Abstract
Lens epithelium-derived growth factor/p75 (LEDGF/p75) is a transcriptional coactivator involved in stress response, autoimmune disease, cancer and HIV replication. A fusion between the nuclear pore protein NUP98 and LEDGF/p75 has been found in human acute and chronic myeloid leukemia and association of LEDGF/p75 with mixed-lineage leukemia (MLL)/menin is critical for leukemic transformation. During lentiviral replication, LEDGF/p75 tethers the pre-integration complex to the host chromatin resulting in a bias of integration into active transcription units (TUs). The consensus function of LEDGF/p75 is tethering of cargos to chromatin. In this regard, we determined the LEDGF/p75 chromatin binding profile. To this purpose, we used DamID technology and focused on the highly annotated ENCODE (Encyclopedia of DNA Elements) regions. LEDGF/p75 primarily binds downstream of the transcription start site of active TUs in agreement with the enrichment of HIV-1 integration sites at these locations. We show that LEDGF/p75 binding is not restricted to stress response elements in the genome, and correlation analysis with more than 200 genomic features revealed an association with active chromatin markers, such as H3 and H4 acetylation, H3K4 monomethylation and RNA polymerase II binding. Interestingly, some associations did not correlate with HIV-1 integration indicating that not all LEDGF/p75 complexes on the chromosome are amenable to HIV-1 integration.
INTRODUCTION
Lens epithelium-derived growth factor p75 (LEDGF/p75) was first described as a transcriptional coactivator and a component of the general RNA polymerase II transcription machinery (1). LEDGF/p75 protects cells against oxidative stress by activation of stress response genes through interaction with stress response elements, and caspase-mediated cleavage of LEDGF/p75 is known to disrupt or reverse this protection (2–6). In addition to mediating stress response, LEDGF/p75 plays a crucial role in cancer (7). Through its interaction with mixed-lineage leukemia (MLL/menin), LEDGF/p75 is involved in MLL-dependent transcription and leukemic transformation. In agreement, NUP98-LEDGF/p75 fusions were found in human acute and chronic myeloid leukaemia (8–11). LEDGF/p75 is also frequently encountered as an autoantigen in a subset of patients with atopic disorders, mainly atopic dermatitis, and other inflammatory conditions although its function as a potential cause or outcome is not understood (12). It remains to be shown whether the roles of LEDGF/p75 in stress response, cancer and as an autoantigen are interrelated.
A shorter splice variant, p52, is expressed from the same gene as LEDGF/p75 (PSIP1 in humans). While the N-terminal 325 amino acids are identical, LEDGF/p75 has a unique C-terminal region of 205 amino acids. Based on the conserved N-terminal PWWP domain, both isoforms are classified as Hepatoma-derived growth factor-related proteins (HRPs). Together with the AT-hooks and the nuclear localization signal found in the N-terminal part of the protein, the PWWP domain tethers LEDGF/p75 to the chromatin (13,14). The C-terminal moiety of LEDGF/p75 contains the integrase binding domain. Next to binding cellular proteins like JPO2, pogZ, menin/MLL and Cdc7-ASK (7,15–18), this domain was shown to interact with the integrase protein of human immunodeficiency virus type 1 (HIV-1) and other lentiviruses (19–24). HIV-1 integration into the host cell genome displays a strong bias towards active genes, disfavoring promoter regions and CpG islands (25–28). Knockdown or knockout of LEDGF/p75 abolishes this integration pattern, supporting the hypothesis that LEDGF/p75 binds active genes and tethers the HIV pre-integration complex to these sites via its interaction with integrase (27,29,30). Contradictory to this, cell biology data on LEDGF/p75 predict promoter association (4,6,31,32). To find an answer to this discrepancy, we mapped chromatin binding of LEDGF/p75 by DamID technology and showed that this profile is reminiscent of that of HIV-1 integration. LEDGF/p75 binds active genes disfavoring promoter regions. Correlation with more than 200 genomic features reveals an association with markers of active chromatin. Intriguingly, some markers associate with LEDGF/p75 binding but not with HIV-1 integration suggesting that not all LEDGF/p75 chromatin interactions are amenable to HIV-1 integration.
MATERIALS AND METHODS
Lentiviral vector plasmid construction and vector production
The lentiviral transfer plasmid pLgw-EcoDam-V5-MCS was a kind gift from Dr Bas van Steensel (The Netherlands Cancer Institute, NKI, the Netherlands) and encodes C-terminally V5-tagged Escherichia coli Dam methylase (EcoDam) under the control of the HSP (heat shock protein) promoter. The LEDGF/p75 coding region was PCR-amplified using primers F: 5′-ggg gac aag ttt gta caa aaa agc agg ctt cac tcg cga ttt caa acc tgg-3′ and R: 5′- ggg gac cac ttt gta caa gaa agc tgg gtc cta gtt atc tag tgt aga atc c-3′ (Invitrogen Gateway® recombination region in italics and LEDGF/p75 homology region underlined) and cloned N-terminally to the V5-EcoDam according to the manufacturer’s protocol (Invitrogen, Merelbeke, Belgium) yielding plasmid pLgw-EcoDam-V5-LEDGF/p75. The integrity of the construct was verified by DNA sequencing. Lentiviral vectors were prepared as described previously (33). The pLNC LEDGF/p75-IRES-Bsd and pLNC LEDGF/p75D366A-IRES-Bsd plasmids for rescue of HIV replication were described before Ref. (34).
Cell culture and transient transfection
HeLaP4 and 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco-BRL, Merelbeke, Belgium) supplemented with 10% fetal calf serum (Sigma, Bornem, Belgium) and 20 μg/ml gentamicin (Gibco-BRL) (further referred to as DMEM complete) at 37°C and 5% CO2 in a humidified atmosphere. For western blotting 5 × 105 293T cells were transfected using 3 µg of plasmid DNA complexed with lipofectamin as described by the manufacturer (Invitrogen) and incubated overnight. HeLa CCR5 A3 cells with a stable LEDGF/p75 knockdown were described before Ref. (34).
Fluorescence laser scanning microscopy
Cells were grown in LabTek II glass chamber slides (VWR International, Haasrode, Belgium) and transfected after 24 h with pmRFP-IN (35) and pLgw-EcoDam-V5-LEDGF/p75 or pLNC LEDGF/p75-IRES-Bsd. The 24-h post-transfection cells were fixed with 4% formaldehyde in PBS for 10 min. Immunohistochemistry staining of endogenous LEDGF/p75 was performed using rabbit anti-LEDGF/p75 antibody (Bethyl, Montgomery, TX, USA). EcoDam-LEDGF/p75 fusion protein was detected using mouse anti-V5 antibody (Sigma). Alexa 488-labelled secondary antibodies were used for detection. Nuclear DNA was stained with 0.5 µg/ml DAPI (Molecular Probes, Merelbeke, Belgium). Confocal microscopy was performed using an LSM 510 meta unit (Zeiss, Zaventem, Belgium). All images were acquired in the multi-track mode. Alexa 488 was excited at 488 nm (by AI laser), mRFP at 543 nm (by HeNe laser) and DAPI at 790 nm (by Spectra-Physics Mai Tai laser). After the main beam splitter (HFT KP 700/543 for mRFP, HFT UV/488/543/633 for eGFP and HFT KP650 for DAPI) the fluorescence signal was divided by a secondary dichroic beam splitter (NFT 490 for eGFP and NFT 545 for mRFP) and detected in the separate channels using the appropriate filters (BP 500–-550 for Alexa 488, BP 565–615 for mRFP and BP 435–485 for DAPI).
Western blotting
Protein samples were separated on 10% SDS–PAGE and electroblotted onto polyvinylidene difluoride membranes (PVDF; Bio-Rad). Membranes were blocked with milk powder in PBS/0.1% Tween20 and detection was carried out using mouse anti-V5 antibody (Sigma-Aldrich). Visualization was performed using chemiluminescence (ECL+, Amersham, Diegem, Belgium) using anti-mouse antibodies coupled to HRP (Dako, Denmark).
DamID and DNA tiling array hybridization
Expression of the EcoDam-LEDGF/p75 fusion protein and the EcoDam-only control protein and preparation of gDNA was performed essentially as described (36). Briefly, 5 × 105 HeLaP4 cells were plated in six-well dishes. The next day, three wells each were transduced overnight with 1.5 ml of a lentivector preparation (12.5 × 103 pg p24/ml) expressing either EcoDam-LEDGF/p75 or EcoDam-only under the control of the HSP promoter or left untreated. After transduction, the medium was replenished and cells were allowed to grow for 48 h under normal cell culture conditions (37°C, 5% CO2, DMEM complete). After 48 h, cells were collected by trypsinization and gDNA was prepared using the GenElute Mammalian Genomic DNA purification kit (Sigma-Aldrich) according to the manufacturer’s protocol. Amplification of methylation-specific PCR fragments was performed as described (36) with minor modifications to comply with the hybridization protocol of the Affymetrix ENCODE 2.0R tiling arrays. For this purpose, PCR amplifications were performed in the presence of dNTP (25 mM each) and dUTP (5 mM). This allowed for the subsequent digestion of the 200–2000 bp PCR fragments into fragments of ∼66 bp using UDG/APE enzymes. The correct length of the fragments was confirmed by electrophoresis using the Agilent Bioanalyzer (Agilent, Diegem, Belgium). Three biological replicates of EcoDam-LEDGF/p75 and EcoDam-only samples each were hybridized with Affymetrix GeneChip ENCODE 2.0R DNA tiling microarrays (Affymetrix, Santa Clara, CA, USA) and subsequently analyzed according to the manufacturer’s protocol.
DamID analysis
The MAT (Model-based Analysis of Tiling arrays) algorithm was used to identify genomic regions of statistically significant enrichment of LEDGF/p75 binding (EcoDam-LEDGF/p75 versus EcoDam-only samples) (37). For each array, MAT linearly fits the baseline probe behavior in function of the probe sequence and copy number and uses this model to standardize the readout t-value for each probe. A sliding window approach was used to define enriched regions (BandWidth: 300 bp; MaxGap: 300 bp; MinProbe; 10; significance cut-off P < 10−5). Other parameters or algorithms (TAS, Tiling Analysis Software; Affymetrix) were also tested and yielded similar results (See also Supplementary Figure S1). The MAT score amounts to the average t-value in the probe-centered window and across replicates after trimming of the upper and lower t-value deciles, multiplied by the square root of the number of observation points the MAT score is based on. The control MAT score was subtracted from that for LEDGF/p75. Significantly enriched regions were recorded in .bed format for further analysis.
Data analysis
The RefSeq data set and all Encyclopedia of DNA Elements (ENCODE) data sets were obtained from the UCSC website (http://genome.ucsc.edu/). The HIV-1 integration set was downloaded from the Bushman Lab website (http://microb230.med.upenn.edu/) (28). All data analysis was performed with in-house-written Python scripts (http://www.python.org/). To determine the distribution of LEDGF/p75 in transcription units (TUs), each base of a LEDGF/p75 island was given the overall island score. This score was divided into bins according to their relative distance to the transrciption start site (TSS). If two TUs overlapped in the opposite strand, the same score was added to both bins. If two or more TUs overlapped in the same strand, the weight of the score was divided by the number of TUs and put in its corresponding bin. An identical approach was followed for the HIV integration sites. In this case, each integration site received a unit weight. To analyze the distribution of LEDGF/p75 around the TSS the same approach was used. In case an LEDGF/p75 positive base was inside one TU and directly upstream of a second TU, the weight of a base was assigned to the former TU since LEDGF/p75 islands do not overlap with TSSs.
Correlation with transcriptional activity
Two publicly available HG-U133A Affymetrix HeLa cell line expression array data sets (GSM156764 from data set GSE9750 and GSM156764 from data set GDS2623) were obtained from the NCBI GEO data set browser (www.ncbi.nlm.nih.gov/geo/). Model-based expression index (MBEI) values were calculated as a measure of the average expression level of individual genes in HeLa cells using the dCHIP algorithm for the combined data sets (38). For each represented gene, the highest expression index was included for further analysis. Genes present in the ENCODE regions were subsequently binned in function of their expression level and the percentage of LEDGF/p75 containing genes was calculated for each bin.
Cross-correlation with ENCODE regions
To calculate the cross-correlation curves between an ENCODE track of interest and the LEDGF/p75 track, the LEDGF/p75 track was shifted along the ENCODE track from −10 kb to +10 kb and the cross-correlation was calculated for each 250 bases (lags) (See Supplementary Figure S3). For cross-correlation with the HIV-1 integration sites, the integration data were blurred to the mean size of the LEDGF/p75 island with scores declining from the island center to the borders. To test for significance, 600 random LEDGF/p75 and integration tracks of equal size as the observed tracks were generated. To generate random LEDGF/p75 islands tracks, islands of the same size and MAT score were picked at a randomly chosen location in the ENCODE region. To generate the matched HIV-1 integration site tracks for each experimental integration site at an observed distance from the cloning restriction site, a control site was selected within the ENCODE regions at the same distance of a randomly chosen restriction site. The 2.5 through 97.5 percentile interval for control Pearson’s cross-correlation coefficients with the ENCODE track of interest was calculated at lag 0. Coefficients of random tracks at lower or higher lags did not statistically differ from lag 0 in a few representative trials and were further omitted to minimize processing time.
Cytological analysis of HeLaP4 and Jurkat cells
Genomic DNA from Jurkat, HeLaP4 and from peripheral blood mononuclear cells from a healthy individual were extracted and labeled with Cy5 or Cy3 using the Bioprim Labeling kit (Invitrogen) according to the manufacturer’s protocol. Probes were hybridized on Syndrome Plus v2 array chips (Oxford Gene Technology, Yarnton, UK) using the Oligo aCGH hybridization kit (Agilent) according to the manufacturer’s protocol. Chips were scanned on an Agilent microarray scanner and analyzed using Agilent Feature Extraction software and Cytosure software (Oxford Gene Technology). Cy5-labeled Jurkat or HeLaP4 DNA was analyzed together with Cy3-labeled control DNA on the same chip. The analysis for each cell line was repeated with switched dyes.
RESULTS
Expression and functional validation of the EcoDam-LEDGF/p75 fusion protein
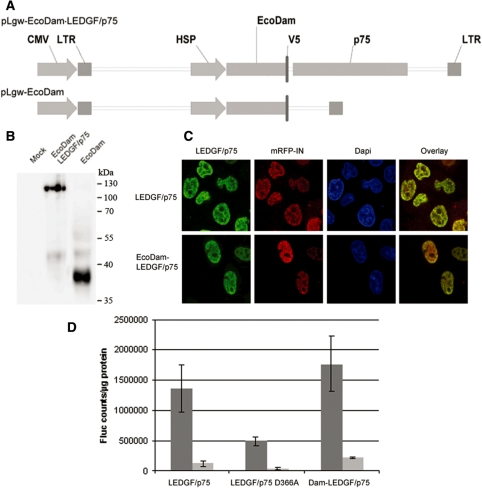
To determine the LEDGF/p75 chromatin interaction profile we used DamID technology, which relies on the expression of a fusion between the EcoDam and a chromatin binding protein of interest (36,39). Chromatin binding results in methylation of neighboring GATC sites that are specifically amplified. The resulting probes are used for microchip analysis. Expression of the EcoDam-LEDGF/p75 fusion protein is driven by an inducible HSP promoter and encoded in a lentiviral vector (Figure 1A). Stable HeLaP4 cells were generated (referred to as HeLaP4 EcoDam-LEDGF/p75) together with an EcoDam cell line (HeLaP4 EcoDam) to control for aspecific methylation. Under normal cell culture conditions (37°C), the HSP promoter hardly supports expression of the EcoDam-LEDGF/p75 fusion or EcoDam protein. Low expression minimizes aspecific chromatin binding and limits the effect of the exogenous protein on the cellular environment. Accordingly, expression in these cell lines was undetectable via immunocytochemistry or western analysis (data not shown). However, transient transfection of lentivector transfer plasmids could confirm expression of EcoDam-LEDGF/p75 and EcoDam by western blot analysis (Figure 1B). EcoDam-LEDGF/p75 displayed the typical, dense, fine-speckled subnuclear distribution pattern of wild-type LEDGF/p75 as observed by confocal microscopy (Figure 1C). Moreover, like wild-type LEDGF/p75, EcoDam-LEDGF/p75 co-localized with HIV-1 integrase (23). Full functionality of the EcoDam-LEDGF/p75 fusion protein was evidenced in an HIV-1 rescue experiment (Figure 1D). HeLa CCR5 A3 cells stably depleted for LEDGF/p75 (34) were back-complemented with the EcoDam-LEDGF/p75 expression construct. As a control, wild-type LEDGF/p75 and a non-interacting LEDGF/p75 mutant (D366A) (40) were used to back-complement the cells. The EcoDam-LEDGF/p75 fusion protein could rescue infection with HIV-1 Fluc to the same extent as wild-type LEDGF/p75.
Figure 1.
The EcoDam-LEDGF/p75 fusion protein behaves like endogenous LEDGF/p75. (A) Schematic representation of the pLgw-EcoDam-V5-LEDGF/p75 transfer plasmid expressing the EcoDam-V5-LEDGF/p75 fusion protein, and the pLgw-EcoDam-V5 transfer plasmid expressing the EcoDam-V5 control protein. CMV, cytomegalovirus promoter; LTR, long terminal repeat; p75, LEDGF/p75. (B) 293T cells were transiently transfected with the respective transfer plasmids. Total cell lysate was prepared and analyzed by western blotting using an anti-V5 antibody. (C) HeLa CCR5 A3 cells depleted for LEDGF/p75 (34) were transfected with a LEDGF/p75 (upper panels, green) or an EcoDam-V5-LEDGF/p75 expression plasmid (lower panels, green) together with an mRFP-integrase expression plasmid (red). DAPI was used to stain the DNA. An overlay of the green and red panels is shown. (D) HIV-1 replication is rescued by the EcoDam-LEDGF/p75 fusion protein. HeLa CCR5 A3 cells were transfected with the indicated constructs. The 24-h post-transfection cells were infected with two dilutions of an HIV-1 clone encoding firefly luciferase (NL4.3 fLuc) (dark and light gray). Luciferase expression was measured 48 h post-infection. Fluc counts upon transfection of a control plasmid encoding an EcoDam fusion to CBX1 (51) were subtracted. LEDGF/p75 D366A, HIV integrase interaction deficient LEDGF/p75 (40). The experiment was done in triplicate and repeated four times. A representative experiment is shown. Error bars indicate the SD of the triplicate values.
LEDGF/p75 associates with transcription units, downstream of the transcription start site
LEDGF/p75 chromatin binding regions were determined by hybridization to ENCODE DNA tiling arrays (41). The ENCODE project characterized 44 regions covering about 1% of the human genome, comprising loci of specific interest, such as the HOXA cluster, together with randomly selected regions. More than 200 specific tracks have been published that record distribution of various epigenetic modifications and transcription factor binding sites in these regions, explaining our choice for ENCODE in this study. The correlation of many of these tracks with HIV-1 integration has been studied before (28). We performed and validated three independent DamID experiments in HeLaP4 cells. The MAT algorithm identified 644 LEDGF/p75 chromatin binding islands from the combined data sets and corresponding controls (37). Islands were heterogeneous with regard to their size and LEDGF/p75 binding intensity (MAT score): sizes ranged between 415 and 2941 bases, with an average of 1025.03 and an SD of 361.43 bases; MAT scores varied between 1.82 and 9.89, with a mean score of 3.18 ± 1.05. One by one comparison of the experiments showed an overlap of ±50% of the islands (Supplementary Table S1). Around 25% of the islands were common to all three experiments and around 40% were unique. Different parameters were tested during MAT analysis to define the LEDGF/p75 islands (See ‘Materials and Methods’ section and Supplementary Figure S1). Downstream analysis yielded the same results as the LEDGF/p75 islands used in this article.
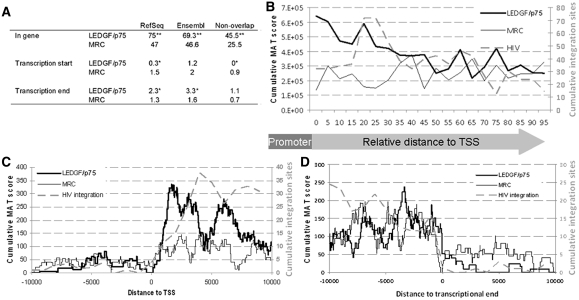
Random control sites were generated computationally. Per experimental site 10 sites were computed, matched with respect to the size and MAT score at a randomly chosen location in the ENCODE region [matched random control, (MRC)]. In the analyses that follow, the distribution of experimental LEDGF binding sites is compared to that of the MRC sites. The total RefSeq TUs coverage of ENCODE regions is 45.7%, which is about 10% higher than for the whole genome. Seventy-five percent of the LEDGF/p75 islands were found in RefSeq TUs as compared to 47% of the MRC islands (Figure 2A). Significantly, fewer LEDGF/p75 islands (0.3%) than random (1.5%) overlapped with the TSS (P < 0.05, χ2-test). Conversely, significantly more LEDGF/p75 islands (2.3%) than random (1.3%) coincided with the transcription termination site (P < 0.05, χ2-test). Similar results were obtained for Ensembl TUs (Figure 2A). Given the fact that many TUs overlap, we repeated the analysis for the 268 non-overlapping genes in the ENCODE regions; none of the LEDGF/p75 islands comprised a TSS (Figure 2A, non-overlap).
Figure 2.
LEDGF/p75 binds TUs downstream of the TSS. (A) Distribution of LEDGF/p75 islands around TUs. The percentage of LEDGF/p75 islands in TUs and overlapping with TSS or transcription ends is shown for RefSeq and Ensembl TUs and for non-overlapping RefSeq TUs. As a control, a random data set of LEDGF/p75 islands was generated 10 times the size of the original data set (MRC). *P < 0.05, **P < 10−27 as compared to random as measured by a χ2-test. (B) LEDGF/p75 binding in TUs was plotted relative to the TSS (LEDGF/p75). All genes were divided in 20 equal bins and the MAT score of each LEDGF/p75 positive base was assigned to the corresponding bin. For each LEDGF/p75 island, 10 new islands were picked at a random position within the ENCODE region having the same size and score as the parental island (MRC). As for LEDGF/p75 islands, the random islands were plotted relative to the TSS. Values were divided by 10 for comparison with the LEDGF/p75 islands. HIV-1 integration sites in TUs (28) were assigned to the corresponding bin relative to the TSS (HIV integration). If LEDGF/p75 or integration sites could be assigned to more than one TU, the weight was divided over all TUs. (C) Distribution of LEDGF/p75, MRC and HIV-1 integration sites around the TSS. Scores for LEDGF/p75 and random islands were accumulated for each base in a 20 kb window around the TSS. All TUs were analyzed in 5′–3′ orientation. Data values of the random curve were divided by 10 to be comparable with LEDGF/p75 islands. For HIV-1 integration sites, the 20 kb window was divided into 1-kb bins. Each integration site was assigned to the corresponding bin. Since LEDGF/p75 islands almost never overlap with the TSS, a positive base found in the TU was fully assigned to this TU and not divided over potential neighboring units. (D) Distribution of LEDGF/p75, random islands (MRC) and HIV-1 integration sites around the transcriptional end. Analysis was performed as described in C.
Taken together, these data demonstrate that LEDGF/p75 preferentially associates with TU downstream of the TSS. Interestingly, 86% of the HIV integration sites in the ENCODE region were also located in a TU (28). Significantly, fewer RefSeq TUs were bound by LEDGF/p75 (28.4%) than expected from MRC (37.9%) (P < 0.0001, χ2-test), suggesting that LEDGF/p75 preferentially targets selected subsets of genes. However, gene ontology analysis did not show significantly favored or disfavored functional GOstat categories in comparison with all ENCODE genes (42). Furthermore, the GIMSAM search engine failed to detect a binding site that was significantly enriched as compared to MRC (43).
To take a closer look at the distribution of LEDGF/p75 binding sites within TUs, LEDGF/p75 islands were binned relative to their position in the TU and the cumulative MAT score for each bin was plotted (Figure 2B). The probability of LEDGF/p75 binding gradually decreases towards the transcription termination site. The downward trend was also observed not taking into account the MAT scores, but only the presence of LEDGF/p75 (data not shown). Linear regression qualified this negative trend as significant (P < 0.0001, Fisher’s z). Analysis of the mean MAT score per bin or the distribution of island sizes over the TU did not show significant differences (P > 0.1, Fisher’s z). This trend was not observed for MRC islands too (P = 0.13, Fisher’s z). As a consequence, the amount of islands is significantly higher at the 5′-site of TUs. Next, we binned MAT scores in function of their absolute distance to the TSS and plotted the MAT score sums for the bins within 10 kb around the TSS. The probability of LEDGF/p75 binding is low upstream of the TSS (Figure 2C), but rises steeply immediately downstream of the TSS. Wherever islands mapped to multiple, partially overlapping genes, we preferentially assigned islands to genes that completely encompass the island, because unequivocally assigned LEDGF/p75 islands almost never include a TSS. As a consequence, the MAT score sums of the MRC islands also increased downstream of the TSS. Importantly, while the profiles are similar upstream of the TSS and near the TSS, the rise of the LEDGF/p75 profile downstream of the TSS is about three times higher than that of MRC, again indicating an enrichment of LEDGF/p75 downstream of the TSS. Since the probability of LEDGF/p75 binding decreases towards the transcription termination site, LEDGF/p75 binding around the transcriptional end was indistinguishable from MRC (Figure 2D). In conclusion, LEDGF/p75 preferentially associates with chromatin downstream of the TSS and binding decreases towards the transcriptional end.
LEDGF/p75 chromatin binding correlates with transcriptional activity of the transcriptional unit bound
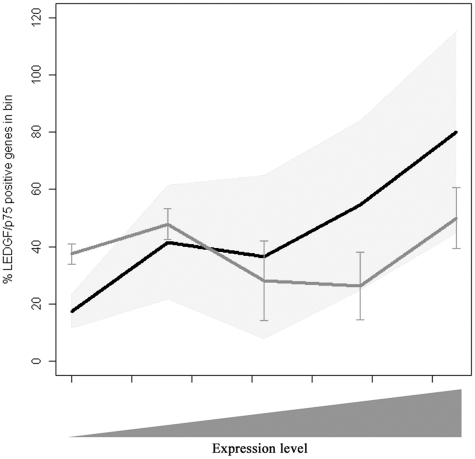
HIV-1 integration correlates positively with transcriptional activity of the targeted TU (26). To study a possible association between transcriptional activity (derived from the NCBI GEO data set) and LEDGF/p75 chromatin binding, ENCODE genes were binned in function of their transcriptional activity and the percentage of LEDGF/p75 positive genes for each bin was calculated (Figure 3). LEDGF/p75 binding was shown to correlate significantly with gene activity (multinomial Fisher’s exact test, P < 0.05), while no correlation was demonstrated for the MRC (P = 0.8).
Figure 3.
LEDGF/p75 chromatin binding correlates with transcriptional activity. Genes in the ENCODE region were divided into bins according to their expression level. The percentage of LEDGF/p75 positive genes in each bin is shown in black. The 95% confidence interval is shown (gray shading). As a control, 10 MRC sets of equal size as the original data were computed and divided over the bins (gray line). The mean ± SD is shown.
HIV-1 integration is targeted to LEDGF/p75 chromatin binding islands
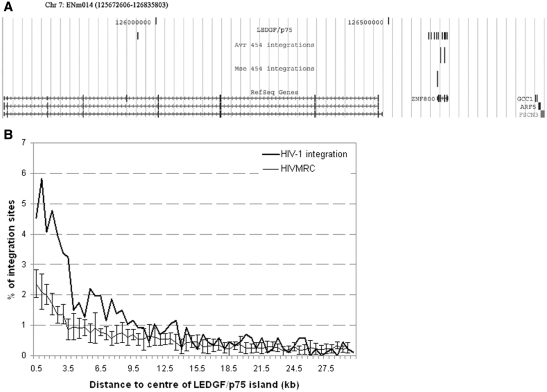
HIV-1 integration is targeted to active TUs, and knockdown or knockout of LEDGF/p75 has been reported to disrupt this targeting preference (27,29,30), suggesting a direct role for LEDGF/p75 in HIV-1 targeting. In addition, by replacement of the N-terminal chromatin binding domains of LEDGF/p75 by alternative chromatin binding domains, retargeting of viral integration was recently achieved (34,44,45). More than 800 HIV-1 integration sites were identified in ENCODE regions in Jurkat T cells (28). A quantitative model that predicts HIV-1 integration sites based on genomic features determined several ENCODE regions to contain more or less integration sites than expected (28,46). For instance, ENCODE region ENm014 on Chromosome 7 carries three integration sites, all located within the same TU (ZNF800; Figure 4A). Strikingly, we also found a major cluster of LEDGF/p75 islands in ZNF800. For a more exhaustive study of the association of HIV-1 integration sites and LEDGF/p75 chromatin binding sites, we generated MRC integration sites (HIV MRC). To this end, for each experimental integration site at an observed distance from the cloning restriction site, 10 control sites were computed within the ENCODE regions each at the same distance of randomly chosen restriction sites. This approach controls for recovery bias due to cleavage by restriction enzymes (46,47). Of 861 HIV-1 integration sites, 4.65% were found in the LEDGF/p75 islands, as compared to only 2.43 ± 0.45% of the HIV MRC sites (P < 0.01, Fisher’s exact test). We then assigned HIV integration sites to the nearest LEDGF/p75 island and computed the percentage of integrations within absolute distance bins from the center of the LEDGF/p75 islands (Figure 4B). Zooming in on the center of LEDGF islands (<3.5 kb from the center), HIV-1 integrates three times more frequently near the center of LEDGF/p75 islands than HIV MRC. Beyond 3.5 kb from the island center, the enrichment attenuates and becomes indistinguishable from that of HIV MRC, around 10 kb from the center. Since the HIV integration data and the LEDGF/p75 chromatin binding data were obtained in two different cell lines, possible biases due to a different expression profile or different chromosomal content of the cells had to be controlled for. To analyze whether different expression levels could influence the results, HeLa and Jurkat expression profiles were derived from the NCBI GEO data set and compared. Chromatin regions with genes showing more than 10-fold difference in expression were omitted from the analysis. Also, for this data set, integration preferentially took place in the neighborhood of LEDGF/p75 islands, ruling out that observed differences were due to different expression profiles (Supplementary Figure S2a). To analyze the status of the ENCODE regions in Jurkat and HeLaP4 cells, genomic DNA was extracted and analyzed on OGT Syndrome Plus v2 array chips. These chips are especially designed to detect genome-wide chromosomal aberrations. The results showed that Jurkat cells are near diploid. Only one of the 44 ENCODE regions contained a small deletion. HeLa cells contain more chromosomal aberrations although at least one copy of all ENCODE regions was present. Fourteen out of 44 ENCODE regions showed duplications or deletions in one allele. As most of these regions seemed to be mosaic, it was impossible to determine an exact copy number. For this reason, we removed all non-diploid ENCODE regions and reanalyzed the correlation between HIV integration sites and LEDGF/p75 binding (Supplementary Figure S2b). Also under these conditions, the association of LEDGF/p75 with genuine HIV integration sites remained tighter than that with MRC integration sites.
Figure 4.
HIV-1 integration is targeted to LEDGF/p75 chromatin binding islands. (A) Schematic representation of LEDGF/p75 islands and HIV-1 integration sites in ENCODE region ENm014 (http://genome.ucsc.edu/). This ENCODE region contains significantly less integration sites than predicted (28). All integration events took place around the same TU that also contains LEDGF/p75 islands. GCC1, GRIP and coiled-coil domain containing 1; ARP5, actin-related protein 5 homolog; FSCN3, fascin 3. (B) The region surrounding the middle of LEDGF/p75 islands was divided in 0.5 kb bins. Integration sites were assigned to the closest LEDGF/p75 island and placed into the corresponding bin. The percentage of integration sites in each bin is shown (bold line). Ten data sets of matched random integration sites (HIV MRC) of equal size as the HIV-1 integration data set were generated and assigned to the corresponding bin (light line). The SD is shown.
Association of LEDGF/p75 with markers of active transcription
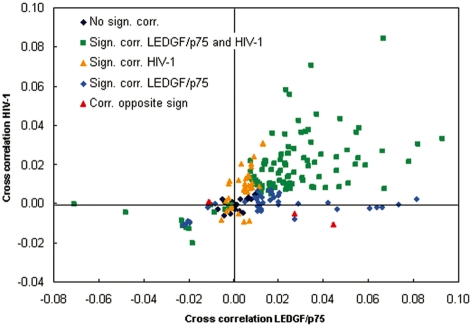
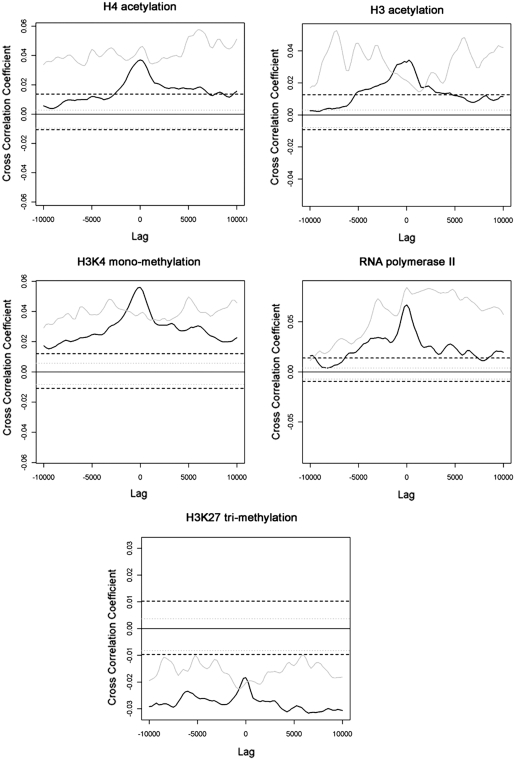
The correlation of HIV-1 integration with ENCODE tracks has been studied before (28). As a logical next step, we performed a similar analysis for the LEDGF/p75 chromatin binding sites. We quantified the cross-correlation of the observed LEDGF/p75 binding sites, known HIV-1 integration sites, and 600 MRC or HIV MRC tracks with 215 ENCODE tracks (41). Sixty-two tracks did not correlate differently with LEDGF/p75 binding sites or the MRC; for 126 tracks a significantly positive and for 27 a significantly negative correlation was obtained. Likewise, HIV-1 integration sites tallied, respectively 69, 125 and 21 tracks in these categories. Sixty-two percent of the ENCODE tracks scored significant correlation coefficients of the same sign for LEDGF/p75 binding and HIV-1 integration sites (green squares; Figure 5), and only 1.8% scored significant correlation coefficients of the opposite sign. This analysis corroborates the proposed role of LEDGF/p75 in targeting of HIV-1 integration. An overview of all tracks can be found in the Supplementary Data (Supplementary Table S2). Cross-correlation enables comparison between corresponding chromatin features, but also between shifted ones as revealed by cross-correlation curves (Supplementary Figure S3). As shown in Figure 6, LEDGF/p75 binding correlated positively with markers of euchromatin and transcriptional activation such as H3 and H4 acetylation, H3K4 monomethylation and RNA polymerase II binding, and negatively with determinants of heterochromatin such as H3K27 trimethylation. Remarkably, although HIV-1 integration also positively correlates with these markers of active transcription as published before (28), HIV-1 integration appeared to be scattered around the LEDGF/p75 peak (Figure 6).
Figure 5.
Cross-correlation of LEDGF/p75 islands and HIV-1 integration with ENCODE tracks. Scatter plot of the correlation of each of the 215 ENCODE tracks with LEDGF/p75 islands and HIV-1 integration sites. Significance was tested by calculating the 2.5 through 97.5 percentile interval of Pearson’s correlation coefficients for 600 HIV-1 and LEDGF/p75 matched random controls with each ENCODE track of interest as described in the ‘Materials and Methods’ section. Most ENCODE tracks with a positive correlation with HIV-1 integration, positively correlate with LEDGF/p75 binding (in green). Black, no correlation with HIV-1 or LEDGF/p75 track; orange, positive or negative correlation with HIV-1 integration, no correlation with LEDGF/p75 binding; blue, positive or negative correlation with LEDGF/p75 binding, no correlation with HIV-1 integration; red, correlation of the HIV-1 and LEDGF/p75 track with the ENCODE track have the opposite sign.
Figure 6.
Shifted cross-correlation of LEDGF/p75 islands and HIV-1 integration with ENCODE tracks. The Pearson’s correlation coefficient between LEDGF/p75 (black) or HIV-1 integration sites (gray) and the indicated ENCODE track was calculated for each lag. Lags indicate the distance the LEDGF/p75 or HIV-1 integration track was shifted as compared to the ENCODE track of interest to calculate the cross-correlation coefficient. Both for LEDGF/p75 islands and HIV-1 integration sites, 600 random tracks of the same size were generated. The correlation coefficient at lag 0 of the depicted ENCODE track with each random track was calculated. The 2.5 through 97.5 percentile interval fell between the dashed lines (black, LEDGF/p75; gray, HIV-1 integration sites). Histones 3 and 4 (H3 and H4) acetylation and Histone 3 Lysine 4 (H3K4) monomethylation are epigenetic markers of active transcription. Histone 3 Lysine 27 (H3K27) trimethylation is a marker of heterochromatin.
Differential association of LEDGF/p75 and HIV-1 integration with ENCODE tracks
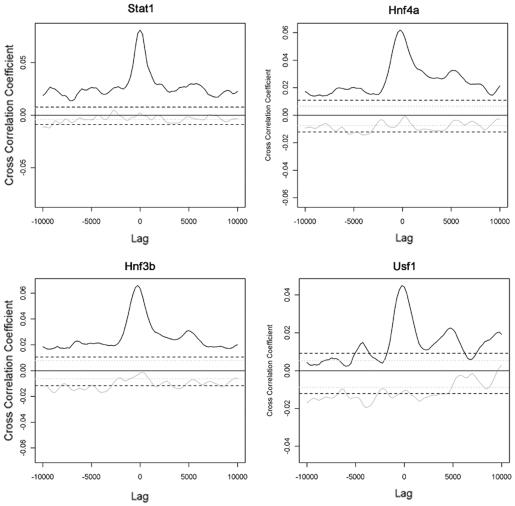
Interestingly, some ENCODE tracks such as Stat1 (signal transducer and activator of transcription), Hnf4a, Hnf3b and Usf1 chromatin binding correlated strongly with LEDGF/p75, but not with HIV-1 integration (Figure 7). After viral induction of interferon-γ, Stat1 is known to translocate from the cytoplasm to the nucleus, where it binds and activates target genes (48). Stat1 is strongly associated with LEDGF/p75 binding but not with HIV-1 integration (Figure 7, upper left panel). The same goes for the functionally related transcription factors, Hnf4a, Hnf3b and Usf1 (49), even though the coefficient of correlation of LEDGF/p75 with these factors is an order of magnitude lower than that between Hnf3b and Hnf4a or Usf1 (data not shown). These examples document for the first time that not all LEDGF/p75 complexes bound to chromatin are accessible for the HIV-1 pre-integration complex to support integration or that integration at these positions is restricted.
Figure 7.
Divergent cross-correlation of LEDGF/p75 islands and HIV-1 integration with specific ENCODE tracks. For an explanation of the cross-correlation curves, see legend to Figure 6. The ENCODE tracks shown have a strong positive correlation with LEDGF/p75, but do not correlate with HIV-1 integration. Stat1, signal transducer and activator of transcription; Hnf3b, hepatocyte nuclear factor 3-β; Hnf4a, hepatocyte nuclear factor 4-α; Usf1, upstream transcription factor 1.
DISCUSSION
Research hints at a role of LEDGF/p75 in stress response [See (12) and references therein]. LEDGF/p75 expression is induced by oxidative stress and LEDGF/p75 itself is believed to activate stress-related genes by binding stress response and heat shock-related elements (4). In this work, we defined the LEDGF/p75 chromatin binding profile in the ENCODE region using DamID. DamID technology is extensively used in Drosophila genomics and is as powerful as ChIP-on-chip to determine chromatin binding profiles (50). While the resolution is the same as for ChIP-on-chip (1–2 kb), DamID has the advantage that neither antibody nor cross-linking is needed. Furthermore, expression of the Dam fusion protein at extremely low level prevents aspecific and saturating methylation levels, as well as disturbance of the normal cellular physiology.
Analysis of the resulting LEDGF/p75 binding islands revealed that 75% of LEDGF/p75 islands were located in TUs although the promoter region itself was disfavored. TUs are not enriched for ‘gatc’ sites as compared to the whole ENCODE region (P < 0.05, Mann–Whitney U-test). Hence, enrichment of the LEDGF/p75 binding islands within TUs cannot be attributed to the ‘gatc’ dependence of the DamID technology, but is indicative of genuine LEDGF/p75 enrichment in these regions. LEDGF/p75 binds downstream of the TSS with a frequency decreasing towards the end of the TU. These results seem at odds with our current understanding of the cellular function of LEDGF/p75, since an activator of expression of stress response genes would be expected to be enriched at promoter sites. Though some studies indicated that LEDGF/p75 binds specific sequence elements associated with the promoter regions of stress response genes (4,6,31,32), others failed to detect sequence-specific LEDGF/p75 binding activity in vitro (14). It can however not be excluded that LEDGF/p75 binds stress response elements during stress conditions. Of note, the ENCODE region does not contain stress-responsive genes. A future DamID experiment with stress-induced cells could shed light on this issue. Nevertheless, at this stage our results demonstrate that LEDGF/p75 chromatin binding is not limited to stress response genes since it amounts to 28% of the ENCODE RefSeq genes. Although the proportion of LEDGF/p75-bound genes might be underestimated due to the strict threshold used to analyze the microarray data, this is significantly less than the 37% for the corresponding control set, suggesting that LEDGF/p75 targets only a specific subset of genes. Though we have to take into account the relatively small amount of genes present in the ENCODE region, Gene Ontology analysis did not support any significant functional enrichment for certain gene product characteristics. Twenty-five percent of the LEDGF/p75 islands located outside of TUs. The mean score and size of this fraction is indistinguishable from that of all islands suggesting that these represent genuine LEDGF/p75 islands. In addition, the mean distance of this fraction to TUs does not differ from random (data not shown). The function of those islands remains to be investigated. At this stage, one can only speculate that these islands may have an important function in the establishment of latent lentiviral proviruses.
As expected and based on the known association between LEDGF/p75 binding and HIV-1 integration, the LEDGF/p75 chromatin interaction profile is reminiscent of that of HIV-1 integration (28). Much alike HIV-1 integration, LEDGF/p75 binding prefers the body of genes, disfavoring the promoter regions and correlating with transcriptional activity. Compared to 75% of the LEDGF/p75 islands, 86% of the ENCODE region-associated HIV integration sites were found in a TU and 30% of all integration sites were found in a window of 3.5 kb around the center of an LEDGF/p75 island, which amounts to a more than 3-fold enrichment over control integration sites. These data corroborate that LEDGF/p75 plays a role in HIV-1 targeting. This apparent window might however be influenced by the resolution and sensitivity of the DamID technology. Moreover, our DamID experiments were carried out in HeLaP4 cells, while the integration data set was derived from a Jurkat T cell line. Nevertheless, controlling our analysis for different expression profiles or chromosomal content between both cell lines did not significantly change the obtained results.
The correlation of the majority of the more than 200 studied ENCODE tracks with the LEDGF/p75 binding profile often mirrors that of HIV integration [this article and (28)]. Overall, LEDGF/p75 binding was associated with markers of active transcription like H3 and H4 acetylation, H3K4 monomethylation and RNA polymerase II binding, but correlated negatively with markers of heterochromatin. The cross-correlation curves with LEDGF/p75 binding sites revealed interesting patterns and some striking differences with HIV integration sites. In most cases, ENCODE tracks with a strong correlation with LEDGF/p75 chromatin binding also showed a high coefficient of correlation with HIV integration. While the correlation with LEDGF/p75 peaked over a relatively small window, that with HIV integration was more spread out around the LEDGF/p75 peak, in line with the window of enrichment of HIV-1 integration straddling LEDGF/p75 binding spots. These results indicate that the DamID resolution is high enough for comparison of LEDGF/p75 chromatin interaction with HIV integration and again suggests that HIV integrates in the wide neighborhood of LEDGF/p75 binding.
Interestingly, our data point out that not all chromatin bound LEDGF/p75 supports effective HIV integration. Indeed, the transcription factors Stat1, Hnf4a, Hnf3b and Usf1 correlated well with LEDGF/p75 binding but not with HIV-1 integration. The chromatin binding profile of this integration incompatible LEDGF/p75 fraction is indistinguishable from that of the complete LEDGF/p75 track (data not shown). HIV-1 integrase interacts with the integrase binding domain of LEDGF/p75, which is known to bind as well to other proteins like Jpo2, pogZ, MLL/menin and Cdc7-ASK (7,15–18). It will be of interest to verify whether the chromatin binding of the LEDGF/p75 fraction that is incompatible for HIV integration correlates with the binding profile of one of these alternative partners. Competition in the binding with LEDGF/p75 may abrogate efficient integration.
In conclusion, the LEDGF/p75 chromatin binding profile corroborates the previously claimed association between LEDGF/p75 binding and HIV-1 integration. Still, other determinants seem to play a role since not all LEDGF/p75 sites support HIV integration, and integration can occur at some distance from the actual LEDGF/p75 chromatin interaction spot. Moreover, our data challenge the current concept on the role of LEDGF/p75 in cell metabolism. It is clear that the function of LEDGF/p75 is not restricted to stress response. The more general chromatin interaction profile is compatible with a global role in transcriptional regulation. Of note, LEDGF/p75 was originally identified as a component of the general transcriptional machinery (1). In this regard, it will be of interest to analyze the expression and chromatin interaction profile of LEDGF/p75 in response to cellular stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CellCoVir SBO grant (60813) of the agency for Innovation by Science and Technology (IWT); the Research Foundation Flanders (FWO) (grant G.0530.08); European Commission grant THINC (HEALTH-F3-2008-201032); Mathilde-Krim postdoctoral fellowship from Amfar (to J.D.R.). Funding for open access charge: THINC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rudy van Eijsden of the KULeuven microarray facility for help with the microarray analysis, the KULeuven Cell Imaging Core for use of the confocal microscope, Prof. Peter Vermeersch, Center for Human Genetics, KULeuven for help with the cytogenetic analysis and Prof. Bas van Steensel, Amsterdam Cancer Institute, The Netherlands for providing the DamID constructs and the discussions on obtained results.
REFERENCES
- 1.Ge H, Si Y, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P, Fatma N, Kubo E, Shinohara T, Chylack LT, Jr, Singh DP. Lens epithelium-derived growth factor relieves transforming growth factor-beta1-induced transcription repression of heat shock proteins in human lens epithelial cells. J. Biol. Chem. 2003;278:20037–20046. doi: 10.1074/jbc.M212016200. [DOI] [PubMed] [Google Scholar]
- 3.Sharma P, Singh DP, Fatma N, Chylack LT, Jr, Shinohara T. Activation of LEDGF gene by thermal-and oxidative-stresses. Biochem. Biophys. Res. Commun. 2000;276:1320–1324. doi: 10.1006/bbrc.2000.3606. [DOI] [PubMed] [Google Scholar]
- 4.Singh DP, Fatma N, Kimura A, Chylack LT, Jr, Shinohara T. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem. Biophys. Res. Commun. 2001;283:943–955. doi: 10.1006/bbrc.2001.4887. [DOI] [PubMed] [Google Scholar]
- 5.Singh DP, Ohguro N, Kikuchi T, Sueno T, Reddy VN, Yuge K, Chylack LT, Jr, Shinohara T, Fatma N, Kimura A. Lens epithelium-derived growth factor: effects on growth and survival of lens epithelial cells, keratinocytes, and fibroblasts. Biochem. Biophys. Res. Commun. 2000;267:373–381. doi: 10.1006/bbrc.1999.1979. [DOI] [PubMed] [Google Scholar]
- 6.Brown-Bryan TA, Leoh LS, Ganapathy V, Pacheco FJ, Mediavilla-Varela M, Filippova M, Linkhart TA, Gijsbers R, Debyser Z, Casiano CA. Alternative splicing and caspase-mediated cleavage generate antagonistic variants of the stress oncoprotein LEDGF/p75. Mol. Cancer Res. 2008;6:1293–1307. doi: 10.1158/1541-7786.MCR-08-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahuja HG, Hong J, Aplan PD, Tcheurekdjian L, Forman SJ, Slovak ML. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75-lens epithelium-derived growth factor (LEDGF) Cancer Res. 2000;60:6227–6229. [PubMed] [Google Scholar]
- 9.Grand FH, Koduru P, Cross NC, Allen SL. NUP98-LEDGF fusion and t(9;11) in transformed chronic myeloid leukemia. Leuk. Res. 2005;29:1469–1472. doi: 10.1016/j.leukres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Hussey DJ, Moore S, Nicola M, Dobrovic A. Fusion of the NUP98 gene with the LEDGF/p52 gene defines a recurrent acute myeloid leukemia translocation. BMC Genet. 2001;2:20–24. doi: 10.1186/1471-2156-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morerio C, Acquila M, Rosanda C, Rapella A, Tassano E, Micalizzi C, Panarello C. t(9;11)(p22;p15) with NUP98-LEDGF fusion gene in pediatric acute myeloid leukemia. Leuk. Res. 2005;29:467–470. doi: 10.1016/j.leukres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Ganapathy V, Daniels T, Casiano CA. LEDGF/p75: a novel nuclear autoantigen at the crossroads of cell survival and apoptosis. Autoimmun. Rev. 2003;2:290–297. doi: 10.1016/s1568-9972(03)00063-6. [DOI] [PubMed] [Google Scholar]
- 13.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 14.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1663–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartholomeeusen K, Christ F, Hendrix J, Rain JC, Emiliani S, Benarous R, Debyser Z, Gijsbers R, De Rijck J. Lens epithelium-derived growth factor/p75 interacts with the transposase-derived DDE domain of PogZ. J. Biol. Chem. 2009;284:11467–11477. doi: 10.1074/jbc.M807781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartholomeeusen K, De Rijck J, Busschots K, Desender L, Gijsbers R, Emiliani S, Benarous R, Debyser Z, Christ F. Differential interaction of HIV-1 integrase and JPO2 with the C terminus of LEDGF/p75. J. Mol. Biol. 2007;372:407–421. doi: 10.1016/j.jmb.2007.06.090. [DOI] [PubMed] [Google Scholar]
- 17.Hughes S, Jenkins V, Dar MJ, Engelman A, Cherepanov P. Transcriptional co-activator LEDGF interacts with Cdc7-activator of S-phase kinase (ASK) and stimulates its enzymatic activity. J. Biol. Chem. 2010;285:541–554. doi: 10.1074/jbc.M109.036491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- 19.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 20.Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 21.Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- 22.Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 24.Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 27.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 30.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fatma N, Singh DP, Shinohara T, Chylack LT., Jr Transcriptional regulation of the antioxidant protein 2 gene, a thiol-specific antioxidant, by lens epithelium-derived growth factor to protect cells from oxidative stress. J. Biol. Chem. 2001;276:48899–48907. doi: 10.1074/jbc.M100733200. [DOI] [PubMed] [Google Scholar]
- 32.Sapoznik S, Cohen B, Tzuman Y, Meir G, Ben-Dor S, Harmelin A, Neeman M. Gonadotropin-regulated lymphangiogenesis in ovarian cancer is mediated by LEDGF-induced expression of VEGF-C. Cancer Res. 2009;69:9306–9314. doi: 10.1158/0008-5472.CAN-09-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geraerts M, Michiels M, Baekelandt V, Debyser Z, Gijsbers R. Upscaling of lentiviral vector production by tangential flow filtration. J. Gene Med. 2005;7:1299–1310. doi: 10.1002/jgm.778. [DOI] [PubMed] [Google Scholar]
- 34.Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J. Virol. 2006;80:11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 37.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl Acad. Sci. USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 40.Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 41.ENCODE consortium The. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 42.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 43.Ng P, Keich U. GIMSAN: a Gibbs motif finder with significance analysis. Bioinformatics. 2008;24:2256–2257. doi: 10.1093/bioinformatics/btn408. [DOI] [PubMed] [Google Scholar]
- 44.Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, et al. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc. Natl Acad. Sci. USA. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silvers RM, Smith JA, Schowalter M, Litwin S, Liang Z, Geary K, Daniel R. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum. Gene Ther. 2010;21:337–349. doi: 10.1089/hum.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl Acad. Sci. USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100059. 2006.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Steensel B, Henikoff S. Epigenomic profiling using microarrays. Biotechniques. 2003;35:346–356. doi: 10.2144/03352rv01. [DOI] [PubMed] [Google Scholar]
- 51.Vogel MJ, Guelen L, de Wit E, Peric-Hupkes D, Loden M, Talhout W, Feenstra M, Abbas B, Classen AK, van Steensel B. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006;16:1493–1504. doi: 10.1101/gr.5391806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.