Abstract
Genomic sequence comparisons between individuals are usually restricted to the analysis of single nucleotide polymorphisms (SNPs). While the interrogation of SNPs is efficient, they are not the only form of divergence between genomes. In this report, we expand the scope of polymorphism detection by investigating the occurrence of double nucleotide polymorphisms (DNPs) and triple nucleotide polymorphisms (TNPs), in which two or three consecutive nucleotides are altered compared to the reference sequence. We have found such DNPs and TNPs throughout two complete genomes and eight exomes. Within exons, these novel polymorphisms are over-represented amongst protein-altering variants; nearly all DNPs and TNPs result in a change in amino acid sequence and, in some cases, two adjacent amino acids are changed. DNPs and TNPs represent a potentially important new source of genetic variation which may underlie human disease and they should be included in future medical genetics studies. As a confirmation of the damaging nature of xNPs, we have identified changes in the exome of a glioblastoma cell line that are important in glioblastoma pathogenesis. We have found a TNP causing a single amino acid change in LAMC2 and a TNP causing a truncation of HUWE1.
INTRODUCTION
While all human genomes are extremely similar to one another, there is variability that allows for the uniqueness of each individual. This variability can take the form of copy number variation, chromosomal rearrangements, or nucleotide polymorphisms. The overwhelming majority of recent studies of human variability have utilized microarray technology because of their relatively cheap cost and ready availability. For example, Genome Wide Association Studies (GWAS) have been performed for numerous diseases (1), with varying levels of success. In a GWAS study, numerous individuals with a specific disease or trait and ethnically matched controls are profiled using a microarray for single nucleotide polymorphisms (SNPs). SNP allelles that are more prevalent in the affected individuals relative to the controls are considered to be associated with illness. For a few diseases, such as age related macular degeneration (2), common SNPs with large effects on risk have been identified. In other cases, even when large sample sizes were utilized, the risk alleles identified by GWAS were only able to explain a small percentage of disease heritability (3).
One potential reason for the disappointing results of GWAS studies is because of their limitation to SNPs. The microarray platforms are designed to robustly identify single nucleotide variations (4), but they are not effective in detecting variations involving more than one consecutive nucleotide. If two sequences are identical except for two adjacent nucleotides being altered (e.g. one sequence has AC and the other sequence has GT), this cannot be effectively measured using a microarray. Additionally, considerations regarding DNA melting temperature and the exclusion of repetitive sequences restrict the probes that can be used on a microarray (5).
Recently, high-throughput DNA sequencing techniques (6,7) have been developed and have begun to replace microarrays for genome analysis studies (8). These sequencing techniques are free of the single nucleotide mismatch and melting temperature restrictions of microarrays. In addition, sequencing can produce a more comprehensive picture of a genome, than the particular features included on a given microarray. By analyzing raw sequencing reads, multiple nucleotide polymorphisms can be studied just as easily as SNPs.
We have used the raw sequencing data from two complete genomes, the Venter/HuRef Genome (9) and the Chinese Genome (10), and eight complete exomes (11), to analyze nucleotide polymorphism beyond the single nucleotide level. We have aligned the sequencing reads to the human reference genome and identified thousands of loci with polymorphisms of 2 or 3 nt. These polymorphisms are denoted as double nucleotide polymorphisms (DNPs) and triple nucleotide polymorphisms (TNPs) (see Figure 1 for examples). For simplicity, as a group, SNPs, DNPs and TNPs are identified as xNPs. These xNPs do not include indels where nucleotides are found to be inserted or deleted in one sequence relative to another sequence. We focus on xNPs where the sequence length remains the same, but one, two or three nucleotides are changed. Indels in human genomes and exomes have been extensively characterized in (12) and (11).
Figure 1.
An example of a SNP, a DNP and a TNP between two DNA sequences.
While SNPs are certainly an important source of variation between human genomes, there are a few reasons why DNPs and TNPs have a greater propensity to be involved in disease causing mutations. First, SNPs have a strong propensity to be synonymous (13) whereby they change the nucleotide sequence, but do not alter the amino acid sequence due to the wobble allowed by the genetic code. These synonymous changes are usually silent and do not effect the phenotype, but there are notable exceptions (14,15). In contrast, a DNP or a TNP would effect multiple positions in a codon. Secondly, a SNP can at most result in the change of one amino acid, whereas a DNP or a TNP can change the residue at two adjacent positions and cause a more dramatic change.
Before looking for the xNPs in genomic sequence, we first computationally determined their predicted effects on amino acid sequence under an assumption of randomness (Table 1). For example, given all possible permutations of nucleotides in a codon, 24% of SNPs would be expected to result in a synonymous mutation due to nonspecificity in the genetic code. On the other hand, a DNP or a TNP would randomly produce a synonymous mutation only 9 or 0.4% of the time, respectively. The rare possibility of a synonymous TNP can only occur when it overlaps two codons and changes both of them in a way that they still code for the same amino acid. As also displayed in Table 1, when a DNP causes an amino acid change, it is much more likely to be a single change rather than a double. Similarly, a TNP has a greater chance of causing one amino acid change than two changes, but the ratio is smaller. These theoretical results support the premise that DNPs and TNPs can be important sources of genomic variation, and our analysis of real data will be compared against these predicted results.
Table 1.
Theoretical calculations of the percentages of each type of change that will be cause by SNPs, DNPs and TNPs
| Type of xNP | Number of nucleotides changed | Percentage of xNPS resulting in the same amino acids, % | Stop codon read through (from stop to coding), % | Percentage of changes resulting in stop codons (premature stop), % | Percentage of XNPs resulting in the change of: |
|
|---|---|---|---|---|---|---|
| 1 Amino acid, % | 2 Amino acids, % | |||||
| SNPs | 1 | 24 | 4 | 4 | 68 | N/A |
| DNPs | 2 | 0.90 | 7 | 6 | 78 | 8 |
| TNPs | 3 | 0.40 | 8 | 7 | 51 | 33 |
We have found that in the human genome there is a considerable amount of variation with regard to DNPs and TNPs. For the two complete genomes that we analyzed, we found tens of thousands of DNPs and thousands of TNPs throughout the genome. As with all genomic variation, the majority of this variation was found outside of coding regions. Even so, a substantial amount of xNPs are found within coding exons and they have a strong potential to be involved in disease pathology. In order to test this hypothesis, we have applied our technique to the analysis of an exome from a glioblastoma cell line. In this exome, we have found xNPS causing amino acid changes and a truncated protein in genes whose mis-expression have been previously found in glioblastoma.
MATERIALS AND METHODS
Theoretical calculations
For SNPs, each codon was iterated through, and each position in the codon was changed to one of the three possible different nucleotides. The percentage of changes that caused amino acid changes or no change were tallied. For DNPs and TNPs, two adjacent codons were used and every possible set of two (for DNP) and three (for TNP) changes were performed. In order to allow for the querying of each position in each codon, the last positions of the second codon were wrapped onto the first codon. To illustrate, the six positions in the two codons from 5′ to 3′ will be listed as integers from 1 to 6, such that the list of TNPs is: 123, 234, 345, 456, 561, 612.
Sequencing data and alignment
The Chinese genome data was obtained from (10) as raw FASTQ sequencing reads and only those paired ended reads that were 35 bp in length were used in the analysis. The Venter/HuRef raw sequencing reads were obtained from (9). Since these sequencing reads were from an Applied Biosystems 3730xl they were much longer than 36-bp Illumina reads. To allow for comparison, the long reads were cut into non-overlapping 3-bp reads. The eight exome sequences were from (11). The glioblastoma exome sequence was from (16).
The sequences were aligned using the Bowtie (17) alignment program and three mismatches were allowed. The reference genome used was hg18. The consensus repeat elements were taken from the UCSC genome browser annotations (18) and the genes used were the CCDS gene set (19).
xNP determination
After the reads were aligned to the genome, any single base mismatch was counted as a putative SNP and two or three consecutive mismatches within reads were marked as putative DNPs or TNPs respectively. For each putative xNP, the number of sequencing reads coding for the xNP or the reference sequence were tallied. The following criteria were used for calling an xNP: if there were no reads matching the reference at that position, there needed to be a minimum of three reads supporting the xNP, and the xNP would be called as homozygous. If there were reads matching the reference, two requirements needed to be met to call a heterozygous xNP: First, there needed to be at least three xNP supporting reads at that location. Second, a binomial distribution was computed at each genomic position, based on the total number of reads at that location and a 50% allele probability. A heterozygous xNP was called if the number of xNP-reads was at least half of the total number of reads at that location minus twice the standard deviation of the binomial distribution. This threshold allowed for <5% false negative rate of calling heterozygotes.
Analysis
The functional categorization of genes with xNPs was performed using DAVID (20). The lethality analysis of the xNPs was performed using Polyphen version 1.1.7 (21) and SIFT version 4.0.3 (22). Additionally, we analyzed the xNPs using PANTHER version 6.1 (23). For a substantial number of the polymorphisms, PANTHER was not able to give a prediction of the probability of it being deleterious. This is because the amino acid substitution occurred in a part of the protein that was not covered by the multi-sequence alignments underlying the predictions. This is a known shortcoming of PANTHER (23). Overall, the percentages of polymorphisms predicted to be deleterious by PANTHER were much lower than the percentages from both Polyphen and SIFT. A strong cause of this was polymorphisms not being scored and therefore not having a possibility of being predicted to be damaging. We therefore have not reported the PANTHER predictions.
RESULTS
Determination of xNPs in the genomes
In order to analyze xNPs in complete human genomes, we selected the Venter (9) and the Chinese (10) genomes as examples for our analysis. The sequencing reads for each of the genomes were aligned to the genome reference hg18 (‘Materials and methods’ section). For each alignment, up to three mismatches were allowed in order to capture SNPs, DNPs or TNPs. Any two adjacent mismatches were marked as a DNP; while three adjacent mismatches indicated a TNP.
The number of xNPs found throughout the genome and their locations are shown in Table 2. For each genome and type of xNP, the total number of xNPs and the number that are homozygous are listed. Since the SNPs for these two genomes have been previously determined, we compared our results to the published counts. For the Venter genome, 3.2 million SNPs were reported (9), as compared to our finding of 2.89 million SNPS. The Chinese genome was reported to have 3.07 million SNPs (10), while our method yielded 3.69 million. It should be noted that these differences in SNP counts are in the expected directions, given the different alignment and SNP calling techniques and thresholds that were utilized (‘Discussion’ section).
Table 2.
Genome-wide distribution of SNPs, DNPs and TNPs for the Chinese and Venter Genomes
| xNP location | Chinese SNPs |
Venter SNPs |
Chinese DNPs |
Venter DNPs |
Chinese TNPs |
Venter TNPs |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Homozygous | Total | Homozygous | Total | Homozygous | Total | Homozygous | Total | Homozygous | Total | Homozygous | |
| Downstream of Genes 5 kb | 103 004 | 39 296 | 75 988 | 40 143 | 1277 | 436 | 935 | 360 | 92 | 41 | 50 | 24 |
| Introns | 904 259 | 367 472 | 695 131 | 378 275 | 9898 | 3646 | 7260 | 2978 | 823 | 334 | 469 | 179 |
| Upstream of genes 5kb | 102 359 | 39 096 | 75 803 | 39 814 | 1187 | 423 | 925 | 380 | 100 | 39 | 59 | 29 |
| Exons | 25 381 | 7935 | 15 079 | 8042 | 164 | 48 | 127 | 45 | 3 | 1 | 6 | 1 |
| Intergenic | 2 547 066 | 1 015 539 | 2 032 962 | 1 030 241 | 32 947 | 10 460 | 27 454 | 8974 | 2154 | 767 | 1362 | 406 |
| Total xNPs | 3 682 069 | 1 469 338 | 2 894 963 | 1 496 515 | 45 473 | 15 013 | 36 701 | 12 737 | 3172 | 1182 | 1946 | 639 |
| Consensus repeats | 1 740 523 | 627 051 | 1 413 418 | 650 635 | 23 384 | 6469 | 21 661 | 5848 | 1304 | 461 | 1055 | 265 |
Overall, the numbers of DNPs and TNPs are greatly reduced relative to the numbers of SNPs. This is expected because the production of a DNP or a TNP requires the mutation of two or three adjacent nucleotides whereas a SNP only requires one change. For all of the xNPs, the greatest percentage occurs in intergenic regions, followed by introns, both of which are non-coding and are expected to have relatively lower levels of consistency across individuals. In contrast, far less than 1% of xNPs occur in coding exons, which are under selective pressure to prevent amino acid mutations. TNPs are almost completely absent from coding exons and there are only three coding TNPs from the Chinese genome and six from the Venter genome. Approximately half of all xNPs were observed in portions of the genome that are defined as repeats by RepeatMasker (24); this is expected since such regions cover 45% of the human genome (25).
xNPs within coding exons
Since coding exons are important regions of the genome for protein production, we focused on the analysis of xNPs in these regions. The xNPs were classified according to whether they caused no amino acid change (neutral/synonymous), caused one amino acid change, caused two amino acid changes, changed from a stop codon to a coding codon (read-through), or changed from a coding amino acid to a stop codon (premature stop). These results are shown in Table 3, and a complete list of each gene that had any xNPs along with the change produced by each type of xNP is shown in Supplementary Table S1.
Table 3.
Effects of SNPs, DNPs and TNPs within the genes of the Chinese and Venter Genomes
| Type of change | Chinese SNPs | Venter SNPs | Chinese DNPs | Venter DNPs | Chinese TNPs | Venter TNPs |
|---|---|---|---|---|---|---|
| 1 Amino acid change | ||||||
| Number | 14 784 | 7092 | 152 | 114 | 0 | 6 |
| Percentage of changes | 58 | 47 | 93 | 90 | 0 | 100 |
| Number of genes affected | 7841 | 4407 | 141 | 98 | 0 | 6 |
| 2 Amino acid changes | ||||||
| Number | N/A | N/A | 8 | 10 | 3 | 0 |
| Percentage of changes | N/A | N/A | 5 | 8 | 100 | 0 |
| Number of genes affected | N/A | N/A | 8 | 10 | 3 | 0 |
| Read through (from stop to coding) | ||||||
| Number | 24 | 6 | 0 | 0 | 0 | 0 |
| Percentage of changes | 0.09 | 0.04 | 0.00 | 0.00 | 0.00 | 0.00 |
| Number of genes affected | 23 | 6 | 0 | 0 | 0 | 0 |
| No change | ||||||
| Number | 10 346 | 7925 | 0 | 2 | 0 | 0 |
| Percentage of changes | 41 | 53 | 0 | 2 | 0 | 0 |
| Number of genes affected | 6211 | 5002 | 0 | 2 | 0 | 0 |
| Premature stop codon | ||||||
| Number | 227 | 56 | 4 | 1 | 0 | |
| Percentage of changes | 0.89 | 0.37 | 2 | 0.79 | 0.00 | 0.00 |
| Number of genes affected | 217 | 50 | 4 | 1 | 0 | |
| Total | 25381 | 15079 | 164 | 127 | 3 | 6 |
We first compared the results with the theoretical calculations from Table 1. For the SNPs, a lower percentage resulted in amino acid changes than would be predicted at random. Theoretically, 68% of SNPs should change one amino acid, whereas this was found for 58 and 47% of the SNPs in the Chinese genome and the Venter genome respectively. This decrease was caused by a greater than predicted number of synonymous SNPs. We predicted that there would be 24% synonymous SNPs and we found that 41 and 53% of the Chinese and Venter genome SNPs, respectively, were synonymous. For both genomes, the synonymous to non-synonymous SNP ratio is around 50–50, as has been previously found for the genomes of multiple species (26–28).
In contrast to the bias from the calculations towards synonymous SNPs, we found a strong bias towards non-synonymous DNPs. For both genomes almost all of the exonic DNPs resulted in an amino acid change. The theoretical calculations predicted 86% of the DNPs causing amino acid changes and we found 98% of the DNPs for each genome causing a change. There were only a small number of exonic TNPs, but these were completely non-synonymous.
For all types of xNPs, the occurrence of both premature stop codons and stop codon read-through was less than predicted. For example, while 7% of DNPs were predicted to change a stop codon to a coding codon and result in stop codon read-through; neither genome had any DNPs producing read-through. Premature stop codons were predicted to result from 4 to 6% of SNPs and DNPs, but they were only found in 2% or less of all such events. These findings are presumably due to selective pressure against the potentially catastrophic results of either a protein truncation or elongation.
Analysis of exonic DNPs and TNPs
We found a 225 DNPs located within 200 genes in the Venter and Chinese genomes. Sixty-six (29.3%) of these DNPs are found in both genomes, and may reflect the presence of variants or errors in the reference genome. In both the Venter and Chinese genomes, over 90% of the DNPs resulted in a single amino acid change. For the Venter genome, 30% of them were predicted to me damaging by both Polyphen (21) and SIFT (22). For the Chinese genome, 35% were predicted by Polyphen to be damaging while 34% were predicted to be deleterious by SIFT. We then used DAVID (20) to functionally annotate the genes containing DNPs. For the Chinese genome, the top Gene Ontology category to describe the DNPs was for the MHC protein complex (Benjamini corrected P = 0.032) and for the Venter genome, no categories were found to be significant.
Both genomes had a small number of TNPs within their exons, with the Venter genome only containing six exonic TNPS and the Chinese genome only containing three exonic TNPs. None of these TNPs are shared between the two genomes. For the Venter exonic TNPs, only one is homozygous, but they all cause a single amino acid change. For these TNPs, four of them are predicted by Polyphen to be either damaging to the protein structure, while two are predicted to be deleterious by SIFT. For the Chinese TNPs, only one is homozygous, but all cause two amino acid changes. Since Polyphen and SIFT only look at single amino acid changes, we were unable to evaluate the double nucleotide changes for their damaging potential. These TNPs were not concentrated in proteins of one type and include proteins that are structural (RDX, KRTAP10-1), signal regulatory (SIRPA), RNA binding (GPATC4), an olfactory receptor (OR5L2), bind to protein kinase A (AKAP3), a metalloprotease (ADAMTS9) and an antigen presenter (HLA-DRB5).
xNPs in sequenced exomes
To further investigate the occurrence of xNPs in human genes and their consistency, we utilized complete exome sequencing data from eight individuals (11). Since only the exomes of these individuals were sequenced, we were not able to quantify xNPs outside of genes. The SNPs, DNPs and TNPs in each exome were determined using the same criteria that were used for our initial two genomes (Venter and Chinese) and the results are shown in Table 4. For each of these exomes, we found an average of 17 164 exonic SNPs per exome which is very close to the reported count (11) of 17 272. We found a smaller number of DNPs and TNPs, with an average of 164 DNPs per exome and five TNPs per exome. Notably, the average number of exonic DNPs was identical to that observed in the Chinese genome, which was sequenced on the same platform (Illumina). We then looked at the pervasiveness of each xNP among the eight exomes (Supplementary Table S2). On average the same SNP, DNP or TNPs was found in 2.5, 1.8 and 1.4 exomes respectively. Despite this low average, there were SNPs and DNPs that were found across all eight exomes. For these loci, the occurrence of a SNP or a DNP relative to the reference genome in eight samples indicates that the reference genome probably does not contain the most common sequence. This is the case for 1450 SNPs (8.4%) and 18 DNPs (11%). These low percentages of pervasive xNPS indicate that the majority of xNPS are true variations between genomes rather than reflecting inaccuracies in the reference genome.
Table 4.
Effects of SNPs, DNPs and TNPs in the eight sequenced exomes
| Type of change | NA12156 |
NA12878 |
NA18507 |
NA18517 |
NA18555 |
NA18956 |
NA19129 |
NA19240 |
Average |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPS | DNPS | TNPs | SNPS | DNPS | TNPs | SNPs | DNPS | TNPs | SNPs | DNPS | TNPs | SNPs | DNPS | TNPs | SNPs | DNPS | TNPs | SNPs | DNPS | TNPs | SNPs | DNPS | TNPs | SNPs | DNPS | TNPs | |
| 1 Amino acid Change | |||||||||||||||||||||||||||
| Number | 7604 | 141 | 4 | 7228 | 158 | 4 | 8540 | 150 | 5 | 8685 | 170 | 5 | 7350 | 121 | 5 | 7073 | 154 | 1 | 8355 | 169 | 5 | 8385 | 172 | 2 | 7903 | 154 | 4 |
| Percentage of changes | 47 | 94 | 80 | 47 | 93 | 80 | 46 | 94 | 83 | 46 | 97 | 63 | 46 | 94 | 83 | 46 | 92 | 100 | 45 | 91 | 100 | 46 | 96 | 100 | 46 | 94 | 86 |
| Number of genes affected | 4758 | 132 | 3 | 4563 | 153 | 4 | 5230 | 141 | 3 | 5229 | 154 | 5 | 4672 | 117 | 5 | 4515 | 143 | 1 | 5145 | 154 | 4 | 5208 | 162 | 2 | 4915 | 145 | 3 |
| 2 Amino acid Changes | |||||||||||||||||||||||||||
| Number | N/A | 5 | 1 | N/A | 7 | 1 | N/A | 6 | 1 | N/A | 3 | 3 | N/A | 5 | 1 | N/A | 9 | 0 | N/A | 9 | 0 | N/A | 8 | 0 | N/A | 7 | 1 |
| Percentage of changes | N/A | 3 | 20 | N/A | 4 | 20 | N/A | 4 | 17 | N/A | 2 | 38 | N/A | 4 | 17 | N/A | 5 | 0 | N/A | 5 | 0 | N/A | 4 | 0 | N/A | 4 | 14 |
| Number of genes affected | N/A | 5 | 1 | N/A | 7 | 1 | N/A | 6 | 1 | N/A | 3 | 3 | N/A | 5 | 1 | N/A | 9 | 0 | N/A | 9 | 0 | N/A | 8 | 0 | N/A | 7 | 1 |
| Read through (from Stop to Coding) | |||||||||||||||||||||||||||
| Number | 10 | 0 | 0 | 7 | 0 | 0 | 17 | 1 | 0 | 12 | 0 | 0 | 8 | 0 | 0 | 4 | 0 | 0 | 9 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 |
| Percentage of changes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.06 | 0 | 0 |
| Number of genes affected | 10 | 0 | 0 | 7 | 0 | 0 | 17 | 1 | 0 | 12 | 0 | 0 | 8 | 0 | 0 | 4 | 0 | 0 | 9 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 |
| No Change | |||||||||||||||||||||||||||
| Number | 8578 | 3 | 0 | 8266 | 2 | 0 | 9994 | 2 | 0 | 10 141 | 1 | 0 | 8565 | 2 | 0 | 8149 | 2 | 0 | 10 170 | 5 | 0 | 9833 | 0 | 0 | 9212 | 2 | 0 |
| Percentage of changes | 53 | 2 | 0 | 53 | 1 | 0 | 54 | 1 | 0 | 54 | 1 | 0 | 54 | 2 | 0 | 53 | 1 | 0 | 55 | 3 | 0 | 54 | 0 | 0 | 54 | 1 | 0 |
| Number of genes affected | 5397 | 3 | 0 | 5220 | 2 | 0 | 6139 | 2 | 0 | 6240 | 1 | 0 | 5365 | 2 | 0 | 5216 | 2 | 0 | 6265 | 5 | 0 | 6094 | 0 | 0 | 5742 | 2 | 0 |
| Premature Stop Codon | |||||||||||||||||||||||||||
| Number | 39 | 1 | 0 | 42 | 2 | 0 | 35 | 0 | 0 | 43 | 2 | 0 | 35 | 1 | 0 | 38 | 2 | 0 | 45 | 2 | 0 | 42 | 0 | 0 | 40 | 1 | 0 |
| Percentage of changes | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0.23 | 1 | 0 |
| Number of genes affected | 38 | 1 | 0 | 42 | 2 | 0 | 34 | 0 | 0 | 42 | 2 | 0 | 33 | 1 | 0 | 37 | 2 | 0 | 44 | 2 | 0 | 41 | 0 | 0 | 39 | 1 | 0 |
| Total Exonic xNPS | 16 231 | 150 | 5 | 15 543 | 169 | 5 | 18 586 | 159 | 6 | 18 881 | 176 | 8 | 15 958 | 129 | 6 | 15 264 | 167 | 1 | 18 579 | 185 | 5 | 18 270 | 180 | 2 | 17 164 | 164 | 5 |
The most common TNP was found in four exomes in KRTAP10-1 gene and it was determined by Polyphen and SIFT to be a benign change. As with the TNPs found in the two full genomes, a significant amount of them result in the change of two adjacent amino acids which cannot be easily evaluated.
To further confirm the findings of xNPs in the exomes, we compared our findings for one exome (NA19240) to the complete genome of that individual that has recently been completed using the Complete Genomics technology (29). In our analysis of the data from the exome sequencing (Table 4), we identified 180 DNPs and two TNPs in coding regions; while using the Complete Genomics data, we identified 155 coding DNPs and five coding TNPs. Seventy of the DNPs and one of the TNPs were found using both techniques. Thus, results of xNP analysis could be to some extent be dependent upon sequencing platform. At the same time, the overall abundance of DNPs and TNPs observed in the human genome and exome appear to be relatively consistent across various sequencing technologies.
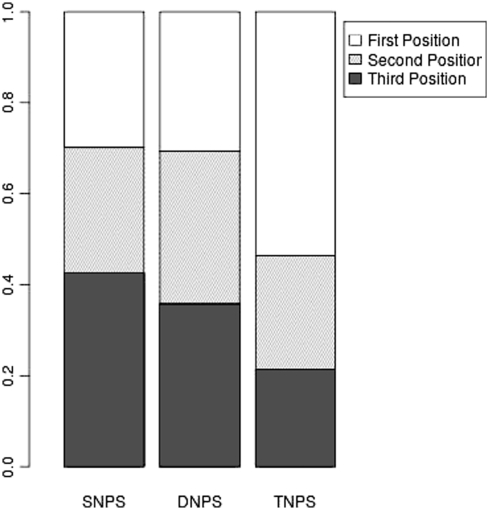
We then looked at the positions within codons where xNPs occur. SNPs should preferentially occur in the third codon position, as has been previously found (26,30). This is because of the wobble nature of the genetic code whereby a change in this position is often silent. DNPs and TNPs have not been previously profiled, but it is expected that a DNP would preferentially occur in either positions 1 and 2 or 2 and 3 of a codon so as not to overlap two codons. Similarly, TNPs would be expected to completely overlap an individual codon. A plot of each type of xNP and the percentage that begin in each position in a codon is shown in Figure 2. As expected, the largest percentage of SNPs occur in the third codon position, but this was not an overwhelming majority (43%). For DNPs, unexpectedly, there appears to be little preference for any codon position. For TNPs, there is a bias towards their beginning in the first codon position (54%) and covering a single codon rather than overlapping two adjacent codons.
Figure 2.
The percentage SNPs, DNPs and TNPs that begin in each codon position.
Nucleotide substitutions can be categorized as either transitions or transversions depending upon the 2 nt involved. There is generally considered to be a strong bias of transitions as compared to transversions in metazoan genomes (31). This was confirmed by our findings for SNPs in the combined set of eight exomes. There were 37 457 transitions and 14 792 transversions observed. For DNPs, and TNPs, the terms of transition and transversions do not directly apply since they are associated with individual nucleotides. Nevertheless, we were able to investigate the positions within a DNP or a TNP as transition or transversions (Table 5). For DNPs, the first position was dominated (66%) by a transition, while there was much less preference at the second position. In contrast, there was a preference among TNPs (46%) for three transversions in a row.
Table 5.
A Tally of each combination of nucleotide changes for DNPs and TNPs
| DNPs |
TNPs |
||
|---|---|---|---|
| Changes | Count | Changes | Count |
| Transition–Transition | 249 | Transition–Transition–Transition | 1 |
| Transition–Transversion | 256 | Transition–Transition–Transversion | 2 |
| Transversion–Transition | 111 | Transition–Transversion–Transition | 1 |
| Transversion–Transversion | 147 | Transition–Transversion–Transversion | 3 |
| Transversion–Transition–Transition | 4 | ||
| Transversion–Transition–Transversion | 2 | ||
| Transversion–Transversion–Transition | 2 | ||
| Transversion–Transversion–Transversion | 13 | ||
xNPs in a cancerous exome
Finally, we applied our analysis to the sequenced exome of the U87 glioblastoma cell line (16). Rather than sequencing a complete exome, this study only sequenced the exons of 5253 cancer associated genes. Using our analysis, we found 53 DNPs and eight TNPs. For the DNPs, four caused a double amino acid change. Of the 49 that caused a single amino acid change, 37% were predicted by Polyphen to be damaging while SIFT determined that 31% would be deleterious. For the TNPs, four caused a double amino acid change and three that caused a single amino acid change. Of the single amino acid changes, all of them were predicted by both Polyphen and SIFT to be damaging. In addition, one TNP caused a premature stop codon in this exome. A summary of the mutations found in each gene are shown in Table 6.
Table 6.
A summary of the DNPs and TNPs found in the glioblastoma U87 exome
| DNPs |
TNPs |
||||
|---|---|---|---|---|---|
| CCDS Name | Gene Name | Result of xNP | CCDS Name | Gene Name | Result of xNP |
| CCDS10326 | ADAMTSL3 | 1 Amino acid change | CCDS11966 | ALPK2 | 1 Amino acid change |
| CCDS10411 | PIGQ | 1 Amino acid change | CCDS1352 | LAMC2 | 1 Amino acid change |
| CCDS11509 | CDC27 | 1 Amino acid change | CCDS4452 | RASGEF1C | 1 Amino acid change |
| CCDS12096 | ZNF555 | 1 Amino acid change | CCDS11905 | FAM59A | 2 Amino acid change |
| CCDS13754 | PRODH | 1 Amino acid change | CCDS2012 | PROM2 | 2 Amino acid change |
| CCDS14006 | ST13 | 1 Amino acid change | CCDS34698 | ZKSCAN1 | 2 Amino acid change |
| CCDS14228 | DMD | 1 Amino acid change | CCDS7328 | PPP3CB | 2 Amino acid change |
| CCDS14596 | ZBTB33 | 1 Amino acid change | CCDS35301 | HUWE1 | Premature stop codon |
| CCDS14607 | STAG2 | 1 Amino acid change | |||
| CCDS14711 | CSAG1 | 1 Amino acid change | |||
| CCDS14713 | MAGEA2 | 1 Amino acid change | |||
| CCDS2057 | IL1RL1 | 1 Amino acid change | |||
| CCDS2397 | BARD1 | 1 Amino acid change | |||
| CCDS30824 | PDE4DIP | 1 Amino acid change | |||
| CCDS30947 | ABL2 | 1 Amino acid change | |||
| CCDS31702 | OR10G4 | 1 Amino acid change | |||
| CCDS32595 | KIAA0100 | 1 Amino acid change | |||
| CCDS33119 | NLRP13 | 1 Amino acid change | |||
| CCDS33432 | TCF15 | 1 Amino acid change | |||
| CCDS33539 | SYNJ1 | 1 Amino acid change | |||
| CCDS33876 | MED12L | 1 Amino acid change | |||
| CCDS34389 | CDSN | 1 Amino acid change | |||
| CCDS34654 | TRIM50 | 1 Amino acid change | |||
| CCDS35277 | SHROOM4 | 1 Amino acid change | |||
| CCDS35277 | SHROOM4 | 1 Amino acid change | |||
| CCDS35305 | FAM104B | 1 Amino acid change | |||
| CCDS35417 | MAGEC1 | 1 Amino acid change | |||
| CCDS35431 | PASD1 | 1 Amino acid change | |||
| CCDS42086 | LRRK1 | 1 Amino acid change | |||
| CCDS42437 | MAPK4 | 1 Amino acid change | |||
| CCDS42535 | ZNF626 | 1 Amino acid change | |||
| CCDS42959 | KRTAP10-6 | 1 Amino acid change | |||
| CCDS42965 | KRTAP12-2 | 1 Amino acid change | |||
| CCDS42965 | KRTAP12-2 | 1 Amino acid change | |||
| CCDS43248 | DSPP | 1 Amino acid change | |||
| CCDS44016 | MAGEA2B | 1 Amino acid change | |||
| CCDS4881 | CUL7 | 1 Amino acid change | |||
| CCDS6415 | CYC1 | 1 Amino acid change | |||
| CCDS7148 | MYO3A | 1 Amino acid change | |||
| CCDS7566 | DUSP5 | 1 Amino acid change | |||
| CCDS7693 | NLRP6 | 1 Amino acid change | |||
| CCDS7927 | DDB2 | 1 Amino acid change | |||
| CCDS7954 | PRG3 | 1 Amino acid change | |||
| CCDS9300 | SACS | 1 Amino acid change | |||
| CCDS9737 | DAAM1 | 1 Amino acid change | |||
| CCDS9766 | HSPA2 | 1 Amino acid change | |||
| CCDS9867 | SNW1 | 1 Amino acid change | |||
| CCDS9928 | SERPINA5 | 1 Amino acid change | |||
| CCDS9998 | JAG2 | 1 Amino acid change | |||
| CCDS11281 | CCL13 | 2 Amino acid change | |||
| CCDS14446 | FAM46D | 2 Amino acid change | |||
| CCDS4269 | PCDH12 | 2 Amino acid change | |||
| CCDS5989 | DLC1 | 2 Amino acid change | |||
In order to determine whether any of these xNPs were in genes that have been previously found to be related to glioblastoma, we conducted Pubmed searches for each of the genes that was found to have a damaging xNP, or an xNP changing the location of a stop codon. We found that a TNP in the LAMC2 gene that results in a single amino acid change L952D which is predicted to be damaging. This gene has been found to be amplified in glioblastomas (32) as well as other cancers (33,34). A TNP in the HUWE1 gene causes a truncation of the protein by the insertion of a premature stop codon reducing its length from 4374 residues to 1668 residues. The HUWE1 gene has been found to be important in brain development, and its deletion has been found to be important in malignant brain tumors (35). In the case of this cell line, HUWE1 is not deleted, but it is truncated and therefore most likely not functional.
DISCUSSION
We have characterized a novel source of genomic variation, DNPs and TNPs, which occur with a frequency of ∼1% of the total number of SNPs. In the two genomes we examined, we found tens of thousands of DNPs and thousands of TNPS. While only a small percentage of these changes are found in coding sequence and directly affect the transcribed protein, the non-exonic xNPs could of course be located in regulatory regions. Although not directly examined in this report, alteration of sequence in a promoter or an enhancer could change the expression dynamics of the associated gene (36).
The coding xNPs, while small in number, could be very important clinically. In order to cause a disease, only a single amino acid change in the genome may be required. Since DNPs and TNPs cause an amino acid change in at least 90% of instances, they could very easily be a cause of a disease. Those DNPs and TNPs that cause two amino acid changes are especially intriguing since they are likely to have a pronounced affect on the protein structure and function. Exonic DNPs and TNPs are approximately 3-fold over-represented amongst amino acid-changing polymorphisms and produce greater than 100 such changes in each normal human genome.
Based upon the average frequency of SNPs and simple probability, DNPs and TNPs should be much rarer than what we found. Assuming the occurrence of a SNP to be ∼1 in 1000 bp (3 million SNPs in a 3 billion base genome) and assuming independence of all SNPs, there should be one DNP every 1 million base pairs (10002), which would total 3000 DNPs in the entire human genome. There should also be one TNP every 1 billion base pairs (10003) which would total three TNPs for the entire human genome. These numbers are vastly lower than the numbers that we observed, supporting the idea that the mutations in a DNP or a TNP are not independent. It has been found that SNPs tend to cluster along the genome rather than being evenly distributed; certain regions of the genome have large amounts of SNPs and other regions of the genome are devoid of SNPs (37). Given a region of the genome with a large amount of SNPs, it is statistically more likely that a DNP or TNP would occur. The occurrence of DNPs and TNPs (as well as clusters of nearby, though non-adjacent SNPs) can be explained by results of polymerase mis-incorporation experiments. In these assays, it was found that if a polymerase incorporates the incorrect nucleotide at a particular location, it increases the likelihood that another nearby nucleotide will be incorporated incorrectly (38,39).
In considering our results, it is important to recognize that the number of variants identified by sequencing can vary as a function of numerous factors, including sequencing platform, read length, depth of coverage, and read alignment parameters (including quality control filters). The present study utilized different alignment parameters from prior whole genome sequencing studies, in order to permit detection of DNPs and TNPs. For example, the Chinese genome (10) was aligned to the reference using the SOAP (40) tool and the paired-end reads were aligned together allowing for two mismatches in each read. In our analysis, we aligned all of the reads using Bowtie (17), because it is very fast at aligning reads and this speed can be further increased by allowing it to use multiple threads on a multiprocessor machine (41). Moreover, we permitted up to three mismatches in each read in order to be able to detect TNPs; if we had used the standard cutoff of two mismatches, any read providing evidence for a TNP would have been discarded as an unmappable read. Given this less stringent filter, it is not surprising that we identified a somewhat larger number of SNPs in the Chinese genome than originally reported (Wang et al. 2008). By contrast, the eight exomes were originally aligned using Maq (Li et al. 2008a) which does not have an explicit cutoff for the number of mismatches allowed; our alignment procedures resulted in an average number of SNPs that was nearly identical to the original report (Shendure et al. 2009). Finally, the original Venter genome analysis (9,42) was based upon the traditional Sanger sequencing and assembly of the Venter genome (43). As such, the SNPs between this genome and the human reference genome were determined by comparing the two genome assemblies de novo. Our analysis of the Venter/HuRef genome was completely different in that we utilized their raw sequencing reads which were truncated into non-overlapping 36-bp reads to simulate Ilumina sequencing reads (‘Materials and Methods’ section). This procedure resulted in a slight under-estimate of the total number of SNPs compared to the Venter Institute report, presumably due to some variation in regions with low-depth of coverage failing to meet our criteria for SNP calling.
As an application of our technique to a real disease, we investigated the U87 glioblastoma cell line. We found two TNPs that cause pathogenic changes in genes that have already been implicated in the disease. Besides these mutations, it is very likely that a further understanding of glioblastoma could be gained from an analysis of the xNPs that were found in genes that have not already been suspected of involvement in glioblastoma.
CONCLUSION
In conclusion, the detection of DNPs and TNPs has not been previously studied, to our knowledge, and would be impractical using microarrays. With the recent advent of high-throughput sequencing and the possibility of sequencing complete exomes (11) and genomes (29), the investigation of DNPs and TNPs should be relatively straightforward. Their identification could be computationally accomplished in a manner as SNPs are called in sequenced genomes. It is hoped that the investigation of DNPs and TNPs in genomes will lead to the identification of causative mutations for genetic diseases that have thus far eluded SNP-based studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01MH084098 to T.L. and P50MH080173 to A.K.M.). Funding for open access charge: R01MH084098.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Rob DeSalle for helpful suggestions concerning the analysis and the writing of the article. The authors would also like to thank John Cholewa for technical assistance.
REFERENCES
- 1.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein DB. Common genetic variation and human traits. N. Eng. J. Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Norton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotech. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 5.Held GA, Grinstein G, Tu Y. Modeling of DNA microarray data by using physical properties of hybridization. Proc. Natl Acad. Sci. USA. 2003;100:7575–7580. doi: 10.1073/pnas.0832500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley D, Balasubramanian S, Swerdlow H, Smith G, Milton J, Brown C, Hall K, Evers D, Barnes C, Bignell H, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shendure J. The beginning of the end for microarrays? Nat. Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 9.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J. The diploid genome sequence of an Asian individual. Nature. 2008;456:60. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwick ME, Cutler DJ, Chakravarti A. Patterns of genetic variation in Mendelian and complex traits. Annu. Rev. Genomics Hum. Genet. 2003;1:387–407. doi: 10.1146/annurev.genom.1.1.387. [DOI] [PubMed] [Google Scholar]
- 14.Kimchi-Sarfaty C, Oh JM, Kim I-W, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A "Silent" Polymorphism in the MDR1 Gene Changes Substrate Specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 15.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 16.Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H, Merriman B, Nelson SF. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:2003–2004. [PubMed] [Google Scholar]
- 21.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004 < http://www.repeatmasker.org>. [Google Scholar]
- 25.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 26.Jimenez-Gomez J, Maloof J. Sequence diversity in three tomato species: SNPs, markers, and molecular evolution. BMC Plant Biol. 2009;9:85. doi: 10.1186/1471-2229-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 28.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 29.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 30.Haga H, Yamada R, Ohnishi Y, Nakamura Y, Tanaka T. Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190 562 genetic variations in the human genome. J. Hum. Genet. 2002;47:605–610. doi: 10.1007/s100380200092. [DOI] [PubMed] [Google Scholar]
- 31.Wakeley J. The excess of transitions among nucleotide substitutions: new methods of estimating transition bias underscore its significance. Trends Ecol. Evol. 1996;11:158–162. doi: 10.1016/0169-5347(96)10009-4. [DOI] [PubMed] [Google Scholar]
- 32.Korshunov A, Sycheva R, Golanov A. Genetically distinct and clinically relevant subtypes of glioblastoma defined by array-based comparative genomic hybridization (array-CGH) Acta Neuropathol. 2006;111:465–474. doi: 10.1007/s00401-006-0057-9. [DOI] [PubMed] [Google Scholar]
- 33.Shou J-Z, Hu N, Takikita M, Roth MJ, Johnson LL, Giffen C, Wang Q-H, Wang C, Wang Y, Su H, et al. Overexpression of CDC25B and LAMC2 mRNA and protein in esophageal squamous cell carcinomas and premalignant lesions in subjects from a high-risk population in China. Cancer Epidemiol. Biomarkers Prev. 2008;17:1424–1435. doi: 10.1158/1055-9965.EPI-06-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith SC, Nicholson B, Nitz M, Frierson HF, Jr, Smolkin M, Hampton G, El-Rifai W, Theodorescu D. Profiling bladder cancer organ site-specific metastasis identifies LAMC2 as a novel biomarker of hematogenous dissemination. Am. J. Pathol. 2009;174:371. doi: 10.2353/ajpath.2009.080538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, D'Arca D, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev. Cell. 2009;17:210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epstein DJ. Cis-regulatory mutations in human disease. Brief. Funct. Genomics. 2009;8:310–316. doi: 10.1093/bfgp/elp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindblad-Toh K, Winchester E, Daly MJ, Wang DG, Hirschhorn JN, Laviolette JP, Ardlie K, Reich DE, Robinson E, Sklar P. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]
- 38.Maki H. Origins of spontaneous mutations: specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu. Rev. Genet. 2003;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- 39.Bebenek K, Roberts JD, Kunkel TA. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J. Biol. Chem. 1992;267:3589–3596. [PubMed] [Google Scholar]
- 40.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng PC, Levy S, Huang J, Stockwell TB, Walenz BP, Li K, Axelrod N, Busam DA, Strausberg RL, Venter JC. Genetic variation in an individual human exome. PLoS Genet. 2008;4:e1000160. doi: 10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.