standard deviation. This provided us a z-score for any sequence, which was converted into a P-value via a standard normal curve.
Annotation of genes and gene ontology (GO) analysis
All FXR-binding sites were assigned to nearest genes based on the Mus musculus NCBI m37 genome assembly (mm9; July 2007). GO analysis of FXR target genes was conducted by using the NIH Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (22). This analysis was used to classify the nearest gene list into functionally related gene groups by using ‘PANTHER Biological Process’ term.
Kolmogorov–Smirnov analysis
The obtained ChIP-seq data was compared with expression microarray data by using a Kolmogorov–Smirnov (KS) plot, a modified method of gene set enrichment analysis (GSEA) (23). The KS plot tests the null hypothesis that the ranks of the genes identified by ChIP-seq is uniformly distributed throughout the FXR expression microarray. A KS plot was obtained by calculating the running sum statistics for our ChIP-seq gene set to observe enrichment in the ranked gene list from expression microarray data.
Co-regulator analysis
To find the binding sites for the co-regulators near the FXR-binding sites, we masked all the FXR IR-1sites (P < 0.001) in the peak sequences with ‘N’, and scanned the masked region ±150 bp for co-regulator binding sites using MEME. We also applied an enumeration-based method, k-mer analysis, to the sequences to search for the co-regulator motifs. For the k-mer analysis, we did each motif length separately, from 6 to 15 bp. We counted every occurance of each k-mer in the peak regions, as well as in a background sequence (1.5 MB of promoter sequence), and calculated z-scores as the statistical significance for its enrichment in the peak sequences compared to a background dataset. The PWM for the motif was used to compare against JASPAR database using STAMP (24).
RESULTS
Identification of novel FXR-binding sites
Hepatic chromatin from 12 C57BL/6 mice was pooled and processed for ChIP with an antibody to FXR or a control IgG as described in ‘Materials and methods’ section and previously (25). The quality of the chromatin and specificity of the antibody were confirmed by comparative ChIP analysis using a known FXR-binding site in the SHP promoter (12,13) with chromatin from WT or FXR-knockout (FXR-KO) mice (Supplementary Figure S1). ChIP using the WT chromatin resulted in a significant enrichment for FXR binding to the SHP promoter whereas the signal was significantly attenuated using FXR-KO chromatin.
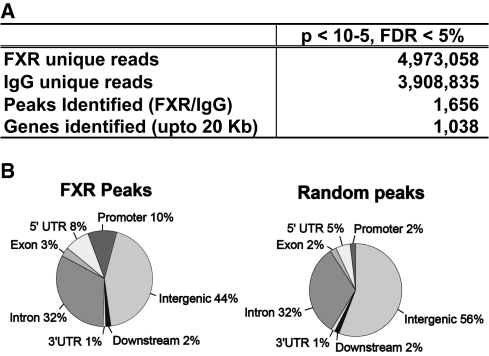
Next, we subjected the ChIPed DNA from WT chromatin to ChIP-seq analysis using the Solexa/Illumina Genome Analyzer. This ChIP-seq method generated a relevant FXR transcription factor binding dataset that contained 4–5 million individual 39-nt sequence reads produced in each run (Figure 1A). The high numbers contribute to high sensitivity and signal-to-noise ratios, and to relative comprehensiveness for the mouse genome (10). To find peaks bound by FXR, we used Model-based Analysis of ChIP-seq (MACS), which was designed to analyze data generated by short read sequencers such as from the Solexa/Illumina Genome Analyzer (20) to first estimate peak size and location (Supplementary Figure S2). Then, using P-value and FDR cutoffs of ≤1 × 10−5 and ≤5% respectively, we identified 1656 genomic sites occupied by FXR (Figure 1A, http://cbcl-1.ics.uci.edu/public_data/FXR). The distribution of FXR-binding regions were predominantly in intergenic regions (44%), and introns (32%) (Figure 1B) with 10% also located within 2-kb 5′ of a transcription start site (TSS) for a known gene. In contrast, when the genomic location for randomly generated peaks of similar size was estimated, only 2% were localized to within 2 kb of a TSS. Thus, the 10% figure for sites within 2 kb of a TSS indicates there is non-random association of FXR-binding sites (P < 0.005) in close proximity to TSS regions (Figure 1B).
Figure 1.
ChIP-seq analysis for FXR binding to DNA in hepatic chromatin. (A) Summary of ChIP-seq analysis by MACS. Given mfold 10 and sonication size (bw) 300 bp, MACS searched 2bw window area across the genome to find genomic peaks with tags more than mfold enriched relative to a random tag genome distribution. The results were obtained using the parameters of P-value cutoff 1×10−5 and FDR 5%. (B) Mapping of FXR-binding regions on genome-wide scale relative to RefSeq mouse genes. The ‘promoter’ and ‘downstream’ are defined as 2 kb of 5′ or 3′ flanking regions. Intergenic region refers to all locations other than ‘promoter’, ‘5′ UTR’, ‘exon’, ‘intron’, ‘3′UTR’ or ‘downstream’.
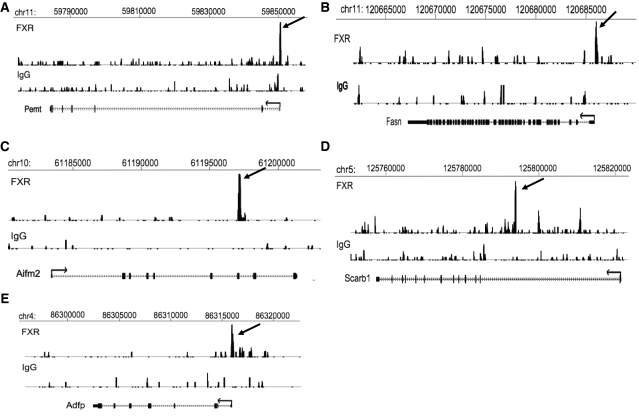
The sequence reads were aligned as a track onto the mouse genome using the University of California at Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu/index.html), and visual inspection of several sites confirmed that the peaks identified by MACS correspond to sites of over-represented sequence tabs. Several examples are shown in Figure 2.
Figure 2.
Representative view of a ChIP-seq peak. The novel FXR binding sites, mapped onto University of California at Santa Cruz (UCSC) genome browser, were identified in several genes presented here. Shown are chromosomal locations according to the July 2007 Mouse Genome Assembly (mm9). Gene associated peaks are noted by the arrows. (A) Pemt (phosphatidylethanolamine N-methyltransferase). (B) Fasn (fatty acid synthase). (C) Aifm2 (apoptosis-inducing factor 2, mitochondrian). (D) Scarb1 (scavenger receptor B-1). (E) Adfp (adipose differentiation related protein).
Motif analysis for FXR binding
FXR-binding specificity has been analyzed in vitro using a binding site enrichment procedure where one-half site was fixed as a nuclear receptor consensus site and random DNA was analyzed in the second half site. Additionally, there have been functional studies of a relatively small set of well characterized FXR target genes. Together, these analyses suggest that the preferred FXR site is an ‘IR-1′ element which is composed of two half sites of the canonical nuclear receptor half site consensus sequence 5′-AGGTCA-3′ oriented in an inverted repeat orientation and separated by a single nucleotide (8,9).
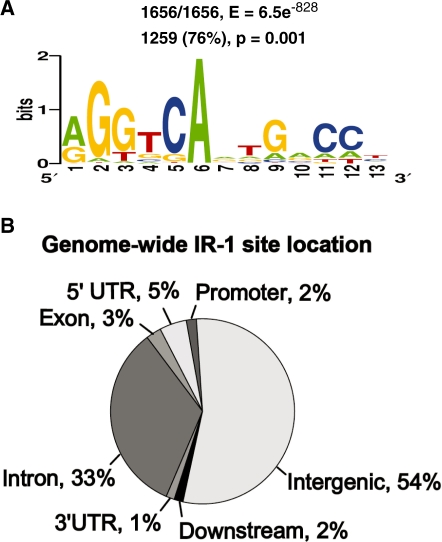
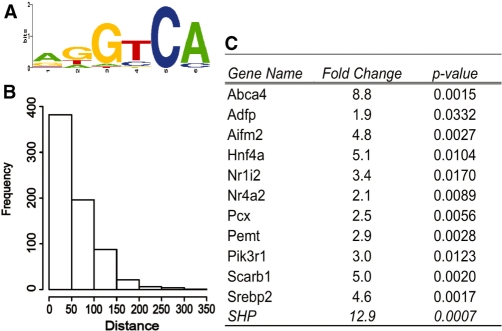
To determine how well these prior studies are predictive for FXR binding on a genome-wide scale, we used the motif finding program MEME (21) to search for enriched motifs in the peaks from our ChIP-seq data set. The highest scoring motif in the analysis was an IR-1 (E-value = 6.5e−828) and all 1656 FXR peaks contained the IR-1 site based on the MEME analysis. The position weight matrix (PWM) for the IR-1 from the MEME analysis was used to scan all our FXR peaks again using a more stringent cutoff (P < 0.001). Using this stringent criterion, an IR-1 was present in 76% (1259/1656, P < 0.001) of the 1656 peaks (Figure 3A). This indicates that our genome-wide analysis of in vivo binding sites is consistent with the IR-1 as the preferred cis-acting element for binding of FXR. It is interesting that the sequence of one-half site is much more strongly enriched than the other. This is similar to the genome-wide binding analyses for PPAR–γ/RXR as well (26,27).
Figure 3.
Motif analysis. (A) Consensus FXR-binding motif Weblogo found within the peaks identified by ChIP-seq using MEME program. The IR-1 motif was found in 76% of the FXR-binding sites when PWM from MEME analysis was used. (B) Genome-wide location of IR-1 sites around mouse genome. Its PWM was obtained by using MEME.
In the MACS analysis of the short sequence reads, a summit for every peak is identified based on combining reads matching to each strand and the summit is defined as the midpoint for the overlapping reads. Theoretically, this is the most likely location of the actual site of FXR–DNA interaction. We calculated the distance from the best IR-1 site in each IR-1 containing peak to the corresponding peak summit. By this analysis, the IR-1 sites were significantly closer to the peak-summits relative to randomly placed motifs, confirming the high accuracy of the ChIP-seq peak mapping technique and providing more confidence that the IR-1 is actually the site of recognition for FXR (Supplementary Figure S3). Most peaks contain one IR-1 element but a significant fraction contains more than one (Supplementary Figure S4). Interestingly, there are over 1.7 million predicted IR-1 sites in the mouse genome with P < 0.001 using the PWM calculated from our data set (Figure 3B), however there are only 1656 (0.09%) that are occupied by FXR in liver chromatin using our stringent cutoff.
Genes located close to FXR binding in liver
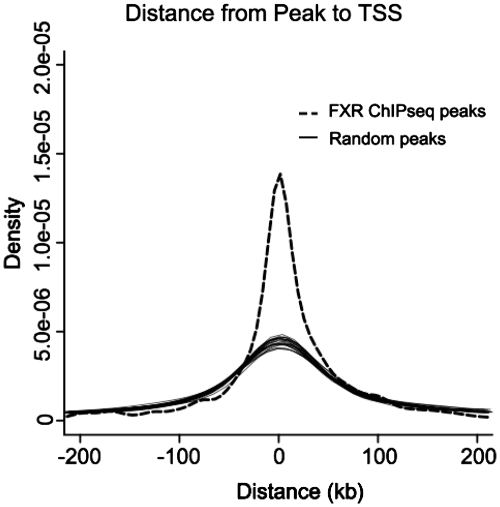
To begin to examine the functional significance of FXR binding on a whole genome scale, the nearest gene was determined for each FXR peak. Then, we examined the distance from the center of each peak to the transcription start site (TSS) of the nearest gene. The FXR-binding peaks were enriched around the TSS compared to a set of randomly generated motifs of similar length (Figure 4).
Figure 4.
Distance from the center of each peak identified as FXR-binding site to the transcription start site (TSS) of the nearest gene. The TSSs were taken from NCBI RefSeq database. Each black line represents a separate set of random sequences of similar length.
There were 1038 genes located within 20 kb of a peak and we analyzed this list using the DAVID Gene Ontology (GO) resource (http://david.abcc.ncifcrf.gov/) and grouped them into enriched broad categories using PANTHER (22). This analysis showed there was a strong enrichment for genes in metabolic processes, and the most significantly enriched genes were associated with metabolism of lipids including fatty acids and cholesterol (Table 1).
Table 1.
Summary of DAVID GO analysis of nearest genes to FXR-binding regions
| Top GO Categories of nearest genes to peaks | |||
|---|---|---|---|
| PANTHER BIOLOGICAL PROCESS | Count | P–value | FDR |
| BP00019: Lipid, fatty acid and steroid metabolism | 58 | 5.48E-08 | 6.97265E-05 |
| BP00036: DNA repair | 86 | 2.46E-07 | 3.12E-04 |
| BP00143: Cation transport | 164 | 1.45E-06 | 1.84E-03 |
| BP00033: Pyrimidine metabolism | 26 | 3.54E-05 | 4.50E-02 |
| BP00141: Transport | 71 | 5.77E-05 | 7.33E-02 |
| BP00008: Tricarboxylic acid pathway | 17 | 1.35E-04 | 0.172142101 |
| BP00104: G-protein mediated signaling | 144 | 1.40E-04 | 0.17745439 |
| BP00286: Cell structure | 137 | 1.44E-04 | 0.182437201 |
| BP00071: Proteolysis | 198 | 2.22E-04 | 0.282634364 |
| BP00276: General vesicle transport | 49 | 3.36E-04 | 0.426752545 |
| BP00063: Protein modification | 115 | 3.66E-04 | 0.465131227 |
| BP00153: Complement-mediated immunity | 25 | 8.04E-04 | 1.017828781 |
| BP00076: Electron transport | 41 | 2.99E-03 | 3.743358089 |
| BP00014: Amino acid biosynthesis | 25 | 3.07E-03 | 3.837211045 |
| BP00013: Amino acid metabolism | 18 | 3.10E-03 | 3.877861993 |
| BP00044: mRNA transcription regulation | 336 | 3.66E-03 | 4.561346276 |
| BP00142: Ion transport | 87 | 7.08E-03 | 8.643459039 |
| BP00005: Glycolysis | 18 | 7.58E-03 | 9.221152371 |
| BP00103: Cell surface receptor mediated signal transduction | 86 | 8.51E-03 | 10.29998477 |
| BP00193: Developmental processes | 78 | 1.11E-02 | 13.186538 |
| BP00298: Glycogen metabolism | 18 | 1.92E-02 | 21.87988309 |
A total of 1038 genes for peaks located within 20 kb of a gene were used to group into enriched functionally important categories using PANTHER.
Validation of FXR-binding peaks
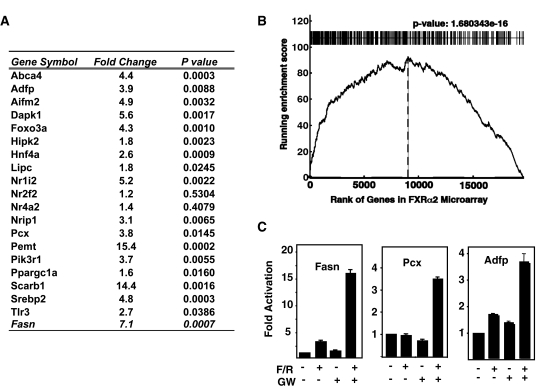
We randomly picked nineteen gene-associated peaks for manual site confirmation with specific primers and quantitative PCR (qPCR). This analysis demonstrated that 17 exhibited at least 1.5-fold enrichment relative to the IgG samples corresponding to an 89% rate of validation (Figure 5A). We also picked PCR validated primers for 11 other genomic target sites that were negative for peak assignment and none of these showed any enrichment in the FXR antibody enriched chromatin (Supplementary Figure S5).
Figure 5.
Peak validation. (A) Manual ChIP confirmation for ChIPed liver DNA by qPCR. Nineteen FXR-binding peaks were randomly selected for validation by gene-specific ChIP qPCR. Fold Change is the fold increase for the signal from DNA enriched by FXR antibody relative to a control IgG. Fasn was used as a positive control. Data were normalized to the housekeeping gene L32. (B) KS plot. The gene list for the ChIP-seq peaks that were located within 20 kb of a known gene was compared for their correlation to a set of genes that were activated by infection of primary mouse hepatocytes with a recombinant adenovirus expressing the constitutive FXRα2-VP16 hybrid protein as described in the text. Genes in the expression microarray were ranked by fold difference (x-axis) and the graph plots the running enrichment score. (C) Promoter activation assay. Luciferase reporter plasmids with promoters from genes from our ChIP-seq dataset were constructed. Pcx (pyruvate carboxylase) and Adfp (adipose differentiation-related protein) were previously unknown to be FXR target genes, and contained putative FXR-binding sites in their proximal promoters and tested for FXR activation by transient transfection. Expression of the reporters was stimulated by GW4064 treatment in cells co-transfected with FXR and RXR expression vectors. Fasn (fatty acid synthase) was used as a positive control.
Correlation between FXR binding and FXR-dependent gene regulation
Next, we analyzed the 1038 genes that were located within 20 kb of an FXR peak by a gene set enrichment analysis (GSEA) using the modified KS test (23). In this analysis, the FXR site proximal genes were analyzed for their distribution in a mRNA microarray expression set where the genes were rank-ordered for differential expression in primary hepaocytes infected with a control adenovirus or a recombinant virus that expresses a constutively active FXR-VP16 fusion protein. The analysis showed a highly significant running enrichment score because the genes identified by ChIP-seq were preferentially located toward the top of the differentially expressed gene list (Figure 5B, P = 1.68e−16). Thus, it is highly likely that the ChIP-seq identified sites correspond to functional sites of FXR action.
To more directly validate the functionality of our FXR-binding sites from ChIP-seq, we analyzed Adfp (adipose differentiation-related protein, −826 to+287) and Pcx (pyruvate carboxylase, −1246 to +110), two genes from our data set that were previously unknown to be FXR responsive, and that contained putative FXR-binding sites in their proximal promoters, by making luciferase reporter constructs that were then analyzed for FXR activation by transient transfection (Figure 5C). When co-transfected with FXRα and RXRα expression vectors, activity of both Pcx and Adfp promoters was stimulated by a combination of FXR/RXR plus the synthetic FXR agonist GW4064 (Figure 5C). This confirms that the promoters for two newly identified putative FXR target genes are directly stimulated by FXR.
Co-regulatory DNA-binding partners of FXR
To determine if there were additional transcription factor-binding sites that might be preferrentially co-enriched with the FXR peaks, we masked the IR-1 element and re-searched the sequence around each FXR peak summit. This revealed that an additional nuclear receptor half-site was present in 896 peaks (71% of 1259, P < 0.001) of the FXR-binding peaks containing IR-1 sites (Figure 6A). Because the sequence of the half site is contained within all of the IR-1 sites we were concerned that the co-enriched half sites might represent ‘weak’ IR-1 elements that failed to reach the P < 0.001 cut off value. However, when we analyzed this by a sequence replacement method we estimated that at least 80% represent true half-sites (Supplementary Figure S6). Interestingly, the IR-1 FXR peak and the additional half-site are located relatively close together with most having the two elements within 50 bases of each other (Figure 6B).
Figure 6.
Co-regulator analysis. (A) Co-regulatory DNA-binding motif identified along with FXR-binding motif. The IR-1 elements in each FXR peak were masked and then FXR-binding peaks were searched for motifs located within 150 bp on either side of each peak. (B) Distance from the best IR-1 site to midpoint of closest additional NR half site. (C) qPCR analysis of 12 peaks for LRH-1 binding. Results are presented as fold change from control as in Supplementary Figure 1.
Monomeric nuclear receptors bind to receptor half-sites and the liver receptor homologue (LRH-1) is an abundant hepatic protein of this class. Additionally, LRH-1 functionally interacts with LXR to activate Cyp7A1 and Fasn promoters in the mouse (12–14).
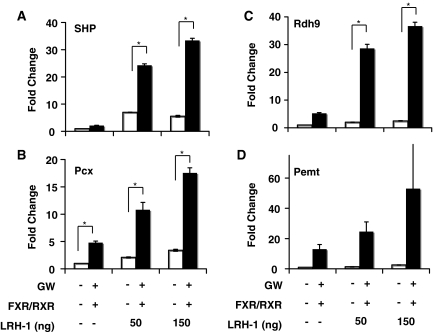
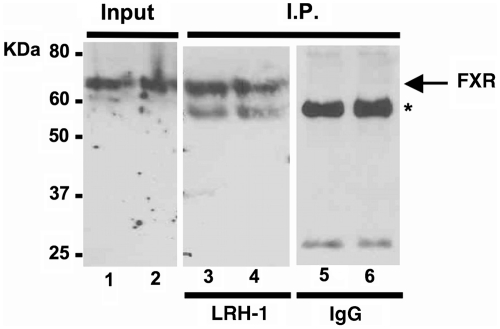
To analyze whether FXR and LRH-1 might also function together, we used three complementary approaches. First, we performed manual ChIP studies for LRH-1 binding to several of the gene promoters predicted to share FXR and LRH-1 sites (Figure 6C). This analysis revealed that LRH-1 also binds close to all of these FXR peak regions. Next, we chose four genes that contained both an FXR-binding peak and also an additional half-site within their proximal promoters. These included Pcx, Rdh9, Pemt and SHP. We prepared luciferase reporter plasmids for each and analyzed their responsiveness to the combination of FXR and LRH-1 in reporter assays. The analysis in Figure 7 shows that each promoter was activated by FXR and the inclusion of the LRH-1 expression vector significantly enhanced the FXR responsiveness on all four promoters in a dose-dependent manner. Lastly, we used a co-immunoprecitipation analysis to evaluate whether the two proteins might directly interact with each other. Liver chromatin was incubated with a control IgG or an antibody to LRH-1 followed by an immunoblotting analysis with an antibody to FXR. The results in Figure 8 show that LRH-1 was specifically precipitated in a complex by the FXR antibody.
Figure 7.
Co-transfection assay for FXR-LRH-1 activation. 293T cells were transfected with luciferase reporter plasmids indicated with or without expression vectors pCMX-FXR2 (100 ng/well), pCMX-RXR (100 ng/well), and LRH-1. After 6 hr of transfection, cells were treated with DMSO or synthetic FXR agonist GW4064 as indicated and cells were harvested after 24 h. Luciferase and -gal assays were then performed on cell extracts. The data are representative of at least three experiments. In the experiment shown, SHP, Pcx, and Rdh9 were analyzed in triplicate and * indicates p < 0.05. In this experiment, Pemt was analyzed in duplicate only. (A) SHP (small heterodimer partner). (B) Pcx (pyruvate carboxylase). (C) Rdh9 (retinol dehydrogenase 9). (D) Pemt (phosphatidylethanolamine N-methyltransferase).
Figure 8.
FXR and LRH-1 proteins interact with each other. Hepatic chromatin was subjected to immunoprecipitation with a LRH-1 antibody (lanes 3 and 4) or control IgG (lanes 5 and 6) and the precipitated material was analyzed by immunoblotting with an antibody to FXR. Duplicate samples were analyzed and the positions for FXR in the starting material is also shown. Input is shown in lanes 1 and 2. The asterisk marks the migration of the IgG heavy chain.
DISCUSSION
In this study, we present the genome-wide profiling of FXR-binding sites in mouse liver chromatin. This ChIP-seq analysis revealed 1656 FXR genomic-binding sites with a high degree of confidence (P < 1 × 10−5, FDR < 5%). Most of the identified FXR-binding sites are located in distal intergenic regions (44%) or introns (32%), with fewer sites localizing to more proximal promoter regions (10%; Figure 1B). The high distribution of binding sites within intergenic and intron regions are consistent with similar reports for other nuclear receptor transcription factors, including PPARγ (26,27), estrogen receptor α (28), and the androgen receptor (29).
However, it should be emphasized that although only 10% of the binding sites were localized to within 2 kb 5′ of a TSS, this is significantly higher than expected based on random localization (Figure 1B). Thus, even though the FXR-binding sites show a broad genome-wide distribution, the 10% indicates significant preference for promoter proximity as well. Additionally, even though most sites were located intragenically or within introns, they do localize close to known TSS (Figure 4).
While there is evidence for agonist dependent changes in binding of some hormone receptors to DNA such as estrogen receptor (ER) in cultured cell models (30), the influences of agonist binding on DNA occupancy for nuclear receptors in vivo in general and for those where endogenous metabolically derived compounds function as agonists in particular is complicated and not clearly understood. In the course of our studies, we first compared the genome-wide association of FXR in livers of a control group fed normal chow versus a group fed chow supplemented with GW4064, currently the synthetic agonist of choice for FXR. While there was a mild induction of gene expression for a handful of known FXR target genes in this analysis, there was no statistically significant difference in genome-wide binding of FXR revealed by this comparison (data not shown). Because the GW4064 agonist is not very potent at stimulating endogenous target genes in vivo, it is not clear if the lack of difference in DNA binding is meaningful. This is an important area of study that we will investigate in the future when more potent in vivo FXR agonists become available.
Our analysis showed that 76% of the peaks contain a stringently identified IR-1 element (P < 0.001; Figure 3A). This discovery both confirms and extends the data from in vitro DNA-binding site selection and the small number of individual gene analyses that have been reported (9). Additionally, the Weblogo obtained from the position weight matrix emphasizes that the bases in one of the half-sites are highly preferred whereas there is relaxed flexibility and less preference observed for the second half site. A similar half-site asymmetry preference was also revealed in the Weblogo for PPARγ/RXR (26,27) where a conserved DR-1 site showed a higher preference for bases in the 3′ half site, which is known to be specifically occupied by the RXR monomer. An explanation for this was likely revealed by the crystal structure for the PPARγ/RXR dimer bound to a DR-1 site (31). The structure revealed that the zinc finger region in the RXR DNA-binding domain makes more half site-specific base contacts compared to the zinc-finger region of PPARγ.
Because the FXR recognition site is a palindrome, the rotational symmetry makes it impossible to assign a specific monomer to each half-site without any more information. However, based on the information above, it is likely that the more highly conserved half site of the IR-1 is bound by RXR.
It is interesting that there are over 1.7 million sites in the mouse genome that match our IR-1 PWM with P < 0.001. The fact that only 1656 (0.09%) were occupied by FXR in our analysis indicates that other local genomic features such as hepatic nucleosome positioning and epigenetic markers that alter chromatin architecture and genomic access influence FXR site occupancy. Additionally, co-occupancy by neighboring DNA-binding factors like LRH-1 likely play a significant role in addition to the primary sequence of the IR-1 motif in defining where FXR is localized in hepatic chromatin.
We compared the list of genes within 20 kb of a FXR peak to a list of genes that were rank-ordered by significance for differential mRNA expression in primary hepatocytes infected with an adenovirus that expresses a constitutively active FXRα–VP16 hybrid protein relative to a control adenovirus. This gene set enrichment analysis (GSEA) was displayed by a KS plot (23) which tests for how well the two data sets correlate with each other. This analysis showed a high degree of correlation between FXR binding and FXR-dependent gene activation (Figure 5B; P = 1.68e−16). Thus, the FXR-binding peaks likely represent functional FXR response elements. It should be noted that the FXR–VP16 fusion protein would activate through genomic sites where wild-type FXR might repress gene expression and there have been reports that FXR binding may repress gene expression (32). Thus, our GSEA provides a good correlation between FXR binding by ChIP-seq and target gene identification but it does not allow us to differentiate between genes that are normally activated or repressed directly through FXR response elements.
After first masking the IR-1 sites we repeated the de novo motif search around the peak summits (±150 bp) to search for putative enriched FXR co-regulatory DNA-binding partners. This analysis revealed that an additional nuclear half site, 5′-AGGTCA-3′, was present close to 71% of the IR-1 containing FXR peaks (Figure 6). It is unlikely that FXR interacts directly with this additional half site because this would have been revealed as a peak summit when the sequence reads were mapped to the genome. We also analyzed directly whether the half-sites might correspond to ‘weak’ IR-1 sites that fell below our statistical threshold. This analysis revealed that at least 80% of the identified half-sites are true half sites (Supplementary Figure S5). It should be noted that FXR has been shown to possibly interact with DNA as a monomer (33) and we cannot rule out with certainty that some of the half-sites might bind a monomeric form of FXR.
Because monomeric nuclear receptors bind to isolated half-sites and LRH-1 is a liver enriched monomeric nuclear receptor, we proposed that LRH-1 would be a good candidate for an FXR co-regulatory protein. Because most FXR-binding sites were localized to introns or intergenic regions it would be difficult to construct reporter genes that retain the native spacing for the proximal promoter together with the intronic/intergenic FXR/RXR response element. Therefore, to analyze LRH-1 as a putative FXR co-regulatory partner we performed gene specific ChIP analysis of 12 peaks predicted to contain LRH-1-binding sites (Figure 6C) and we chose four genes where the FXR/RXR and associated extra half-sites were located within the proximal promoter region for promoter activation assays. In this analysis, all four promoter–reporters confirmed that FXR/RXR activation was significantly enhanced by the addition of an LRH-1 expression vector in the transfection assay (Figure 7). We also showed that FXR and LRH-1 associate with each other directly by co-immunoprecipitation (Figure 8). These three approaches strongly support our hypothesis that FXR and LRH-1 function together to activate hepatic gene expression.
It is interesting to note that there are additional monomeric nuclear receptors that are expressed in the liver such as the reverbs and RORs which are regulated by heme and sterol agonists respectively (34,35) and SF-1 which is very similar to LRH-1 (36). Reverbs and RORs are expressed in reciprocal diurnal patterns in the liver. It will be interesting to analyze these additional monomeric nuclear receptors in future studies for their potential roles as FXR co-regulatory factors that might be associated with a diurnally regulated pattern of FXR activity.
In a GO analysis, the broad category of lipid and fatty acid metabolism was the most significant gene cluster linked to the FXR peak associated genes as expected. However, genes of glycolysis also showed a significant enrichment as well. This is interesting because FXR has been shown to be involved in modulating glucose metabolism associatead with diabetes in mice (3–5). In addition, a number of other broad-based gene clusters related to metabolism, transport and signaling were also significantly enriched. These latter results suggest that FXR may have a much wider role in regulating cellular metabolism than has been proposed to date. Indeed, identification of new FXR targets within these categories may explain the pleiotropic effects on metabolism and cellular physiology noted in both animal studies and patients with bile acid disorders (1,37).
While we were completing the analyses for our study, a comparative evaluation of the genome-wide pattern of FXR binding to hepatic and intestinal chromatin was reported (38). Overall, the binding results are very comparable but the two approaches have some differences in that the synthetic FXR agonist GW4064 was added a few hours before sacrifice by Thomas et al and the sequence mapping and analysis were performed by different methods. Interestingly, Thomas et al also identified an IR-1 element with a co-enriched additional nuclear receptor half site close to the FXR peaks. As mentioned above, we did not observe a consistent difference in FXR binding by the addition of the GW4064 agonist. Thus, we were not surprised that overall the results are similar to ours. In addition to defining the genomic sites for FXR binding, our study goes further and also provides functional evidence that FXR is likely to affect expression of the genes associated with the peaks. We also provide evidence that LRH-1 is an important monomeric nuclear receptor partner for FXR that binds to the co-enriched nuclear receptor half site to co-activate gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grants to T.O. (DK71021) and P.E. (HL68445 and a National Science Foundation grant to X. X. (DBI-0846218); National Institutes of Health/NLM bioinformatics training grant (T1507443) to H.K.C. and A.I. Funding for open access charge: National Institutes of Health (grant DK71021).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of our laboratories for suggestions and comments on this project.
REFERENCES
- 1.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 2.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Edwards PA. FXR signaling in metabolic disease. FEBS Lett. 2008;582:10–18. doi: 10.1016/j.febslet.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Claudel T, Staels B, Kuipers F. The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler. Thromb. Vasc. Biol. 2005;25:2020–2030. doi: 10.1161/01.ATV.0000178994.21828.a7. [DOI] [PubMed] [Google Scholar]
- 5.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlman T, Noonan DJ, Burka LT, McMorris T, Lamph WW, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 7.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J. Biol. Chem. 2000;275:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- 9.Edwards PA, Kast HR, Anisfeld AM. BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J. Lipid Res. 2002;43:2–12. [PubMed] [Google Scholar]
- 10.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 11.Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc. Natl Acad. Sci. USA. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 13.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 14.Matsukuma KE, Wang L, Bennett MK, Osborne TF. A key role for orphan nuclear receptor liver receptor homologue-1 in activation of fatty acid synthase promoter by liver X receptor. J. Biol. Chem. 2007;282:20164–20171. doi: 10.1074/jbc.M702895200. [DOI] [PubMed] [Google Scholar]
- 15.Bennett MK, Seo Y-K, Datta S, Shin D-J, Osborne TF. Selective binding of SREBP isoforms and co-regulatory proteins to promoters for lipid metabolic genes in liver. J. Biol. Chem. 2008;283:15628–15637. doi: 10.1074/jbc.M800391200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo YK, Chong HK, Infante AM, Im SS, Xie X, Osborne TF. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc. Natl Acad. Sci. USA. 2009;106:13765–13769. doi: 10.1073/pnas.0904246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc. Natl Acad. Sci. USA. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbert ML, Zhang Y, Lee FY, Edwards PA. Regulation of hepatic Insig-2 by the farnesoid X receptor. Mol. Endocrinol. 2007;21:1359–1369. doi: 10.1210/me.2007-0089. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey TL. Discovering novel sequence motifs with MEME. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0204s00. Chapter 2, Unit 2.4. [DOI] [PubMed] [Google Scholar]
- 22.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsukuma KE, Bennett MK, Huang J, Wang L, Gil G, Osborne TF. Coordinated control of bile acids and lipogenesis through FXR-dependent regulation of fatty acid synthase. J. Lipid Res. 2006;47:2754–2761. doi: 10.1194/jlr.M600342-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 31.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR-alpha nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claudel T, Inoue Y, Barbier O, Duran-Sandoval D, Kosykh V, Fruchart J, Fruchart JC, Gonzalez FJ, Staels B. Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology. 2003;125:544–555. doi: 10.1016/s0016-5085(03)00896-5. [DOI] [PubMed] [Google Scholar]
- 33.Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, Fruchart JC, Dallongeville J, Hum DW, Kuipers F, et al. Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J. Clin. Invest. 2002;109:961–971. doi: 10.1172/JCI14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat. Struct. Mol. Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Kumar N, Solt LA, Richardson TI, Helvering LM, Crumbley C, Garcia-Ordonez RA, Stayrook KR, Zhang X, Novick S, et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. J. Biol. Chem. 2010;285:5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catalano S, Malivindi R, Giordano C, Gu G, Panza S, Bonofiglio D, Lanzino M, Sisci D, Panno ML, Ando S. Farnesoid X receptor, through the binding with steroidogenic factor 1-responsive element, inhibits aromatase expression in tumor Leydig cells. J. Biol. Chem. 285:5581–5593. doi: 10.1074/jbc.M109.052670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang YD, Chen WD, Moore DD, Huang W. FXR: a metabolic regulator and cell protector. Cell Res. 2008;18:1087–1095. doi: 10.1038/cr.2008.289. [DOI] [PubMed] [Google Scholar]
- 38.Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, Guo GL. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2009;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.