Abstract
We present extensive explicit solvent molecular dynamics analysis of three RNA three-way junctions (3WJs) from the large ribosomal subunit: the 3WJ formed by Helices 90–92 (H90–H92) of 23S rRNA; the 3WJ formed by H42–H44 organizing the GTPase associated center (GAC) of 23S rRNA; and the 3WJ of 5S rRNA. H92 near the peptidyl transferase center binds the 3′-CCA end of amino-acylated tRNA. The GAC binds protein factors and stimulates GTP hydrolysis driving protein synthesis. The 5S rRNA binds the central protuberance and A-site finger (ASF) involved in bridges with the 30S subunit. The simulations reveal that all three 3WJs possess significant anisotropic hinge-like flexibility between their stacked stems and dynamics within the compact regions of their adjacent stems. The A-site 3WJ dynamics may facilitate accommodation of tRNA, while the 5S 3WJ flexibility appears to be essential for coordinated movements of ASF and 5S rRNA. The GAC 3WJ may support large-scale dynamics of the L7/L12-stalk region. The simulations reveal that H42–H44 rRNA segments are not fully relaxed and in the X-ray structures they are bent towards the large subunit. The bending may be related to L10 binding and is distributed between the 3WJ and the H42–H97 contact.
INTRODUCTION
RNA three-way junctions (3WJs) are common elements of structured RNAs that result from the base pairing of three distinct strand segments of the RNA sequence so as to form three helices diverging from a single point in the secondary structure. 3WJs consist of six components: Three helices (designated P1, P2 and P3) and three single-stranded joiner segments (J12, J23 and J31) (1). In most structured 3WJs, two of the helices stack coaxially (Figure 1A). Based on the coaxial stacking patterns and the lengths of the linker segments, 3WJs have been classified into three basic types, A, B and C (1). In type C 3WJs, which are the most common, J31 is longer than J23 and helices P1 and P2 are coaxially stacked. In addition, P1 and P2 are usually directly connected (i.e. J12 has length 0) and P3 is tilted towards P1, close enough for tertiary interactions to form between P1 and P3, some distance away from the junction. The J31 segment generally forms a loop-like structure interacting with the minor groove of P2. In several instances, J31 forms a small hairpin-like RNA motif closed by a single base pair.
Figure 1.
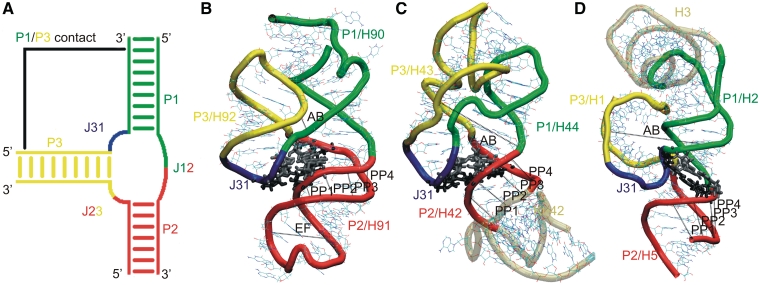
(A) Schematic representation of the components of RNA 3WJs. The P1/P3 contact occurs only in type C 3WJs. Simulated systems: (B) H90–H92 segment of 23S rRNA; (C) H42–H44 segment of 23S rRNA; and (D) H1–H3 and H5 of 5S rRNA. The junction regions are shown in color as in (A). The gray regions in (B) and (C) are included in the simulations, but are formally outside the junction. Black lines (AB, PP1–PP4 and EF) represent the distances describing the motions (see below). Tertiary A-minor I and A-minor II interactions (57,58) (or equivalent interactions) of the junction region (see below) are shown in gray and black licorice representations.
We complement structural studies of RNA 3WJs with explicit solvent molecular dynamics (MD) simulations. While MD is limited by a number of approximations, especially simplifying assumptions of the force field (2–7), it can nonetheless capture the basic intrinsic flexibility of RNA modular motifs and thus help to interpret experimental data (8–17). MD can provide insights into dynamical features of RNA that are not fully apparent from structural studies that typically reveal static averaged structures. Large RNA-based nanomachines like the ribosome work in the regime of high viscosity and very low inertia so that the essential principles of their function are strikingly different from those of macroscopic machines (18–24). They use chemical energy (in the form of GTP) to rectify random thermal fluctuations into directional motion. Thus, during the protein synthesis cycle, each cognate tRNA is transported (along with the bound mRNA) directionally across the interface between the large and small subunits of the ribosome, from the A-site to the P-site and to the E-site. The subunits and their flexible parts also move relative to each other in a coordinated and cyclical manner. These motions are largely driven by stochastic processes, where fluctuations are of utmost importance. MD can shed light on the overall stochastic and anisotropic flexibilities of the RNA building blocks that are important for their function and which usually are not apparent from the structural studies. Simulations can further provide atomistic insights into the structural dynamics (2,4,7,9–11,14,17,25–27).
MD captures motions that occur on the picosecond to sub-microsecond timescale and are therefore subject to low energy barriers. Due to the simulation timescale, simulations of RNA structural modules extracted from larger structures (e.g. 3WJs extracted from the ribosome) show dynamics pertinent to the starting functional geometry of the studied RNA. This geometry may be different from non-functional structures formed in equilibrium solution experiments (28). The fluctuations seen in simulations characterize the intrinsic flexibility of the studied RNA building blocks, which can be used to achieve functional movements. Due to limitations such as uncertainties in the starting structures (which are generally medium resolution, at best) and approximations inherent to the force fields, the simulation results should not be overinterpreted. For the purpose of basic understanding of RNA flexibility, however, the method is robust. We investigate the basic dynamical properties and flexibility of representative RNA 3WJs, specifically type C 3WJ (Figures 1 and 2).
Figure 2.
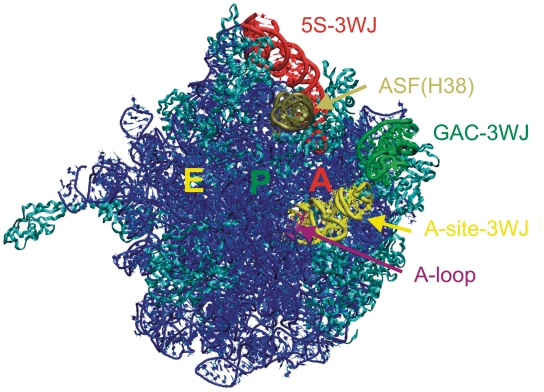
‘Crown view’ of the E. coli 50S ribosomal subunit, viewed from the position of the 30S subunit, with A, P and E tRNA binding sites labeled. In addition to the 3WJs, the A-loop (in purple) and the ASF (comprising H38, in tan) are highlighted. The rest of the RNA is in blue and ribosomal proteins are in cyan.
First, we studied a 76-nt segment containing the 3WJ that comprises helices 90, 91 and 92 (H90, H91 and H92) of 23S rRNA and is located adjacent to the peptidyl transferase center (PTC). H90 corresponds to P1, H91 to P2 and H92 to P3. We call this 3WJ the ‘A-site junction’ because the hairpin loop of H92, the so-called ‘A-loop’ of 23S rRNA, binds the 3′-CCA end of A-site aminoacyl-tRNA (aa-tRNA). H90 is one of the helices composing the multi-helix junction that has been identified as the PTC of 23S rRNA. H90 covalently anchors the A-site junction to the PTC, while H91 stacks coaxially with H90 and projects away from the PTC toward the upper surface of the 50S subunit in the standard view, in which the L1-site, the central protuberance (CP) and the GTPase associated center (GAC) all project upwards (Figure 2). H91 provides a docking site for the short hairpin loop formed by the nucleotides that connect H92 to H90, which corresponds to J31 in Figure 1A. This hairpin loop stacks on H92 and comprises three stacked adenosines (nucleotides 2564–2566 in the Escherichia coli numbering). The conserved bases A2566 and U2562 form a trans H/WC base pair (29) that closes the loop. This short element has also been called H92a. H91 forms several other tertiary interactions, in addition to stacking on H90 and providing a docking site for H92a (J31). The A-site junction provides a potentially flexible point of connection for H92. The A-site loop of H92 helps to position the aminoacyl group in the PTC during the protein synthesis. This is important for robust catalysis (30). The recent crystallographic work suggested that the A-site loop stabilizes the CCA-end of tRNAA and helps to position the aminoacyl group in order to nucleophilically attack the peptidyl tRNA ester (31). Posttranscriptional modification (2′-O-ribose methylation) was detected for U2552 in E. coli (U2587 in Haloarcula marismortui) (32). Such modification may induce a structural change of the adjacent G2553 in E. coli 23S (G2588 in H. marismortui) and may affect the geometry of the A-loop and its interactions with the tRNA in A-site (33). The A-site junction occurs in one of the most crowded regions of the 50S subunit.
We have also investigated the 84-nt RNA element (H42–H44) that includes the 3WJ of the L7/L12 stalk. This junction is present in one of the most exposed protuberances of the ribosome (Figure 2), which undergoes substantial dynamics during the elongation cycle (20,21). The middle part of H42 contains a kink-turn (Kt) (Kt-42). Kts are highly recurrent, flexible hinge-like RNA motifs (14,34–36). The 3WJ is formed by the distal part of H42, H43 and H44. The H43/H44 RNA element comprises the main part of the GAC of 23S rRNA, which binds the GTPase translation factors, including EF-Tu and EF-G at appropriate points in the translation and stimulates GTP hydrolysis in response to appropriate signals to trigger directional motions such as the ratchet-like motion of the subunits (37). Preliminary MD simulation for the H. marismortui H42–H44 element revealed that the H42–H44 23S rRNA region is constructed as a sophisticated RNA-based double-elbow arm (38), which consists of alternating relatively rigid and flexible elements. The first flexible element, closest to the main body of the 50S, is Kt-42, while the more distal one is the 3WJ. The relatively rigid elements are the two arms of H42 flanking the kink and the compact H43/H44 domain (i.e. the GAC per se). In the present study, we decisively extend the simulations of the H42–H44 region, analyze in detail the properties of the 3WJ itself and provide assessment of the effect of binding of the L10 protein to the H42–H44 RNA.
The third 3WJ is found in 5S rRNA. The 5S, albeit being the smallest rRNA molecule, is conserved in all three major phylogenetic domains. It is prominently located at the interface between the large and small subunits (Figure 2), bridging the CP of the 23S rRNA, to which it is attached by ribosomal proteins, with H38. CP and H38 make crucial dynamic contacts to the 30S subunit involved in the ratchet-like motion. The 5S rRNA likely helps to coordinate these motions (39).
MATERIALS AND METHODS
The explicit solvent simulations and analyses were carried out using standard protocols, and the whole method section can be found in Supplementary Data (7,40–51). Most simulations were carried out using the Cornell et al. (52) Parm99 force field (53) and assuming net-neutralizing Na+ atmosphere. This corresponds to ∼0.18–0.25 M cation concentration. Control simulations were also carried out using the Parmbsc0 variant of the force field (54) and using excess salt KCl environment, with cation concentration ∼0.4 M. The simulations are not sensitive to the force field choice and ion atmosphere (see below). For all methodological details including comments on the solute force field and ion atmosphere see Supplementary Data.
List of simulations
Table 1 summarizes key 3WJ simulations (711 ns in total). To avoid cluttering Table 1, some additional 3WJ simulations (totaling ∼250 ns) were not included, but are mentioned in the text or in Supplementary Data (Supplementary Table S1). The fourth column of Table 1 lists the nucleotides included in each simulation, while the sixth column defines the 3WJ regions that are the primary focus of our study. The last column gives the range of instantaneous RMSD between the simulated and starting structures calculated for the 3WJ regions from column 6. Figures 1B–D and 3 summarize 3D and annotated 2D structures of the simulated 3WJs, respectively. The simulations of the A-site 3WJs were carried out without modification of the E. coli U2552 (H. marismortui U2587) nucleotide, as this modification was not expected to substantially affect the overall junction dynamics. However, we have also carried out four simulations (totaling 550 ns) of the A-loop region (H92) of the A-site 3WJ. Two structures included 2′-O-methylated H. marismortui U2587, see Supplementary Data (Supplementary Table S2).
Table 1.
Summary of key simulations carried out in this study
| Name of the simulation | Length (ns) | Organism (PDB code) | Simulated structures (crystal numbering) | Ribosomal helices | 3WJ nucleotides (crystal numbering) | RMSD 3WJa (Å) |
|---|---|---|---|---|---|---|
| Hm_A-site | 100 | H. marismortui (1JJ2) (36) | 2542–2617 | H90–H92 | 2542–2617 | 4.4 ± 0.5 |
| Hm_A-site_H90extb | 30 | H. marismortui (1JJ2) (36) | 2541–2618 | H90–H92 | 2541–2618 | 4.6 ± 0.7 |
| Ec_A-site | 70 | E. coli (2AW4) (62) | 2507–2582 | H90–H92 | 2507–2582 | 4.5 ± 0.8 |
| Ec_A-site_H90extb | 30 | E. coli (2AW4) (62) | 2506–2583 | H90–H92 | 2507–2582 | 4.3 ± 0.8 |
| Ec_GAC_B | 50 | E. coli (2AWB) (62) | 1036–1119 | H42–H44 | 1051–1108 | 1.8 ± 0.2 |
| Ec_GAC_B_ntc | 30 | E. coli (2AWB) (62) | 1036–1119, 2751 | H42–H44 | 1051–1108 | 1.6 ± 0.3 |
| Ec_GAC_4 | 50 | E. coli (2AW4) (62) | 1036–1119 | H42–H44 | 1051–1108 | 1.8 ± 0.2 |
| Ec_GAC_B_ionsd | 50 | E. coli (2AWB) (62) | 1036–1119 | H42–H44 | 1051–1108 | 1.5 ± 0.2 |
| Hm_GAC | 30 | H. marismortui (1JJ2) (36) | 1140–1223 | H42–H44 | 1155–1212 | 2.6 ± 0.3 |
| Hm_GAC_ntc | 30 | H. marismortui (1JJ2) (36) | 1140–1223, 2786 | H42–H44 | 1155–1212 | 2.1 ± 0.3 |
| Hm_GAC_3WJe | 50 | H. marismortui (1JJ2) (36) | 1155–1212 | H42–H44 | 1155–1212 | 2.3 ± 0.3 |
| Hm_GAC_parmbsc0f | 50 | H. marismortui (1JJ2) (36) | 1140–1223 | H42–H44 | 1155–1212 | 2.5 ± 0.3 |
| Hm_GAC_antig | 41 | H. marismortui (1JJ2) (36) | 1140–1223 | H42–H44 | 1155–1212 | 2.0 ± 0.3 |
| 5S_Lh | 50 | H. marismortui (1S72) (91) | 1–75; 106–120 | H1, H2, H3, H5 | 1–28; 54–75; 106–120 | 2.4 ± 0.5 |
| 5S_Si | 50 | H. marismortui (1S72) (91) | 4–21; 59–75; 104–119 | H1, H2, H5 | 4–21; 59–75; 104–119 | 3.2 ± 0.5 |
Some additional simulations are explained in the text.
aInstantaneous RMSD calculated for individual snapshots with respect to the starting structures along the trajectories considering only the 3WJ regions (colored in Figure 1 and listed in the sixth column of this Table).
bOne additional base pair at the end of H90 was included compared to the preceding simulation (U2541/G2618 for H. marismortui and U2506/G2583 for E. coli). This prevents disruption of C2542=G2617 in H. marismortui (C2507=G2582 in E. coli) base pair, which tends to open and consequently interact with G2588 (G2553). G2588 should stay free in MD, because it interacts with tRNA in the ribosomal context.
cOne nucleotide from terminal loop of H97 was included (G2751 and G2786 in E. coli and H. marismortui, respectively). This guanine nucleotide is inserted into the H42 stem and forms key tertiary contact with the otherwise unpaired C1049 (E. coli, cf. Figure 3). This tertiary interaction is CG canonical base pair in the E. coli X-ray structure, while it appears to be rather cis H/WC G2786/C1153 interaction in H. marismortui. The inserted H97 nucleotide in the simulations was modeled according to the respective X-ray structures, with N3-protonated cytosine for the H. marismortui system. The rest of H97 was not included.
dSimulation with excess salt NaCl. The concentration of the Na+ ions was ∼0.35 M. See Supplementary Data for more details.
eSimulation of truncated GAC system; the lower part of H42 including Kt-42 is omitted (Figure 3).
fControl simulation with Parmbsc0 AMBER force field.
gThe geometry of A1189 was changed to anti-conformation instead of the original syn-geometry observed in the H. marismortui X-ray structures.
h5S_L (large)—the structure comprises four helices (H1–H3 and H5). The characteristic P1/P3 contact is formed by cis WC/H A3/G25 and trans SE/SE A3/G21 base pairs (Figure 3).
i5S_S (small)—reduced 5S_L structure with omitted H3. The P1/P3 contact includes only the trans SE/SE A3/G21 base pair, since G25 is now absent (Figure 3).
Figure 3.
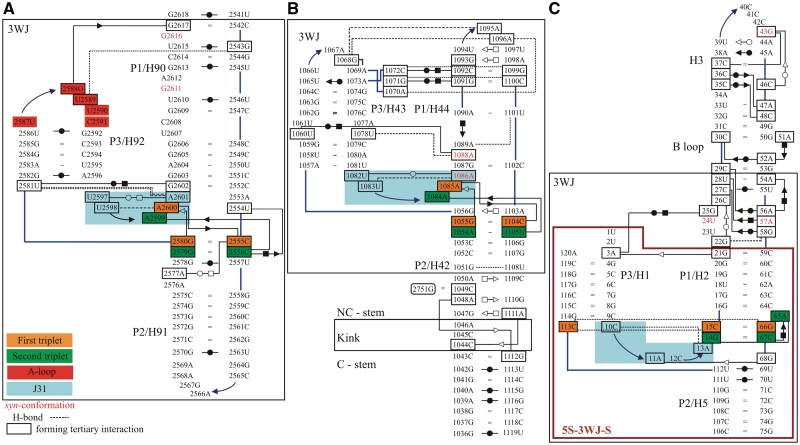
Base pairing in the X-ray structures of the simulated systems using symbols for base pairs by Leontis et al. (29): (A) H90–H92 from the 23S rRNA of H. marismortui (36) (simulation Hm_A-site; cf. Table 1 for the respective simulation names), (B) H42–H44 from the 23S rRNA of E. coli (62) (simulations Ec_GAC_B and Ec_GAC_4) and (C) H1–H3 and H5 from the 5S rRNA of H. marismortui (91) (simulation 5S_L). The large black boxes define the 3WJ parts (3WJ residues, see also Table 1) of the simulated structures. For the H42–H44 system, we also indicate positions of the Kt with its attached C and NC-stems. Control simulation 5S_S was executed for a smaller system, which is marked by the large purple box. The backbone is represented by blue lines, while other single H-bonds are represented by black dashed lines. Residues in black boxes are involved in tertiary interactions. The residues forming J31 parts are demarcated by blue rectangle area. Nucleotides in syn-conformation are written in red. The 3WJs contain essential base triplets, usually represented by A-minor I (first triplet) and A-minor II (second triplet) interactions, which are marked by orange and green rectangles, respectively. In the 5S 3WJ, the A-minor II interaction is replaced by equivalently positioned CCG triple and A-minor I interaction by cis H/SE base pair C67/A65 (Supplementary Figure S2). G2751 in (B) comes from H97 and was included only in some simulations (see Table 1 for detailed description). The E. coli A-site and H. marismortui GAC systems are shown in Supplementary Figure S3. To avoid cluttering, base–phosphate interactions are not shown (92).
Starting structures
All studied structures have the defining characteristics of type C 3WJs (1). Coaxial stacking occurs between P1 and P2 helices (Figure 1). These are H90 and H91 for the A-site 3WJ, H44 and H42 for the GAC 3WJ and H2 and H5 for the 5S 3WJ. In the A-site 3WJ, the characteristic J31 segment (nucleotides H. marismortui U2597–A2601 and E. coli U2562–A2566) interacting with the P2 helix forms a mini-hairpin stem–loop that is closed by a single trans WC/H base pair and a U-turn and therefore resembles a T-loop (55) (Figure 3). In the GAC 3WJ, J31 (nucleotides E. coli U1082–A1086 and H. marismortui C1186–G1190) also forms a mini-hairpin stem–loop, but this one is closed by a single trans WC/WC base pair (Figure 3). It also contains a U-turn motif (56). The J31 loops of both the A-site and the GAC 3WJ contain two conserved stacked adenines forming consecutive cis and trans SE/SE base pairs (also called A-minor type I and II interactions, A-minor motif; Supplementary Figure S1) (57–59) with P2. In the GAC 3WJ of H. marismortui, one of these interactions is not fully formed because A1189 in the J31 loop is modeled in the unusual syn glycosidic conformation instead of anti (38). This might be a refinement error. However, it did not noticeably affect the simulations as the J31/P2 interaction remained stable. Control simulation with anti glycosidic orientation was done. In the 5S 3WJ, J31 comprises nucleotides C10–A13 (H. marismortui) that do not form a characteristic loop structure, while there is only one minor groove interaction there (Figure 3).
The characteristic P1/P3 interactions of type C 3WJ are described in the last column of the Supplementary Table S3. The GAC 3WJ possesses additional P1/P3 contacts (absent in the A-site and 5S 3WJs) further away from the junction involving the middle sections of these helices. In E. coli 23S, these interactions include the cis WC/H C1072/C1092 base pair, the C1072(N4)–G1099(O6) H-bond, the cis WC/H G1071/G1091 base pair, the G1071(O6)–C1100(N4) H-bond and the A1070(N7)–A1096(O2′) H-bond (Figure 3). The equivalent interactions in the H. marismortui structure are given in Supplementary Data.
Parameters for description of hinge- and breathing-like motions of the 3WJs
We selected several distances and angles to monitor the relative motion of the P1 and P3 helices (Figure 1 and Table 2). First, inter-phosphate distance AB reflects motion dominated by interaction between the P1 and P3 stems, which we call breathing-like motion throughout this article. The phosphate atoms defining the AB distance were selected based on visual inspection of the trajectories to include the largest observed fluctuations in the area. All inter-phosphate distances in this article are given as distances from a phosphorus atom center to a phosphorus atom center. Then, the relative motion (hinge-like motion) of the P1/P3 element relative to the P2 stem was described by the ω angle defined by the centers of mass of the following base pairs (considering the base atoms only): (i) the third base pair in P1 above the first triplet; (ii) the C=G base pair in the first triplet; and (iii) the third base pair below the second triplet in P2. In the case of 5S 3WJ, we used the cis WC/WC U70/U111 base pair instead of the C=G base pair of the first triplet. See Table 2 for exact definition.
Table 2.
List of inter-phosphate distances used to describe the main motions of the simulated structures; the last column gives the base pairs defining the ω angle
| 3WJ | AB distance nt(P)–nt(P) | PP1 distance nt(P)–nt(P) | PP2 distance nt(P)–nt(P) | PP3 distance nt(P)–nt(P) | PP4 distance nt(P)–nt(P) | EF distance nt(P)–nt(P) | Hinge angle ω |
|---|---|---|---|---|---|---|---|
| A-site | C2591–G2603 | C2556–G2572 | C2555–G2573 | U2554–G2574 | A2553–C2575 | C2559–A2569 | A2604–U2550 G2580=C2555 G2574=C2559 |
| C2556–U2568 | C2521–U2537 | C2520–G2538 | U2519–C2539 | A2518–C2540 | G2524–A2534 | G2569=C2515 G2545=C2520 C2539=G2524 | |
| GAC | C1182–U1206 | G1210–A1152 | C1209–C1153 | C1208–A1154 | A1207–G1155 | A1194–U1205 G1159=C1208 G1155=C1212 | |
| U1078–C1102 | G1106–A1048 | U1105–C1049 | C1104–A1050 | A1103–G1051 | A1090–U1101 G1055=C1104 G1051/U1108 | ||
| 5S | G8–U18 | U69(P)–C106(C5′)a | G68–C107 | C67–C108 | G66–G109 | C15=G66 U111/U70 C108=G73 |
For A-site and GAC 3WJs, the H. marismortui and E. coli numbers are listed in the upper and lower (italics) rows, respectively.
aC106 is the 3′-end nucleotide of P2, thus, we use C5′ instead of P atom for the PP4 distance.
Further, PP1, PP2, PP3 and PP4 inter-phosphate distances were used to describe the major groove dynamics of the P2 helix. Each PP parameter was chosen to be the shortest distance between phosphorus atoms in the opposite strands of this short RNA duplex over its major groove (Figure 1B–D and Table 2). Finally, EF phosphate–phosphate distance was defined to extract dynamics of the loop in the P2 (H91) in the case of PTC 3WJ (Figure 1B and Table 2). Note that the observed motions are complex and their description by the above-mentioned parameters is simplification.
RESULTS
Hinge- and breathing-like fluctuations dominate in the A-site 3WJ dynamics
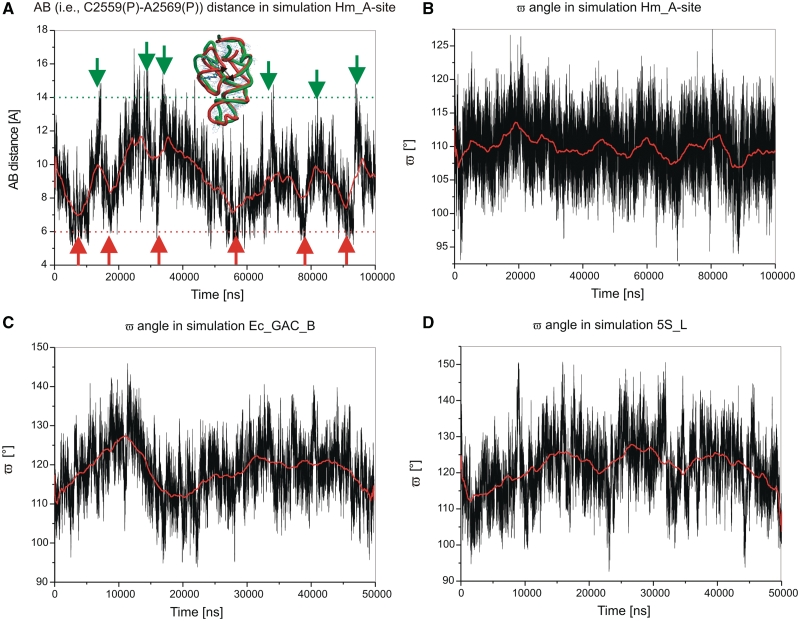
The 3WJ dynamics in the Hm_A-site simulation is characterized by two salient fluctuation motions. Significant fluctuations occur within the P1/P3 stem segment (Figure 4B). The AB distance (Figure 1B) fluctuates between 5 and 17 Å while its X-ray value is 9.0 Å (Table 3, Figure 5A). We call these motions as breathing-like fluctuations, as they modulate the shape of the compact H90/H92 region. This P3 motion is associated with partial opening of the cis WC/WC U2586/G2592 base pair toward the minor groove and opening of the cis WC/WC G2582/A2596 base pair toward the major groove (Supplementary Figure S4). The other base pairs of the P1/P3 region were stable.
Figure 4.
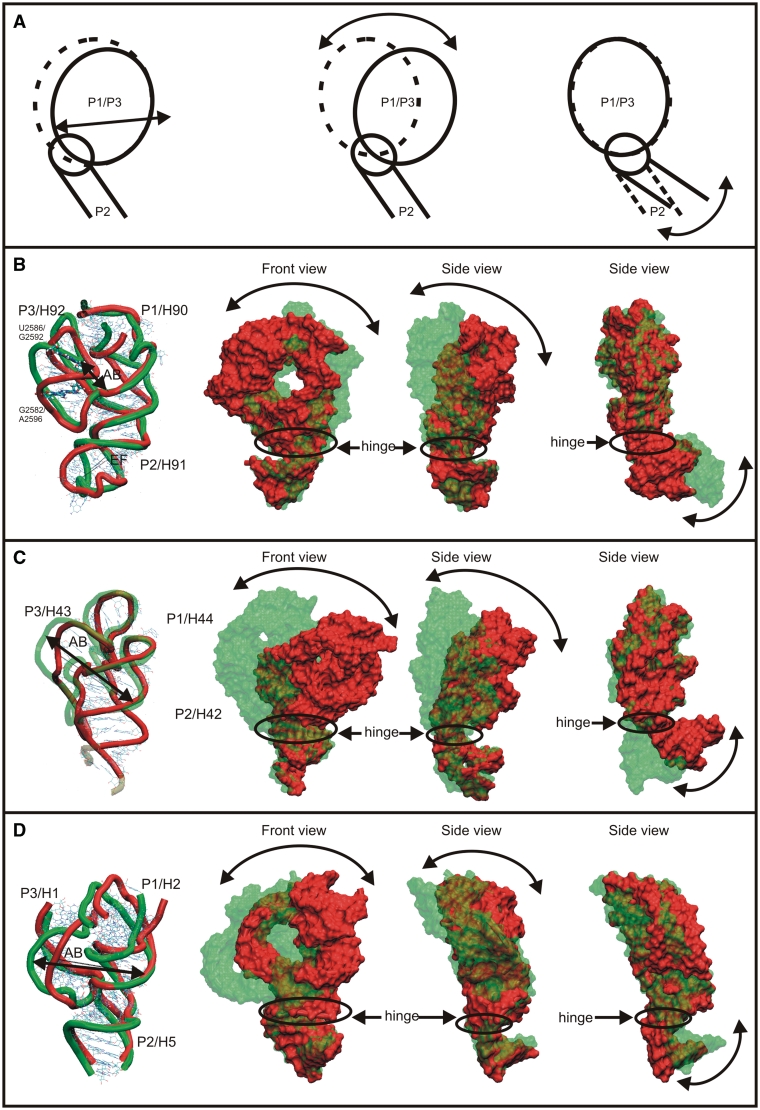
(A) Schematic view of the two dominant motions found in the simulations. (B), (C) and (D) show dynamics of A-site 3WJ (simulation Hm_A-site), GAC 3WJ (simulation Ec_GAC_B) and 5S 3WJ (simulation 5S_L), respectively. Note that (C) includes only the upper part of H42 and does not capture the dynamics of the Kt-42 (14,61). Left—superposition of two substates illustrating the breathing-like motion of P1/P3 regions in MD simulations. The figure is oriented to provide the best view on all three helices. Middle—two views (rotated by 90°) of the overlay of simulated 3WJ structures with maximal amplitudes of the hinge-like motion. The P2 stems are superimposed to visualize the relative motion of the P1/P3 part versus the P2 helix. Right—the same structures as in the middle but superimposed using the P1 helix. PDB files of structures used to visualize the hinge-like dynamics are in the Supplementary Data that also contains graphs showing time courses of representative quantities.
Table 3.
Summary of averaged values, and ranges of measured distances and angles characterizing main movements of studied 3WJsa
| Name of the simulation | Length (ns) | ω (°) |
AB distance (Å) |
RMSD (Å) of 3WJ part | PP1 (Å) |
PP2 (Å) |
PP3 (Å) |
PP4 (Å) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Interval | average | X-ray | Interval | Average | X-ray | Average | X-ray | Average | X-ray | Average | X-ray | Average | X-ray | |||||||||||
| Hm_A-site | 100 | 92–128 | 109 | 109 | 5–17 | 10.6 | 9.0 | 4.4 ± 0.5 | 14.8 | 9.7 | 13.8 | 8.6 | 12.5 | 7.5 | 12.2 | 7.6 | ||||||||
| Hm_A-site_H90ext | 30 | 87–130 | 107 | 109 | 5–17 | 10.8 | 9.0 | 4.6 ± 0.7 | 13.6 | 9.7 | 12.7 | 8.6 | 11.4 | 7.5 | 9.9 | 7.6 | ||||||||
| Ec_A-site | 70 | 87–129 | 108 | 106 | 6–17 | 11.3 | 9.2 | 4.5 ± 0.8 | 17.4 | 10.6 | 14.8 | 9.8 | 12.8 | 8.1 | 11.3 | 7.2 | ||||||||
| Ec_A-site_H90ext | 30 | 90–130 | 107 | 106 | 5–17 | 10.7 | 9.2 | 4.3 ± 0.8 | 14.1 | 10.6 | 13.1 | 9.8 | 11.0 | 8.1 | 11.5 | 7.2 | ||||||||
| Ec_GAC_B | 50 | 93–146 | 118 | 116 | 28–34 | 30.3 | 28.9 | 1.8 ± 0.2 | 15.5 | 9.2 | 18.0 | 9.4 | 19.0 | 10.2 | 16.6 | 9.2 | ||||||||
| Ec_GAC_B_nt | 30 | 94–144 | 117 | 116 | 28–34 | 29.4 | 28.8 | 1.6 ± 0.3 | 13.1 | 9.2 | 15.9 | 9.4 | 17.5 | 10.2 | 15.6 | 9.2 | ||||||||
| Ec_GAC_4 | 50 | 87–150 | 118 | 116 | 28–34 | 29.7 | 28.9 | 1.8 ± 0.2 | 14.6 | 9 | 16.8 | 8.7 | 17.6 | 9.7 | 15.1 | 9.6 | ||||||||
| Ec_GAC_B_ions | 50 | 95–145 | 117 | 116 | 28–33 | 29.6 | 28.9 | 1.5 ± 0.2 | 13.8 | 9.2 | 16.3 | 9.4 | 16.6 | 10.2 | 13.7 | 9.2 | ||||||||
| Hm_GAC | 30 | 101–141 | 121 | 117 | 28–34 | 30.5 | 28.8 | 2.6 ± 0.3 | 12.6 | 9.1 | 14.7 | 10.3 | 17.6 | 10.5 | 16.3 | 9.3 | ||||||||
| Hm_GAC_nt | 30 | 107–147 | 128 | 117 | 28–34 | 30.9 | 28.8 | 2.1 ± 0.3 | 12.9 | 9.1 | 15.5 | 10.3 | 18.5 | 10.5 | 15.8 | 9.3 | ||||||||
| Hm_GAC_3WJb | 50 | 97–150 | 124 | 117 | 27–34 | 29.9 | 28.8 | 2.3 ± 0.3 | N/A | 9.1 | N/A | 10.3 | N/A | 10.5 | 15.6 | 9.3 | ||||||||
| Hm_GAC_parmbsc0 | 50 | 97–149 | 128 | 117 | 24–32 | 28.1 | 28.8 | 2.5 ± 0.3 | 12.9 | 9.1 | 15.3 | 10.3 | 17.3 | 10.5 | 16.7 | 9.3 | ||||||||
| Hm_GAC_anti | 41 | 96–150 | 121 | 117 | 28–34 | 30.0 | 28.8 | 2.0 ± 0.3 | 11.9 | 9.1 | 14.7 | 10 | 17.0 | 11.0 | 15.9 | 9.3 | ||||||||
| 5S_L | 50 | 94–150 | 121 | 117 | 30–40 | 35.9 | 32.5 | 2.4 ± 0.5 | 15.3 | 11.4 | 13.3 | 10.2 | 14.6 | 10.6 | 11.8 | 9.8 | ||||||||
| 5S_S | 50 | 93–150 | 121 | 117 | 30–40 | 35.8 | 32.5 | 3.2 ± 0.5 | 15.8 | 11.4 | 13.3 | 10.2 | 14.5 | 10.6 | 11.5 | 9.8 | ||||||||
See Supplementary Data (Supplementary Figures S7–S22) for development of 3WJ RMSD, ω angle, AB- and PP1-distances for Hm_A-site, Ec_GAC_B and 5S_L simulations.
aSix additional control simulations are given for space reasons in the Supplementary Table S1.
bThe Hm_GAC_3WJ simulation omits a large portion of H42 (Table 1), therefore PP1–PP3 distances are absent in the Table.
Figure 5.
(A) Time development of AB [i.e. C2591(P)…G2603(P)] distance in the Hm_A-site simulation. The geometries in extremes are defined if the AB value is >14 Å or <6 Å; the border is marked in red or green, respectively. (B), (C) and (D) show ω angle in simulations Hm_A-site, Ec_GAC_B and 5S-Ls, respectively. All data were smoothed (in red) taking 5000 consecutive 2 fs points. Many additional graphs are shown in Supplementary Data.
The second motion is a hinge-like fluctuation of P2 relative to the P1/P3 element (Figure 4B). The ω angle describing the position of the P1/P3 helices with respect to the P2 helix was 109° in the X-ray structure and it fluctuated in the course of the simulations in the range of 92° to 128° with the averaged value matching the experimental one (Figure 5B).
Figure 5 demonstrates the fluctuational nature of the motions seen in the simulations. The fluctuations are non-periodic, as expected for stochastic thermal fluctuations. Many more such plots can be found in the Supplementary Data and Supplementary Figures S7–S22. All dependencies suggest non-periodic thermal fluctuations.
Relaxation of inter-phosphate distances
Besides these oscillations, we noticed two unrelated permanent changes. One of them was an increase of the major groove width of H91 (P2) below the junction region, described by the inter-phosphate distances PP1–PP4 (Figure 1B and Table 2). The PP1, PP2, PP3 and PP4 distances were 9.7, 8.6, 7.5 and 7.6 Å, respectively, in the X-ray structure, but after a swift relaxation their averaged MD values increased to 14.8, 13.8, 12.5 and 12.2 Å, respectively. The widening of the groove has no apparent effect on the overall topology of the junction and base pairing.
Small increases of A-RNA major groove widths are common in simulations with the AMBER force field. With the Parm99 and net-neutralizing Na+, we noticed increases of inter-phosphate distances in the range of 1–4 Å in the canonical A-RNA duplexes and 5S rRNA loop E duplex, with respect to several starting oligonucleotide crystal structures (15,27). For these systems, the use of the Parmbsc0 force field and excess salt reduces the inter-phosphate distances by a few angstroms (depending on the system) compared to the Parm99/Na+ simulations. However, then the major groove width often became subtly underestimated compared to experiment (15). Therefore, no unambiguous conclusion could be reached regarding the optimal simulation protocol to reproduce the experimental P…P distances that appear to vary in different experiments.
The major groove widening of H91 in our simulations is larger than for the previously analyzed oligonucleotide X-ray structures. The groove widening contributes to RMSD but does not have any effect on the overall RNA structure, base pairing or tertiary interactions. Similar behavior has been noticed also for some other ribosomal helices when using ribosomal X-ray structures as initial structures for simulation (see blelow). Thus, the experimental P…P distances vary widely, while it appears that some helices in ribosomal structures tend to have narrower major grooves compared to typical values seen in oligonucleotide X-ray structures. We tentatively suggest that in case of the ribosomal helices, the P…P distances could be affected by more substantial overall compensatory effects of positive charges (cations as well as amino acids) in the major groove, i.e. by additional structural context around the RNA—see also discussion section. It is important to point out that the instantaneous P…P distances in simulations are very dynamic, with fast (even sub-nanosecond) thermal fluctuations typically spanning a range of ∼20 Å (cf. Supplementary Figure S9), without being accompanied by any changes of base stacking and pairing. In other words, RNA helical topologies are compatible with a wide range of plausible P…P distances, while the inherently dynamical P…P distances cannot be fully characterized by the averaged (either experimental or MD) single values. Thus, we suggest that the observed expansion of the major groove width does not have serious implications regarding the description of RNA flexibility by our simulations.
Another rearrangement occurred in the upper part of the P1 helix, where G2611 and A2612 irreversibly flipped into the stem. They mutually stacked and G2611 formed new cis WC/WC base pair with U2546. This remote structural development had no effect on the junction. Both bases are flipped out and form tertiary contacts with other parts of the ribosome in the X-ray structures. We further observed fluctuations of the EF distance (Table 2, Figure 1B and Figure 4B). Specifically, there are fluctuations of the terminal loop of P2, its modest rotation (twisting) with respect to the P2 stem, and opening and closing of this loop. This can be considered as the third fluctuation motion of the system, albeit it occurs outside the 3WJ.
Control simulations
We have carried out three additional simulations of A-site 3WJ: 70-ns Ec_A-site simulation of the A-site 3WJ from E. coli, 30-ns Hm_A-site_H90ext simulation and 30-ns Ec_A-site_H90ext simulation. The behavior of all three simulations was almost identical to the first Hm_A-site trajectory (Table 3).
Dynamics of the E. coli GAC 3WJ
The development of simulations of the GAC 3WJs was more complex. Let us first describe the Ec_GAC_B E. coli simulation. We observed similar hinge-like dynamics of the GAC 3WJ as for the A-site 3WJ, albeit with a considerably larger magnitude. The ω angle is 116° in the crystal structure, its averaged MD value is 118° and the range of fluctuations is 93°–146° (Figure 4C and 5C).
The second main fluctuation motion occurred in the P1/P3 and is similar, but not identical, to the A-site 3WJ P1/P3 breathing-like dynamics. This is probably due to the contacts between the middle parts of the P1 and P3 helices absent in the A-site 3WJ. Instead of the significant motion within P1/P3 seen in A-site 3WJ, the GAC 3WJ rather shows backbone deformations of the P1 and P3 helices. The AB distance (Figure 4C and Table 2) fluctuated between 28 and 34 Å while the X-ray value is 30.3 Å (Supplementary Figure S12).
Relaxation of the L11 stalk region
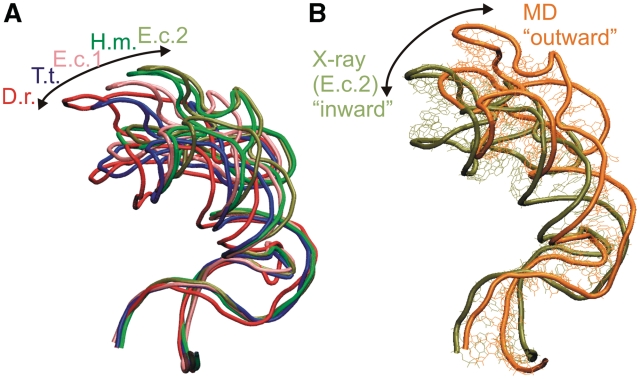
The GAC region (known also as L11 or L7/L12 stalk or protuberance) adopts a set of ‘inward’ and ‘outward’ geometries with respect to the A-site of the large ribosomal subunit in the available X-ray structures (60) (Figure 6A). In line with the literature, the words inward and outward refer to the relative position or movement of the upper part of the H42–H44 rRNA toward and away from the body of the large ribosomal subunit (see Supplementary Figure S28). It was suggested that the observed H42–H44 structural variability may be affected by the hinge-like and twisting flexibility of Kt-42 (14,38,61), by twisting of the lower part of H42 (the C-stem of the Kt; 62), by flexibility of the 3WJ (38), or by non-canonical base pairs and RNA interactions at the base of H42 (60). The experimental structures do not furnish an unambiguous answer (60).
Figure 6.
(A) Superposition of H42–H44 region (including the complete H42 with Kt) from five crystal structures [Deinococcus radiodurans (D.r.)-1NKW (93), T.t.-3D5B (94), E.c.1-2AW4 (62), H.m.-1JJ2 (36) and E.c.2-2AWB (62)] illustrates the range of geometries in the X-ray structures. The body of the large ribosomal subunit, which is not shown is on the left—see Supplementary Figure S28. (B) Comparison of H42–H44 X-ray structure from E. coli. (using the open 2AWB structure) and relaxed averaged geometry (31–40 ns) from Ec_GAC_B simulation. The superposition is done using residues C1044–G1051 and U1108–A1111 in NC-stem of H42. MD simulations prefer more open (outward) geometries compared to the range of geometries seen in the experiment. We were not able to find a clear structural descriptor of the motion, similar to the experimental studies (60). It may be caused by summation of a number of small effects.
In the Ec_GAC_B simulation, the major groove width of the A-RNA between the 3WJ and the Kt increased. The averaged MD PP1, PP2, PP3 and PP4 distances were 15.5, 18.0, 19.0 and 16.6 Å, respectively, while the corresponding X-ray values were 9.2, 9.4, 10.2 and 9.2 Å, respectively. As explained above, some increase in P…P distances is common in simulations and usually not accompanied by any changes of the overall topology. However, in the Ec_GAC_B simulation, the structure relaxes to achieve an even more outward-directed geometry compared to the range of experimental structures (Figure 6B), along the inward–outward path defined by the experimental structures. This indicates that a straightening in the region between the junction and the Kt occurs during the simulation. This behavior suggests that in the initial, observed structure, the RNA is bent compared to its intrinsically preferred geometry, revealed by the simulation. The straightening should be unrelated to the relaxation of the inter-phosphate distances, since the later effect occurs also in simulations with no apparent helix straightening (see the A-site 3WJ simulations above). Unfortunately, it is not straightforward to describe the movement in more detail. The resolution of the X-ray structures is rather low, while the comparison between X-ray and simulated structures is further biased by the relaxation of P…P distances and their large thermal fluctuations (in the range of ∼20 Å) in simulations. It is therefore difficult to make a quantitative superposition of different structures. We suggest that in the experimental structures, the H42–H44 rRNA segment is modestly bent towards the large ribosomal subunit compared to its fully relaxed arrangement due to interactions with surrounding ribosomal elements such as protein L10 (see ‘Discussion’ section). The fully relaxed geometry is then preferred by the simulation of the isolated H42–H44 segment. Most likely, the bending is smoothly distributed over the whole region reaching from the 3WJ down to the Kt so that its localization is difficult. Importantly, the outward shift (opening) of the simulated structure for the E. coli system is not accompanied by increase of the 3WJ bending ω angle. This indicates that the straightening of the structure occurs mostly somewhere below the junction.
Control simulations
Additional simulations Ec_GAC_4, Ec_GAC_B_nt and Ec_GAC_B_ions considered the second independent X-ray structure, insertion of tertiary E. coli G2751 nucleotide from H97 to form canonical base pair with C1049 (Figure 3) and excess salt environment. The junction dynamics, straightening of the whole system and relaxation of P…P distances are basically unchanged (Table 3). Specifically, the excess salt simulation reduced the P…P distances only by ∼2 Å compared to the remaining simulations. Thus the presented results are robust.
To sum up, the stochastic dynamics of the GAC 3WJ from E. coli is similar to the A-site 3WJ, however, the magnitude of the hinge-like fluctuations is considerably larger. The simulations further suggest that the H42–H44 rRNA element is modestly bent by some external force towards the body of the large subunit in the experimental structures.
Dynamics of the H. marismortui GAC 3WJ
We have carried out five 30–50 ns simulations of H42–H44 from H. marismortui (Table 1), which have slightly different behavior compared to the E. coli simulations. The breathing-like dynamics of H. marismortui resembles the equivalent E. coli motion (Table 3). The fluctuational hinge-like dynamics of both systems is also similar. However, the H. marismortui simulations show a somewhat larger initial repositioning of the upper part of the structure (H43/H44 versus H42) followed by usual stochastic 3WJ fluctuations around the equilibrium geometry. The ω angle increased from the X-ray value of 117° to the averaged MD values of 121°–128° (Table 3). Thus, besides straightening of the double helix between the 3WJ and Kt-42, there is also a relaxation localized right at the junction. In the ribosome, this would result in a shift of the GAC rRNA further away from the body of the large subunit. No changes of base pairing were seen in the junction region. A 50-ns simulation with the Parmbsc0 force field did not produce any significant differences in the simulation behavior.
We could not identify the cause of this difference between the E. coli and H. marismortui simulations. We carried out a control simulation of GAC 3WJ from H. marismortui with modeled anti-conformation of A1189 (Hm_GAC_anti) to form a full A-minor I tertiary triplet interaction. The triplet was stable while the dynamics of the structure was very similar to the other H. marismortui GAC simulations. The average ω angle was 121°, i.e. still slightly larger than in the X-ray structure.
Earlier, we reported a single H42–H44 H. marismortui simulation with much larger initial outward relaxation, albeit in the same direction (38). The averaged 3WJ ω angle increased to 148°. The 3WJ then fluctuated around this geometry with a similar range and direction of hinge-like fluctuations as in all other simulations. However, the two G/A base pairs (not optimally paired in the experimental structure) in the NC-stem of Kt-42 were lost (Supplementary Figure S6) and there was an excessive widening of the major groove of the A-RNA region below the 3WJ. We now consider this behavior as a possible but less frequent simulation development. In the Supplementary Data, we present several simulations aimed to better understand its origin. In the ‘Discussion’ section, we suggest that the absence of the surrounding ribosomal elements (such as L10 protein) in the simulation reduces the structural stability of the H42 NC-stem and can occasionally lead to its perturbation in simulations.
Dynamics of 5S 3WJ
We carried out a 50-ns simulation of a fragment of H. marismortui. The 5S rRNA (5S_L) includes the 3WJ and the adjoining helices H1 (P3), H2 (P1) and H5 (P2), as well as part of H3, which is attached to H2. The simulation revealed hinge-like oscillations between the H2/H1 segment and H5 (Figure 4D; cf. also Supplementary Figures S15 and S16) similar to hinge-like motions of the preceding junctions. The ω angle oscillates from 94° to 150° with average value of 121° (Figure 5D), being slightly larger than the X-ray value of 117°. Further, we observed an oscillatory motion within the P1/P3 (H2/H1) element, captured by the AB distance fluctuating in the range of 30–40 Å (the X-ray value is 32.5 Å). It can be described primarily as rearrangement of the backbone path within the P1 (H3) element (Figure 4D). This motion is similar to breathing-like motion of GAC 3WJ. The simulation revealed the usual widening of the major groove of P2 (Table 3) with no visible change of the P2 position. The 50-ns simulation 5S_S confirmed the results.
Essential dynamics analysis and normal mode analysis
The above results are based on careful analysis of full trajectories. However, there are several popular alternative methods to study flexibility of biomolecules (16,48,50,51,63–67). Essential dynamics analysis (EDA) aims to extract significant concerted motions from MD trajectories, i.e. to separate significant motions from noise (16,48). EDA is an implementation of linear principle component analysis (PCA). EDA is thus based on diagonalization of the covariance coordinate matrix, which yields a set of eigenvectors and eigenvalues. The eigenvectors show the directions of motions of the atoms and the eigenvalues correspond to the displacement variances of the particular eigenvectors. Normal mode analysis (NMA) is harmonic vibrational analysis based on the starting (experimental) geometry. The method has two variants. In all-atom NMA, the molecule is described by the full atomistic potential (the same as used in the simulations). The molecule is gradiently optimized to get a local minimum close to the experimental structure and then NMA is performed (63,66,67). Typically, solvent effects are mimicked with a distance-dependent dielectric constant. Alternatively, NMA can be performed within the framework of a coarse-grained (CG) structure–energy description of the molecule (50,51,64–67).
We have applied all these methods to the H42–H44 GAC system. The results are described in more detail in the Supplementary Data. We suggest that for the present systems none of the above methods can replace complete analysis of full-simulation trajectories. The EDA method, although presently widely used as a convenient method to visualize trajectories, is based on substantial approximations inherent to the PCA method, which are often not properly taken into consideration. Thus, EDA is prone to overinterpretation. First, EDA assumes linearity (68). However, the real motions can show substantial non-linear correlations that bias the linear PCA. PCA also assumes that the principal components are orthogonal. In addition, it assumes that the mean and the variance fully describe the data distributions, which requires Gaussian or another exponential type of distribution. If these conditions are not fulfilled, the analysis can be biased. Finally, EDA requires high ‘signal-to-noise’ ratio. Therefore, EDA appears more suitable to analyze trajectories of systems that undergo structural transitions than trajectories aimed to highlight mere stochastic thermal fluctuations of flexible molecules. The analysis should be done by grouping the leading (presumably noise-free) components together to visualize the noise-free dynamics. It is less appropriate (albeit often seen in the literature) to try to interpret the individual modes separately. As shown in the Supplementary Data, although the EDA basically reflects those motions seen in the trajectories, the results are not unambiguous and the individual components indeed do not clearly separate hinge motions from breathing motions. Therefore, EDA cannot replace careful monitoring of the full trajectories.
The all-atom NMA analyzes the motions around the minimum-energy conformation as they would be within harmonic approximation. (In other words, the all-atom NMA is formally equivalent to EDA of simulations with harmonically approximated force field, so the same limitations of EDA mentioned above apply also to all-atom NMA.) In our calculations, all-atom NMA indicates only very limited motions, which in our opinion do not resemble the MD trajectories. With this method, the system is confined tightly to the initial relaxed local minimum structure for which the harmonic vibrational analysis is executed. In addition, harmonic vibrational analysis for such large systems as studied here is very computationally demanding and does not bring any advantage over the full simulations in this respect.
Finally, CG NMA at first sight captured the inward–outward dynamics of the H42–H44 rRNA segment. However, closer inspection (Supplementary Data) revealed that the CG NMA description is incorrect. First, the NMA approach does not capture the initial relaxation of the system, as it treats the starting structure as the minimum. Second, the dynamics predicted by CG NMA is totally unrealistic. It predicts the opening–closing motion to be sharply localized as hinge between just two base pairs in the center of the NC-stem (i.e. the P2 helix) between the Kt and the 3WJ. In simulations, the P2 stem shows smooth distribution of the bending over its entire length with no hinge. In addition, CG NMA depicts the Kt and 3WJ to be the stiffest parts of the structure, while the simulations identify both these elements as very flexible elbow-like elements. This occurs because CG description assumes that the flexibility is a collective property that depends on the overall shape of the molecule and its compactness, but is insensitive to structural details. These assumptions are not satisfied for the present system. Thus, CG NMA predicts the leading modes as hinging and torques localized in the center of the structure, where the structure is the least compact. Flexibility of Kt-42 and 3WJ is missed since these are more compact regions due to the presence of extensive tertiary interactions and bulged nucleotides. Our finding agrees with earlier study by Cui et al. (67) which showed that while CG NMA is useful for globular biomolecules it may fail for non-globular RNA molecules.
Additional results
Supplementary Data describe simulations of H92 analyzing the posttranscriptional modification (2′-O-ribose methylation) in U2587 in H. marismortui (U2552 in E. coli) and analysis of local substates of A-minor II interaction in A-minor motif in A-site and GAC 3WJ simulations.
DISCUSSION
The ribosome is a large biomolecular nanomachine. Stochastic motions and fluctuations represent important components of its functional dynamics (18–24). In biomolecular machines, thermal motions are converted to directional functional movements via release of chemical energy. Thermal fluctuations are mediated by the specific intrinsic flexibilities of the individual molecular building blocks and their dynamical communication with the surrounding elements. Distinct modular RNA 3D motifs possess dynamical and elastic properties ranging from relatively rigid elements with specific shapes (for example, sarcin–ricin motifs) to highly flexible elements that can be considered as genuine molecular elbows (for example, Kts) (11,17,27,38).
MD simulation is essentially a single-molecule modeling technique currently capable of mimicking the 10 to ≥100-ns room temperature dynamics of solvated RNA molecules, utilizing well-defined experimental structures as the starting coordinates. Although the simulations are limited by force field approximations (2,5,7,69–71), they reflect key features of the structural dynamics of RNA modules.
The 10 to ≥100-ns simulations access structural arrangements that are separated from the starting geometry by small energy barriers of the order of the thermal energy. The most extreme simulation fluctuations define the borders of the low-energy region as they correspond to geometries where the energy increases steeply so as to reverse the motion. The observed fluctuations reflect the intrinsic flexibility of the molecule around its starting (experimental) geometry, i.e. the intrinsic low-energy deformation modes of the molecules that can cooperate with the surrounding ribosomal elements to achieve the functional dynamics. Fluctuations occurring in MD simulations were earlier used to derive quantitative sequence-dependent elastic models of double helices (11,72).
Common flexibility of type C 3WJs
We studied three functionally significant ribosomal type C RNA 3WJs (1). The analysis is primarily based on fifteen 30–100-ns 3WJ simulations with a combined length of 711 ns (Table 1) based on the respective X-ray structures of the H. marismortui and E. coli large ribosomal subunits. We have further carried out ∼250 ns of control 3WJ simulations and 550-ns simulations of the isolated A-loop of the A-site 3WJ.
All three junctions possess two salient modes of intrinsic flexibility (i.e. flexibility that is inherent to the particular RNA architecture), which appear to be common for the type C 3WJs. The first one is anisotropic (directional) dynamics, which is best described as hinge-like motion between the P2 stem and the P1/P3 element. The second mode is dynamics within the P1/P3 element, which vary from system to system. We call it breathing-like motion, as the fluctuations modulate the shape of the compact P1/P3 region. It does not result in movements between distal parts of the molecule.
A-site 3WJ
Figure 4B depicts the hinge-like and breathing-like flexibility for the A-site 3WJ, while Figure 2 shows its position in the large ribosomal subunit. H90 attaches the A-site 3WJ to the rest of the ribosome and is embedded in the interior of the 50S subunit. H91 (P2), in contrast, extends upward and toward the GAC, roughly parallel and interacting with both H89, which is also attached to the PTC at its base, and with the H95 containing the sarcin–ricin loop (SRL), which also forms part of the GAC along with the H42–H44 region (Supplementary Figure S26). The A-loop hairpin (the terminal part of H92, i.e. P3) which binds the 3′-CCA end of A-site tRNA is oriented at the PTC, away from P2 and the 3WJ by tertiary interactions between P3 and P1. Figure 4B (right) visualizes the hinge-like flexibility when superimposing H90 (P1) to show the dynamics of P2 (H91) and P3 (H92) with respect to fixed H90. This is probably the most relevant view when considering the attachment of this 3WJ to the ribosome via H90. Accommodation is the process by which the 3′-end of cognate aa-tRNA moves into the PTC and correctly binds to the A-loop after being released by EF-Tu following GTP hydrolysis. Targeted MD simulation study of accommodation of cognate tRNA suggested that the 3′-end of aa-tRNA is guided toward the A-loop by a corridor formed by elements of 23S (73). The tRNA comes consecutively in contact with over 20 universally conserved RNA bases (73). We propose that flexibility of the A-site 3WJ may contribute to smooth cognate tRNA accommodation, while the flexibility may be also tuned so as to suppress near cognate tRNAs’ accommodation, as near cognate tRNAs may use a somewhat different path for accommodation.
The GAC 3WJ
The GAC 3WJ is composed of the H42–H44, with Kt-42 in the middle of H42. The GAC 3WJ and the rest of H42 are the RNA components of the L7/L12 (or L11) stalk. This stalk forms a side protuberance of the large ribosomal subunit (Figure 2 and Supplementary Figure S28), which further includes L10 and L11 proteins to which two or three very dynamical L7/L12 dimeric protein complexes are attached. The L7/L12 stalk interacts with the EF-Tu + GTP + tRNA complex and with EF-G during the proteosynthesis. The GAC 3WJ is rather an external RNA element attached to the body of the subunit via H42. It shows a range of positions in the available X-ray structures (Figure 6) consistent with flexibility inferred from cryo-EM studies (20,21,74).
GAC contact with H89
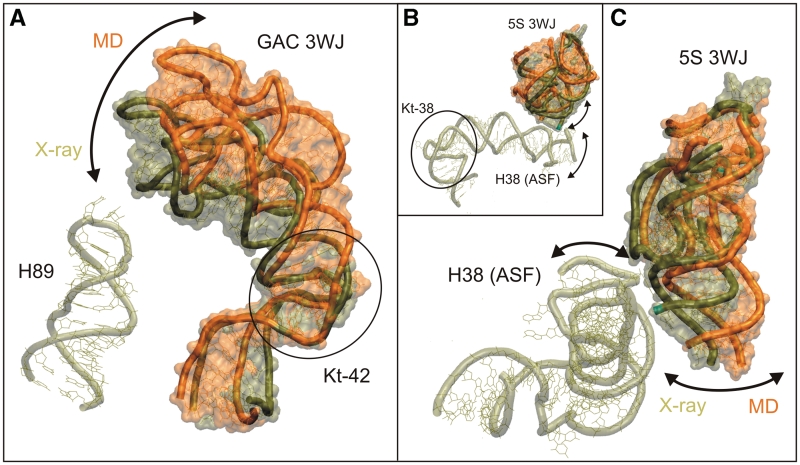
When the H42–H44 domain is in the overall ‘closed’ conformation, the tip of the hairpin loop of H89 can fit into the groove defined by the docking of the hairpin loops of H43 and H44, which lock together to form the compact folded conformation of the H42–H44 domain (Figure 7A). Such contact could mediate dynamical signaling of conformational changes in the ribosome (75), but is only seen approximately and only in the 2AW4 crystal structure of vacant E. coli 70S ribosome, which has no tRNAs present (62). Even in this case, the closest inter-atomic distance observed is only 3.5 Å and involves A1074(O3′) and C2475(O1P) H-bond acceptor atoms (Supplementary Figure S27). In other crystal structures, the distance between the H89 and GAC RNA varies widely (it even exceeds 10 Å in some cases). Nevertheless, genuine 3WJ thermal fluctuations along the path identified in our simulations could lead to dynamical H89/GAC interactions, provided all other interactions are fixed. At first sight, the initial outward relaxation of the structure in simulations (Figure 6) indicates a preferred movement in the opposite direction. This relaxation (see below), however, reflects the lack of the full ribosomal context in the simulation and does not directly include the 3WJ. Thus, assuming absence of this relaxation in the full structural context, 3WJ fluctuations (Figure 4C) could easily reach to H89.
Figure 7.
(A) Comparison of H42–H44 X-ray structure from E. coli [tan, using the more outward 2AWB (62) geometry, see also Figure 6] and the relaxed averaged geometry (31–40 ns, in orange) from the Ec_GAC_B simulation. The superposition is done using residues C1044–G1051 and U1108–A1111 in the NC-stem of H42. MD simulations prefer more open geometries compared to the X-ray structures. Nevertheless, movements toward H89 would be along the low-energy deformation path and thus are easily achievable, especially when assuming that the RNA is restricted by the H42/H97 interaction (Figure 8). (B) Superposition of the 5S RNA junction from the H. marismortui crystal structure 1S72 (91) (in tan) and the averaged geometry (in 22–23 ns, in orange) in the 5S_L simulation done over residues 16–28 and 57–64 in H2. Further, the relative position of ASF (also in tan) with its Kt-38 elbow region is shown. A recent simulation study (90) indicates that significant part of the directional dynamics of the ASF may stem from the flexible Kt-38. The arrows show the fluctuations consistent with simulations of the H38 elbow region and of the 5S rRNA 3WJ. (C) Detailed view on the 5S 3WJ and the bottom part of the ASF. The anisotropic flexibility of the H5 in 5S 3WJ revealed by the MD simulations complements the anisotropic flexibility of the ASF elbow region.
Interaction with H97
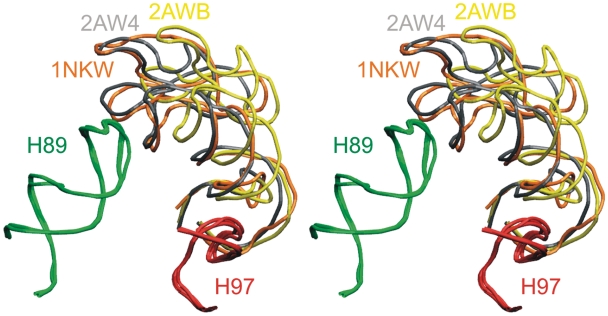
Another interaction that affects the GAC RNA position is the insertion of G2751 in E. coli (G2786 in H. marismortui) from H97 into the NC-stem of H42 right above the Kt-42, where it pairs with C1049 in E. coli (C1153 in H. marismortui) (Supplementary Figure S26). We have carried out a search using Ribostral (76), which shows that this interaction is H42(C)–H97(G) in all three domains of life (for more details see Supplementary Data). The importance of this interaction is also supported by a recent mutational study (77). We could not include this interaction into the simulations. We only carried out simulations with insertion of a single G2751 nucleotide (neglecting its attachment to H97). It does not affect the dynamics of the isolated H42–H44 within the ‘resolution limits’ of our technique. In reality, the full interaction between H97 and H42 may help to fix the overall position of the GAC RNA in 3D space; however, reliable simulation of this situation would not be straightforward. Visual inspection indicates that the H97/H42 tertiary base pairing should primarily restrain movements rather perpendicular to the observed inward–outward (closing–opening) path defined by the experimental structures and preferred by simulations (Figures 6 and 8). The 3WJ is localized above the H42/H97 contact. Thus, the 3WJ intrinsic fluctuations should not be affected by this contact and the 3WJ should provide enough flexibility for the GAC RNA even if the H42/H97 contact is entirely fixed. Indeed, when we use H97 as a fixed reference point, the positional variability of GAC in the available X-ray structures originates above the H42/H97 contact (Figure 8). The experimentally observed positional variability of the GAC RNA (Figure 6) could be due to flexibility of the 3WJ and deformation of the region below it.
Figure 8.
Stereoview of the superimposed H97 (in red) and H89 (in green), together with the GAC position from the crystal structures of T. thermophilus (1NKW, in orange) and E. coli (2AWB, in yellow and 2AW4, in gray) ribosomes. It is notable that the H89 and H97 are almost perfectly superimposable, while the structural difference between the GAC positions appears to originate above the H42–H97 contact point. This clearly indicates flexibility of the GAC 3WJ, entirely in line with the simulation results.
Relaxation of the H42–H44 structure and the effect of L10 binding
The structures of some rRNA elements are deformed compared to their intrinsically preferred geometries by the surrounding ribosomal components (11,38,78). Our simulations indicate that such deformation is present between Kt-42 and the GAC 3WJ. The experimental structures show a wide range of positions (Figure 6), sampling a set of more inward and more outward structures with respect to the A-site of the large subunit and H89. The simulations relax to even more open geometries of the H42–H44 region compared to the experimental structures. We suggest that the deformation (initial bend) of the RNA below the junction is real and is caused by the surrounding ribosomal components. Unfortunately, due to disorder, most of the protein stalk components (mainly the L10 protein that directly interacts with the rRNA and the attached L7/L12 dimers) are invisible for contemporary structural experiments (79). A notable exception is the latest X-ray structure of Thermus thermophilus 70S ribosome, in which the EF-G trapped in posttranslocational state sufficiently immobilizes the L12 proteins to visualize them (80). Unfortunately, this structure does not show the positions of protein side chains, which precludes performing simulations.
There is just one X-ray structure of H. marismortui 50S that captures almost completely the binding of the extended N-terminal domain of L10 to the GAC rRNA. The L10 N-terminal domain reaches from the 3WJ to the Kt-42 (81). The bulged-out base A1150 of the Kt-42 is sandwiched between Ile12 and Arg69 of L10 (81). This specific protein/RNA complex was suggested to be rather weak (81). We have performed one 50-ns and two 25-ns simulations (for more details see Supplementary Data and Supplementary Tables S1 and S5) of the H42–H44 H. marismortui rRNA segment with bound L10 N-terminal domain. In the 50-ns simulation, the rRNA segment remained basically in the X-ray geometry with no initial relaxation (Supplementary Figure S29). The averaged ω angle was 114°, i.e. identical to the value of 114° in this particular X-ray structure. It appears that L10 binding could be responsible for (or significantly contribute to) the observed bend of the H42–H44 rRNA. Interestingly, L10 inserts its Arg63 into the major groove of the RNA to directly bridge the backbones of G1211 and A1152. Consequently, L10 binding improves the agreement between computed and observed P…P distances across the major groove. The X-ray PP1–PP4 distances are 9.4, 10.9, 10.4 and 10.0 Å, respectively, while the averaged MD values are 7.5, 10.5, 13.2 and 12.5 Å, respectively, visibly shorter compared to simulations carried out without the bound L10. Importantly, the L10 binding does not have any effect on the 3WJ dynamics. The range and direction of hinge-like fluctuations as well as breathing-like dynamics are similar as in the other simulations. The two shorter simulations confirm this picture. Inclusion of L10 brings the simulation behavior of the H42–H44 very close to the X-ray values, prevents the initial relaxation of the system, and does not affect the range and directionality of thermal fluctuations of the 3WJ substantially. It is thus possible that structural communication between L10 and the rRNA may contribute also to the flexibility of the rRNA visualized by the X-ray structures (Figure 6).
As noted above, the GAC position is certainly affected by the H42/H97 interaction. However, the position of the H97 with respect to the GAC RNA indicates that it should not cause the observed bending of the GAC RNA (see above and Figure 8). Finally, the L7/L12 proteins may also affect the overall position of the stalk; however, presently available experimental data do not allow one to make any specific suggestions about their role.
The simulation data indicate that the H42–H44 rRNA region is capable of swift large-scale movements along a series of positions from the inward (closed) orientation seen in some X-ray structures to considerably more outward (open) geometries that are beyond the range observed in currently available X-ray structures. Larger outward motions would probably require that some of the interactions described above (such as L10 binding) are either dynamical or temporarily disrupted. GAC rRNA could undergo large-scale, rapid, thermal fluctuations or structural adaptations to facilitate gliding of the tRNA or elongation factors into their functional destinations. The flexibility of the H42–H44 region can also play a role in dynamical signaling throughout the ribosome, for example, via the above-noted H42/H97 and potential GAC/H89 contacts. It may also support functional dynamics of the L7/L12 protein complex. Possibly, the range of experimental GAC positions (Figure 6A) still does not fully capture the range of functional dynamics of the L7/L12 stalk RNA.
5S 3WJ
The 5S rRNA is the smallest ribosomal RNA molecule, bridging the CP and the ASF (H38) of the 50S subunit (Figure 2), which form crucial dynamic inter-subunit bridges (82). Saturation mutational studies suggest that ASF could mediate the allosteric transmission of information from the decoding center via the B1a/B1b bridges to the elongation factor binding site GAC (83,84).
The H1/H2/H3/loop C stem of 5S rRNA is firmly anchored to the 50S CP via extensive interactions with proteins L5 and L18. One side of L5 interacts extensively with H3 (H3 is the continuation of H2, Figure 3) and loop C of 5S rRNA. The other side of L5 interacts with the ‘P-site finger’ (H84) of 23S rRNA. Loop E, in the H4/H5 stem of 5S rRNA, which is located on the other side of the 3WJ, makes extensive contacts with the ASF. ASF is jutting out from the 50S subunit to form conserved dynamical bridge (B1a) with proteins S13 and S19 in the 30S subunit (85). During the ‘ratchet-like’ inter-subunit motion that occurs during translocation, the tip of the ASF loses contact with S13 and forms a new contact with S19 in the 30S subunit. The ASF contacts the elbow region (D-loop/T-loop) of A-site tRNA. If the ASF is shortened and cannot contact B1a, translocation is stimulated, providing evidence that ASF serves to attenuate translocation, e.g. to prevent frameshifting (83,84,86,87).
The 5S rRNA 3WJ possesses hinge-like flexibility between the P1/P3 unit (H2/H1) and P2 (H5) (Figure 4D). We suggest that flexibility at the 3WJ could allow the ‘P2 arm’ (including loop E/H4) of 5S to remain attached to the ASF through tertiary interactions involving loop E, while the ASF undergoes up and down motions relative to the main body of the 50S subunit (viewed with the CP facing upwards) during ratchet motions (74,88). It was suggested that the movement of the ASF is needed to allow peptidyl-tRNA to move from the A-site to the P-site (14). Thus, we propose that motions involving the 5S rRNA 3WJ help to coordinate the motions of the ASF relative to the L5 protein that is anchored in the CP. Both L5 and the tip of the ASF form crucial, dynamic bridges to the 30S subunit (82,86,89). Figure 7 indeed shows that if one considers H2 anchored to the CP, then the anisotropic hinge-like flexibility of the 5S 3WJ complements flexibility of the elbow segment at the base of ASF (90).
Concluding remarks
Our simulations show that isolated RNA 3WJs possess two dominant types of thermal fluctuations: global anisotropic hinge-like fluctuations between their P2 and P1/P3 segments; and more localized fluctuations within their P1/P3 regions (Figures 4–7). The maximal fluctuations occur multiple times during the trajectories and are non-periodic, as expected for stochastic thermal fluctuations. It is possible to roughly estimate, assuming Arrhenius kinetics that the most extreme ps snapshot realized during 100-ns simulation has activation energy of ∼8 kcal/mol. Note that describing the 3WJ fluctuations by just two movements is a simplification. In reality, the simulated molecules show many other fluctuations, as is common for biomolecules. Nevertheless, the two movements represent the characteristic thermal fluctuation signature for type C 3WJs. Full simulation trajectories can be obtained from the authors upon request.
It is interesting to compare the 3WJ fluctuations with those of other RNAs. For example, fluctuations of long A-RNA helices are isotropic (identical in all directions) and the bending curvature is distributed along the whole structure, as was visualized in detail in Ref. (11). On the other side, RNA Kts show directional (anisotropic) elbow-like fluctuations visibly localized in the kink area. In the case of Kts, a significant part of the dynamics could be assigned to local fluctuations of the A-minor interaction between the C- and NC-stems of Kt, which acts as the pivoting point (14,38,61). In contrast, although the 3WJ hinge-like fluctuations are visibly localized to the junction regions, we did not find any apparent pivoting local interaction. The fluctuations are thus distributed over several nucleotides. Kt and type C 3WJ differ significantly in their topologies. While for Kts, the hinge motions occur within a V-shaped structure between permanently kinked and unstacked arms that change the RNA path, the hinge-like dynamics of the 3WJs occur between coaxially stacked helices.
How to interpret the simulation results? First, the simulations provide direct assessment of the range of fast (ps to 100-ns timescale) thermal fluctuations of molecules that are either isolated or their fluctuations are not limited by the nearby ribosomal elements. For example, if fluctuations of GAC 3WJ are not attenuated by other interactions, the H43/H44 region would repeatedly make transient contacts with H89 (see above). Nevertheless, it is likely that adjacent ribosomal elements restrain the fluctuations of the studies elements, which may be even entirely immobilized. Even in this case, the results do have relevance because the fluctuations seen for isolated systems define the intrinsic flexibility of the molecules. In other words, if the isolated 3WJ shows anisotropic thermal hinge-like fluctuations, then the molecule possesses a hinge-like flexibility mirroring the fluctuations. This flexibility is intimately experienced by all surrounding elements in larger assemblies. The simulations are relatively short and do not show any rearrangement of the studied RNAs compared to the starting X-ray structures. So, we characterize flexibility of their single substates that correspond to their geometries as realized in the ribosome. We have analyzed the H42–H44 system also by all-atom and coarse grained NMA and demonstrate that NMA does not reproduce the simulation results.
Ribosome function requires complex, coordinated large-scale motions of its components to take place. Based on the results, we suggest that 3WJs flexibility can contribute to the functional flexibility with no requirements for rearrangements of base pairing. RNA junctions, besides structuring RNA molecules, can act as flexible elements in RNA architectures. In addition to the general description of flexibility of type C RNA 3WJs, we also provide specific results for three ribosomal 3WJs. We demonstrate that flexibility of 5S 3WJ complements flexibility of ASF ‘elbow’ segment (90) and flexibility of GAC 3WJ coincides with observed set of inward–outward GAC geometries seen in the X-ray structures. Both findings suggest that the 3WJ flexibilities are coupled with flexibilities of surrounding elements and contribute to ribosome function.
The main limitation of our approach is that we simulate the RNA elements in isolation. Nevertheless, such simulations best capture the intrinsic molecular flexibilities. Evidently, the role of the context can be significant, as demonstrated by our preliminary simulations of the H42–H44 rRNA–L10 protein complex. The protein binding appears to stabilize the rRNA in the experimental geometry that is modestly bent compared to the fully relaxed H42–H44 geometry. However, L10 binding does not substantially affect the 3WJ fluctuations. Unfortunately, including the ribosome context into simulations in a realistic manner is not always possible, since it is difficult to dissect models that would not be further coupled with the rest of the ribosome. Simulations of the whole ribosome would not necessarily solve the problem. Due to the complexity of the ribosome and the limited resolution, it would be difficult to achieve a really satisfactory initial relaxation of the system (all the RNA, protein, solvent and ion particles). Lack of perfect relaxation would bias extraction of spontaneous fine thermal movements in unrestrained simulations that occur on a scale of few kcal/mol. The simulations would be obscured by high-energy motions and large forces caused by imperfect relaxation of the system (7). The available simulation of tRNA accommodation thus utilized targeted MD, where the tRNAs were forced from the initial to the final positions through the ribosome along a predefined path (73). So, we suggest that despite all the limitations, large-scale simulations of isolated RNA elements constitute a viable approach that is capable to provide useful information that complements other experimental, bioinformatics and computational approaches.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant Agency of the Academy of Sciences of the Czech Republic (grants IAA400040802, IQS500040581 and KJB400040901); Grant Agency of the Czech Republic (grants 203/09/ H046 and 203/09/1476); Ministry of Education of the Czech Republic (LC06030 and MSM0021622413); Academy of Sciences of the Czech Republic (grant nos AV0Z50040507 and AV0Z50040702); National Institutes of Health (2 R15GM055898-04); National Science Foundation (Research Coordination Network Grant No. 0443508). Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDowell SE, Spackova N, Sponer J, Walter NG. Molecular dynamics simulations of RNA: an in silico single molecule approach. Biopolymers. 2007;85:169–184. doi: 10.1002/bip.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall KB. RNA in motion. Curr. Opin. Chem. Biol. 2008;12:612–618. doi: 10.1016/j.cbpa.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auffinger P, Hashem Y. Nucleic acid solvation: from outside to insight. Curr. Opin. Struct. Biol. 2007;17:325–333. doi: 10.1016/j.sbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Cheatham TE. Simulation and modeling of nucleic acid structure, dynamics and interactions. Curr. Opin. Struct. Biol. 2004;14:360–367. doi: 10.1016/j.sbi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Banas P, Jurecka P, Walter NG, Sponer J, Otyepka M. Theoretical studies of RNA catalysis: hybrid QM/MM methods and their comparison with MD and QM. Methods. 2009;49:202–216. doi: 10.1016/j.ymeth.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ditzler MA, Otyepka M, Šponer J, Walter NG. Molecular dynamics and quantum mechanics of RNA: conformational and chemical change we can believe in. Acc. Chem. Res. 2010;43:40–47. doi: 10.1021/ar900093g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villa A, Wöhnert J, Stock G. Molecular dynamics simulation study of the binding of purine bases to the aptamer domain of the guanine sensing riboswitch. Nucleic Acids Res. 2009;37:4774–4786. doi: 10.1093/nar/gkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auffinger P, Bielecki L, Westhof E. Symmetric K+ and Mg2+ ion-binding sites in the 5 S rRNA loop E inferred from molecular dynamics simulations. J. Mol. Biol. 2004;335:555–571. doi: 10.1016/j.jmb.2003.10.057. [DOI] [PubMed] [Google Scholar]
- 11.Reblova K, Lankas F, Razga F, Krasovska MV, Koca J, Sponer J. Structure, dynamics, and elasticity of free 16S rRNA helix 44 studied by molecular dynamics simulations. Biopolymers. 2006;82:504–520. doi: 10.1002/bip.20503. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Ma BY, Shapiro BA. Binding interactions between the core central domain of 16S rRNA and the ribosomal protein S15 determined by molecular dynamics simulations. Nucleic Acids Res. 2003;31:629–638. doi: 10.1093/nar/gkg149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li PTX, Vieregg J, Tinoco I. How RNA unfolds and refolds. Annu. Rev. Biochem. 2008;77:77–100. doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- 14.Razga F, Koca J, Sponer J, Leontis NB. Hinge-like motions in RNA kink-turns: the role of the second A-minor motif and nominally unpaired bases. Biophys. J. 2005;88:3466–3485. doi: 10.1529/biophysj.104.054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besseova I, Otyepka M, Reblova K, Sponer J. Dependence of A-RNA simulations on the choice of the force field and salt strength. Phys. Chem. Chem. Phys. 2009;11:10701–10711. doi: 10.1039/b911169g. [DOI] [PubMed] [Google Scholar]
- 16.Orozco M, Perez A, Noy A, Luque FJ. Theoretical methods for the simulation of nucleic acids. Chem. Soc. Rev. 2003;32:350–364. doi: 10.1039/b207226m. [DOI] [PubMed] [Google Scholar]
- 17.Spackova N, Sponer J. Molecular dynamics simulations of sarcin-ricin rRNA motif. Nucleic Acids Res. 2006;34:697–708. doi: 10.1093/nar/gkj470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ninio J. Multiple stages in codon-anticodon recognition: double-trigger mechanisms and geometric constraints. Biochimie. 2006;88:963–992. doi: 10.1016/j.biochi.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Blanchard SC, Kim HD, Gonzalez RL, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra K, Frank J. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu. Rev. Biophys. Biomol. Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- 21.Frank J, Spahn CMT. The ribosome and the mechanism of protein synthesis. Rep. Prog. Phys. 2006;69:1383–1417. [Google Scholar]
- 22.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 23.Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spirin AS. Ribosome as a molecular machine. FEBS Lett. 2002;514:2–10. doi: 10.1016/s0014-5793(02)02309-8. [DOI] [PubMed] [Google Scholar]
- 25.Krasovska MV, Sefcikova J, Reblova K, Schneider B, Walter NG, Sponer J. Cations and hydration in catalytic RNA: molecular dynamics of the hepatitis delta virus ribozyme. Biophys. J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auffinger P, Bielecki L, Westhof E. The Mg2+ binding sites of the 5S rRNA loop E motif as investigated by molecular dynamics simulations. Chem. Biol. 2003;10:551–561. doi: 10.1016/s1074-5521(03)00121-2. [DOI] [PubMed] [Google Scholar]
- 27.Reblova K, Spackova N, Stefl R, Csaszar K, Koca J, Leontis NB, Sponer J. Non-Watson-Crick basepairing and hydration in RNA motifs: molecular dynamics of 5S rRNA loop E. Biophys. J. 2003;84:3564–3582. doi: 10.1016/S0006-3495(03)75089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shankar N, Kennedy SD, Chen G, Krugh TR, Turner DH. The NMR structure of an internal loop from 23S ribosomal RNA differs from its structure in crystals of 50S ribosomal subunits. Biochemistry. 2006;45:11776–11789. doi: 10.1021/bi0605787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunelle JL, Youngman EM, Sharma D, Green R. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA. 2006;12:33–39. doi: 10.1261/rna.2256706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen MA, Kirpekar F, Ritterbusch W, Vester B. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA. 2002;8:202–213. doi: 10.1017/s1355838202013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widerak M, Kern R, Malki A, Richarme G. U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene. 2005;347:109–114. doi: 10.1016/j.gene.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 34.Razga F, Spackova N, Reblova K, Koca J, Leontis NB, Sponer J. Ribosomal RNA kink-turn motif - a flexible molecular hinge. J. Biomol. Struct. Dyn. 2004;22:183–193. doi: 10.1080/07391102.2004.10506994. [DOI] [PubMed] [Google Scholar]
- 35.Goody TA, Melcher SE, Norman DG, Lilley DMJ. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 38.Razga F, Koca J, Mokdad A, Sponer J. Elastic properties of ribosomal RNA building blocks: molecular dynamics of the GTPase-associated center rRNA. Nucleic Acids Res. 2007;35:4007–4017. doi: 10.1093/nar/gkm245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouvela EC, Gerbanas GV, Xaplanteri MA, Petropoulos AD, Dinos GP, Kalpaxis DL. Changes in the conformation of 5S rRNA cause alterations in principal functions of the ribosomal nanomachine. Nucleic Acids Res. 2007;35:5108–5119. doi: 10.1093/nar/gkm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, Ferguson D, Seibel G, Kollman PA. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 41.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, et al. Amber 8. San Francisco, CA: University of California; 2004. [Google Scholar]
- 42.Duke RE, Pedersen LG. PMEMD 3. Chapel Hill: University of North Carolina; 2003. [PubMed] [Google Scholar]
- 43.Darden T, York D, Pedersen L. Particle mesh Ewald – an N.LOG(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 44.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 45.Harvey SC, Tan RKZ, Cheatham TE. The flying ice cube: velocity rescaling in molecular dynamics leads to violation of energy equipartition. J. Comput. Chem. 1998;19:726–740. [Google Scholar]
- 46.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, et al. Amber 9. San Francisco, CA: University of California; 2006. [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 48.Amadei A, Linssen ABM, Berendsen HJC. Essential Dynamics of Proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 49.Mongan J. Interactive essential dynamics. J. Comput. Aided Mol. Des. 2004;18:433–436. doi: 10.1007/s10822-004-4121-z. [DOI] [PubMed] [Google Scholar]
- 50.Zheng WJ, Doniach S. A comparative study of motor-protein motions by using a simple elastic-network model. Proc. Natl Acad. Sci. USA. 2003;100:13253–13258. doi: 10.1073/pnas.2235686100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 53.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 54.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, Laughton CA, Orozco M. Refinenement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krasilnikov AS, Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutell RR, Cannone JJ, Konings D, Gautheret D. Predicting U-turns in ribosomal RNA with comparative sequence analysis. J. Mol. Biol. 2000;300:791–803. doi: 10.1006/jmbi.2000.3900. [DOI] [PubMed] [Google Scholar]
- 57.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lescoute A, Westhof E. The A-minor motifs in the decoding recognition process. Biochimie. 2006;88:993–999. doi: 10.1016/j.biochi.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 59.Tamura M, Holbrook SR. Sequence and structural conservation in RNA ribose zippers. J. Mol. Biol. 2002;320:455–474. doi: 10.1016/s0022-2836(02)00515-6. [DOI] [PubMed] [Google Scholar]
- 60.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr. Opin. Chem. Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Razga F, Zacharias M, Reblova K, Koca J, Sponer J. RNA kink-turns as molecular elbows: hydration, cation binding, and large-scale dynamics. Structure. 2006;14:825–835. doi: 10.1016/j.str.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 62.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 angstrom resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 63.Zacharias M. Comparison of molecular dynamics and harmonic mode calculations on RNA. Biopolymers. 2000;54:547–560. doi: 10.1002/1097-0282(200012)54:7<547::AID-BIP70>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 64.Tama F, Wriggers W, Brooks CL. Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J. Mol. Biol. 2002;321:297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- 65.Tama F, Valle M, Frank J, Brooks CL. Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc. Natl Acad. Sci. USA. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fulle S, Gohlke H. Molecular recognition of RNA: challenges for modelling interactions and plasticity. J. Mol. Recognit. 2010;23:220–231. doi: 10.1002/jmr.1000. [DOI] [PubMed] [Google Scholar]
- 67.Van Wynsberghe AW, Cui Q. Comparison of mode analyses at different resolutions applied to nucleic acid systems. Biophys. J. 2005;89:2939–2949. doi: 10.1529/biophysj.105.065664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kramer MA. Nonlinear principal component analysis autoassociative neural networks. AIChE J. 1991;37:233–243. [Google Scholar]
- 69.Cheatham TE, Young MA. Molecular dynamics simulation of nucleic acids: successes, limitations, and promise. Biopolymers. 2000;56:232–256. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 70.Fadrna E, Spackova N, Stefl R, Koca J, Cheatham TE, Sponer J. Molecular dynamics simulations of guanine quadruplex loops: advances and force field limitations. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zacharias M. Simulation of the structure and dynamics of nonhelical RNA motifs. Curr. Opin. Struct. Biol. 2000;10:311–317. doi: 10.1016/s0959-440x(00)00089-0. [DOI] [PubMed] [Google Scholar]
- 72.Lankas F, Sponer J, Hobza P, Langowski J. Sequence-dependent elastic properties of DNA. J. Mol. Biol. 2000;299:695–709. doi: 10.1006/jmbi.2000.3781. [DOI] [PubMed] [Google Scholar]
- 73.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl Acad. Sci. USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao HX, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, et al. Study of the structural dynamics of the E-coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 75.Sergiev PV, Lesnyak DV, Burakovsky DE, Kiparisov SV, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA. Alteration in location of a conserved GTPase-associated center of the ribosome induced by mutagenesis influences the structure of peptidyltransferase center and activity of elongation factor G. J. Biol. Chem. 2005;280:31882–31889. doi: 10.1074/jbc.M505670200. [DOI] [PubMed] [Google Scholar]
- 76.Mokdad A, Leontis NB. Ribostral: an RNA 3D alignment analyzer and viewer based on basepair isostericities. Bioinformatics. 2006;22:2168–2170. doi: 10.1093/bioinformatics/btl360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miyoshi T, Uchiumi T. Functional interaction between bases C1049 in domain II and G2751 in domain VI of 23S rRNA in Escherichia coli ribosomes. Nucleic Acids Res. 2008;36:1783–1791. doi: 10.1093/nar/gkm1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanbonmatsu KY. Alignment/misalignment hypothesis for tRNA selection by the ribosome. Biochimie. 2006;88:1075–1089. doi: 10.1016/j.biochi.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 79.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 80.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kavran JM, Steitz TA. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J. Mol. Biol. 2007;371:1047–1059. doi: 10.1016/j.jmb.2007.05.091. [DOI] [PubMed] [Google Scholar]
- 82.Kiparisov S, Petrov A, Meskauskas A, Sergiev PV, Dontsova OA, Dinman JD. Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2005;274:235–247. doi: 10.1007/s00438-005-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rakauskaite R, Dinman JD. An arc of unpaired “hinge bases” facilitates information exchange among functional centers of the ribosome. Mol. Cell. Biol. 2006;26:8992–9002. doi: 10.1128/MCB.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sergiev PV, Kiparisov SV, Burakovsky DE, Lesnyak DV, Leonov AA, Bogdanov AA, Dontsova OA. The conserved A-site finger of the 23 S rRNA: just one of the intersubunit bridges or a part of the allosteric communication pathway? J. Mol. Biol. 2005;353:116–123. doi: 10.1016/j.jmb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 85.Gao N, Frank J. A library of RNA bridges. Nat. Chem. Biol. 2006;2:231–232. doi: 10.1038/nchembio0506-231. [DOI] [PubMed] [Google Scholar]
- 86.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The a-site finger in 23 S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 87.Piekna-Przybylska D, Przybylski P, Baudin-Baillieu AS, Rousset JP, Fournier MJ. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J. Biol. Chem. 2008;283:26026–26036. doi: 10.1074/jbc.M803049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Agirrezabala X, Lei JL, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol. Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the ribosome at 5.5 angstrom resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 90.Reblova K, Razga F, Li W, Gao H, Frank J, Sponer J. Dynamics of the base of ribosomal A-site finger revealed by molecular dynamics simulations and cryo-EM. Nucleic Acids Res. 2009;38:1325–1340. doi: 10.1093/nar/gkp1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 92.Zirbel CL, Sponer JE, Sponer J, Stombaugh J, Leontis NB. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res. 2009;37:4898–4918. doi: 10.1093/nar/gkp468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 94.Laurberg M, Asahara H, Korostelev A, Zhu JY, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.