Abstract
Heavy metals have the potential to engage in strong bonding interactions and can thus function in essential as well as toxic or therapeutic capacities. We conducted crystallographic analyses of heavy cation binding to the nucleosome core particle and found that Co2+ and Ni2+ preferentially associate with the DNA major groove, in a sequence- and conformation-dependent manner. Conversely, Rb+ and Cs+ are found to bind only opportunistically to minor groove elements of the DNA, in particular at narrow AT dinucleotide sites. Furthermore, relative to Mn2+ the aggressive coordination of Co2+ and Ni2+ to guanine bases is observed to induce a shift in histone–DNA register around the nucleosome center by stabilizing DNA stretching over one region accompanied by expulsion of two bases at an opposing location. These ‘softer’ transition metals also associate with multiple histone protein sites, including inter-nucleosomal cross-linking, and display a proclivity for coordination to histidine. Sustained binding and the ability to induce structural perturbations at specific locations in the nucleosome may contribute to genetic and epigenetic mechanisms of carcinogenesis mediated by Co2+ and Ni2+.
INTRODUCTION
Heavy metals represent an important class of highly toxic environmental pollutants (1–8). As, Cd, Cr, Ni, Co, Cu, Fe and Pt contaminants can arise as industrial by-products, which have mutagenic and carcinogenic potential. The metal cations are capable of catalyzing production of reactive oxygen species as well as eliciting a variety of alterations in macromolecular structure and activity that impair cellular function. Although DNA damage and mutation are apparently contributing factors, recent work is pointing towards epigenetic changes and alterations to DNA repair processes as key mechanisms in metal-mediated carcinogenesis. In any case, pathways involved in generating the cancerous state can be metal-specific, and thus many questions remain regarding the relative importance of different structural and chemical modifications in producing the disease state.
The pathologic potential of heavy metals relates closely to their ability to form strong bonding interactions. The ‘hard and soft acids and bases’ (HSAB) theory describes the capacity for metals cations and ligands to engage in bonds with partial covalent character (9). Thus, hard metals, such as Mg2+ and Ca2+, form low-affinity, purely ionic interactions with hard ligands, which are generally oxygen atoms in vivo. On the other hand, very soft metals, such as Cd2+, Cu+ or Pt2+, are highly polarizable and can engage in extremely stable coordinate-covalent bonds with suitably soft sulfurous or nitrogenous ligands. Inside the cell, such bonding potential translates to sustained site occupancy and the ability to elicit profound conformational changes in proteins and nucleic acids. Moreover, many soft cations are redox active and have elevated toxicity attributes by generating reactive oxygen species (1–4).
By forming high-affinity interactions with ring nitrogen atoms of DNA bases, soft metals are generally disruptive to right-handed double helical structure. For instance, transition metals can induce the B- to Z-form transition at much lower concentrations compared to Mg2+ (10). Soft metal cations also lower the melting temperature of duplex DNA—an effect opposite to that of Mg2+, Ca2+ or Mn2+ (11).
Although slightly softer than Mg2+ or Ca2+, Mn2+ is considered a hard ion, meaning its association with proteins and nucleic acids is still governed largely by ionic forces (12). Nonetheless, we found that Mn2+ binds with high site selectivity to nucleosomal DNA, with discrimination towards both sequence and conformation (13,14). At the other end of the spectrum, we also observed that platinum anticancer agents form site-selective DNA and histone adducts in the nucleosome core (15). This suggests that many of the toxic metals have distinct site preferences in interacting with nucleosomes.
Given the ever-increasing industrial usage of toxic metals, especially in developing nations, the importance for a detailed understanding of this family of carcinogens is on the rise (8). Furthermore, improved knowledge of metal-mediated cancer pathways would help us to develop more safe metal-based therapeutic agents. In order to shed light on the contribution of their interaction with chromatin towards carcinogenesis, we conducted a study of heavy metal binding in the nucleosome core.
MATERIALS AND METHODS
Nucleosome core particle crystal preparation
Nucleosome core particle (NCP) was prepared from recombinant Xenopus laevis histones and a 147-bp DNA fragment using established methodologies (16,17). Crystals were grown as described previously and stabilized in a harvest buffer of 37 mM MnCl2, 40 mM KCl, 20 mM K-cacodylate (pH 6.0), 24% (v/v) MPD and 2% (w/v) trehalose (17).
Co2+-NCP and Ni2+-NCP crystals were prepared by rinsing Mn2+-stabilized crystals with a modified harvest buffer, in which the 37 mM MnCl2 was replaced by an equimolar amount of either CoCl2 or NiCl2. Crystal buffer volume was reduced to ∼200 µl, followed by the addition of 1 ml of Co2+ or Ni2+ substitute buffer and equilibration with gentle mixing over ∼10 min. The crystal buffer was subsequently reduced again to ∼200 µl, and the process repeated 10 times in total. This gives a theoretical dilution of Mn2+ concentration at >60 million-fold. Elimination of Mn2+ from the crystals was confirmed by atomic absorption spectroscopy, which indicated a Mn2+ concentration below the detection limit of the instrument, at <10 nM.
Preparation of Cs+-NCP and Rb+-NCP crystals was conducted using the same protocol as described above for Co2+/Ni2+. For these monovalent cation buffer substitutions, however, only the 40 mM KCl and 20 mM K-cacodylate (pH 6.0) harvest buffer components were replaced with the respective Cs+ or Rb+ equivalents.
Structure solution and analysis
Single crystal X-ray diffraction data were recorded as described previously (18) at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) using the PILATUS detector on beam line X06SA (Co2+-NCP and Ni2+-NCP) and a Mar225 CCD detector on beam line X06DA (Cs+-NCP and Rb+-NCP). Data were processed with MOSFLM (19) and SCALA from the CCP4 suite (20).
The histone–DNA model of NCP147 (pdb code 1KX5; 17) was used for structure solution by molecular replacement. Structural refinement and model building were carried out with routines from the CCP4 suite (20). Co2+/Ni2+ occupancy and B-factor values were refined against the anomalous difference data using Mlphare. The initial occupancy values, on an arbitrary scale, were normalized with respect to the fully occupied divalent metal binding site at the histone–histone interparticle interface (14).
DNA conformational analysis was conducted using CURVES (21). Graphic figures were prepared with PyMOL (DeLano Scientific LLC, San Carlos, CA, USA).
RESULTS
Candidates for heavy metal-nucleosome study
In order to find suitable candidates for characterization of heavy metal binding, we screened a variety of cations, including Co2+, Ni2+, Cu2+, Zn2+, Cd2+, Hg2+ and Pb2+, in crystallization and crystal buffer exchange trials of the NCP. High-resolution diffracting NCP crystals are grown in Mn2+-containing buffers (17), and we did not obtain well diffracting crystals grown in the presence of alternate divalent cations. Moreover, replacement of Mn2+ with a substitute divalent metal through buffer exchange of pre-grown NCP crystals generally gave rise to a substantial reduction in diffraction quality. Conversely, exchange with Co2+- or Ni2+-containing buffers resulted in only minor disordering of NCP crystals (Table 1). In addition, we were able to introduce Cs+ and Rb+, in place of K+, into preformed NCP crystals with only modest losses in diffraction quality.
Table 1.
NCP147 data collection and refinement statistics
| Co2+ | Ni2+ | Cs+ | Rb+ | |
|---|---|---|---|---|
| Data collectiona | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a / b / c (Å) | 106.5/109.9/183.4 | 106.1/109.6/183.1 | 106.5/109.7/182.6 | 106.5/109.7/182.2 |
| α/β/γ (°) | 90.0/90.0/90.0 | 90.0/90.0/90.0 | 90.0/90.0/90.0 | 90.0/90.0/90.0 |
| Resolution (Å) | 2.44–52.6 | 2.65–51.0 | 3.15–94.1 | 2.30–60.0 |
| Highest shell (Å) | 2.44–2.57 | 2.65–2.79 | 3.15–3.32 | 2.30–2.42 |
| Rmerge, % | 6.3 (44.2) | 6.4 (49.6) | 9.1 (47.7) | 5.0 (47.8) |
| I / σI | 17.0 (1.0) | 19.7 (2.0) | 17.8 (3.1) | 21.1 (3.2) |
| Completeness (%) | 91.6 (59.5) | 98.2 (89.2) | 99.8 (98.5) | 90.3 (58.6) |
| Redundancy | 4.7 (2.1) | 5.6 (4.3) | 7.0 (5.7) | 6.7 (4.6) |
| Wavelength (Å) | 1.60 | 1.48 | 1.93 | 0.81 |
| Refinement | ||||
| Resolution (Å) | 2.44–52.6 | 2.65–51.0 | 3.15–94.1 | 2.30–60.0 |
| No. of reflections | 72 231 | 60 265 | 36 839 | 84 087 |
| Rwork/Rfree, %/% | 23.9/28.2 | 23.2/27.6 | 21.6/23.8 | 24.2/26.7 |
| No. of atoms | 12 228 | 12 241 | 12 207 | 12 200 |
| Protein | 6160 | 6169 | 6156 | 6156 |
| DNA | 6021 | 6021 | 6021 | 6021 |
| Ion | 47 | 51 | 30 | 23 |
| B-factors (Å2) | 84 | 86 | 76 | 60 |
| Protein | 56 | 59 | 52 | 40 |
| DNA | 112 | 113 | 100 | 81 |
| Ion | 98 | 117 | 98 | 70 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.010 | 0.010 | 0.009 | 0.010 |
| Bond angles (°) | 1.43 | 1.50 | 1.46 | 1.47 |
aData sets are based on single crystal diffraction, and values in parentheses are for the highest-resolution shell.
To identify sites of metal binding, we relied on the anomalous scattering generated by heavy atoms. This special form of X-ray scattering from the lowest energy electron orbitals is maximal at the atom type-specific absorption edge (22). For collection of Co2+-, Ni2+- and Rb+-NCP data, the X-ray wavelength was tuned to the respective metal absorption edges (Table 1). The absorption edges of Cs, however, occur at inaccessibly short and long X-ray wavelengths. For collection of Cs+-NCP data, we therefore tuned to a longer wavelength, at which the anomalous signal from Cs+ is relatively strong, while that of Mn2+ (present in the buffer of the monovalent cation substitutions) is at a local minimum. Data collection at such long wavelengths (‘soft’ X-rays) does, however, result in a dramatic reduction in achievable resolution (23).
Co2+ and Ni2+ binding induce shifting of histone–DNA register
Co2+ and Ni2+ ion locations were based on peak positions in the anomalous difference map having magnitudes greater than four times the r.m.s variation in electron density (>4σ). A total of 43 and 47 sites were identified for Co2+ and Ni2+, respectively, of which the vast majority are associated with the DNA (Table 2 and Figure 1A). Moreover, most of the DNA-associated sites correspond to metal coordination to the major groove, with a small fraction of Co2+/Ni2+ binding as apparent metal hydrates in the major or minor grooves. This indirect mode of association entails interaction of the metal water ligands with mostly hydrogen bond acceptor groups of the DNA (24).
Table 2.
Summary of metal binding sitesa
| Type | Mn2+ | Co2+ | Ni2+ | Cs+ | Rb+ |
|---|---|---|---|---|---|
| Total | 45 | 43 | 47 | 12 | 5 |
| DNA | 43 | 37 | 37 | 9 | 3 |
| major | 18 | 27 | 27 | ||
| major-MH | 5 | 7 | 7 | ||
| minor | 9 | 3 | |||
| minor-MH | 17 | 3 | 3 | ||
| PO4 | 3 | ||||
| Histone | 2 | 6 | 10 | 3 | 2 |
| His | 3 | 3 | |||
| His+Asp/Glu | 1 | 2 | |||
| Asp/Glu | 1 | 2 | 5 | ||
| peptide C=O | 1 | 3 | 2 |
aInteraction with DNA entails direct coordination or association of the metal hydrate (MH).
Protein binding is characterized as coordination involving histidine, acidic side chains or a combination of the two amino acid types, or coordination exclusively with peptide carbonyl groups.
Data for Mn2+ association are from an earlier study (14).
Figure 1.
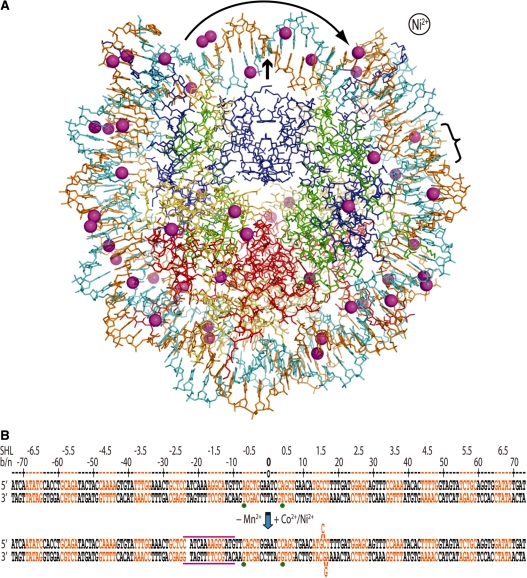
The presence of Co2+ or Ni2+ induces a shift in histone–DNA register and distortions in the double helix. (A) The Ni2+-NCP structure viewed along the particle pseudo 2-fold axis (small arrow). The 47 Ni2+ ions appear as magenta spheres, the two DNA strands are orange and cyan and histone proteins are shown in blue (H3), green (H4), yellow (H2A) and red (H2B). Ni2+ or Co2+ association causes displacement of a bp from one particle half to the opposing half (curved arrow; bracket indicates location of base flip-out distortion). This is also apparent from asymmetry in metal binding about the dyad axis. (B) Minor and major groove-inward sections of NCP147 are orange and black, respectively. The base or nucleotide numbering scheme (b/n) is relative to Mn2+-NCP (top) and corresponds to the 5′ (−) to 3′ (+) direction of either DNA strand in the duplex (SHL = superhelix location, turns from center). A gap in the sequence represents a shift in histone-bp register from DNA stretching (approximate expanse of distortion, magenta lines), which is accompanied by base expulsion at the 1.5-turn location in Co2+-NCP and Ni2+-NCP. Green dots represent two strong peaks in the anomalous difference map that correspond to Co2+/Ni2+-guanine coordination in the refined model (bottom), whereas they coincide with inappropriate binding sites in the Mn2+-NCP model (top).
Compared to the high-resolution structure of the NCP in the presence of Mn2+ (Mn2+-NCP; 17), those of Co2+ and Ni2+ are nearly identical, with the exception of the central ∼51 bp. The r.m.s deviation in atomic position from superposition of the outer 96 bp (all DNA atoms of position ±26 to the terminus in each particle half) of Mn2+-NCP with Co2+-NCP or Ni2+-NCP are only 0.89 Å and 0.93 Å, respectively. Moreover, there are no significant local DNA conformational changes apparent from Co2+/Ni2+ binding in these non-central regions.
In contrast to the non-central sections, the DNA around the nucleosome center displays a shift in register with respect to the histone octamer in the presence of Co2+ or Ni2+ (Figure 1B). The change in register is apparent from the electron density, which indicates DNA stretching in one particle half, causing displacement of a bp through the center and resulting in expulsion of two bases in the opposing particle half. In this manner, the ‘extra’ bp generated by stretching is ultimately displaced by flipping out two bases from the double helical stack, thus not altering histone–DNA register downstream.
An additional indication of the occurrence and directionality of the register shift comes from the presence of two 8–10σ peaks in the anomalous difference map, which are initially centered on the methyl groups of two thymine bases (positions −4 and 7) in the unrefined, starting model (corresponding to the Mn2+-NCP structure; see Figure 1B). These are not possible metal coordination sites, indicative of conformational change in the DNA, and only bp shifting in one direction brings potential coordinating bases, purines, into appropriate register.
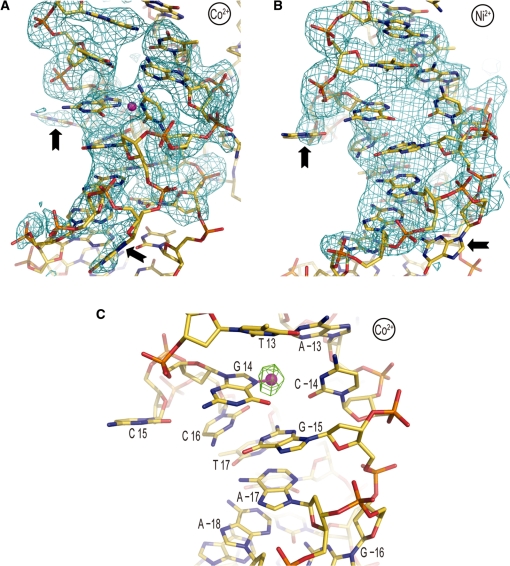
The two bases flipped out (underlined) of the double helix coincide with an AGGCA/TGCCT element situated 1.5 double helical turns from the nucleosome center (Figure 2). The location of the distortion at a GG=CC dinucleotide allows a guanine and a cytosine base from the two different bp to engage in Watson–Crick pairing as a result of the single base dislocations from opposing strands over the 2 bp. Although there is a significant degree of disorder in this region, the same flip-out configuration is consistent with both the Co2+ and Ni2+ data. Furthermore, the higher-resolution Co2+ data give clear indication of metal-guanine (position 14) coordination at the site of the deformation, in which non-canonical base pairing appears to occur (see Figure 2C). In any case, a 3.8σ peak at this same guanine-N7 position is also evident in the anomalous difference map for Ni2+-NCP.
Figure 2.
Co2+ or Ni2+ binding promotes base expulsion from the double helical stack at the 1.5-turn location. (A and B) Co2+-NCP (A) and Ni2+-NCP (B) display flipping out of cytosine 15 and guanine −16 (arrows). An Fo−Fc electron density map (2.5σ), in which ATGCCTT/AAGGCAT nucleotides ±12 to ±18 were omitted from the respective models, is displayed superimposed on the structures. (C) Heavy divalent metal binding appears to directly promote the double helix deformation. Co2+ (magenta sphere) is observed to coordinate to the N7 atom of guanine 14, which is situated in the ‘minor groove’ by virtue of assuming the syn glycosidic conformation. An anomalous difference electron density map (3.5σ) is shown superimposed on the Co2+-NCP model.
The combination of stretching and base expulsion here yields an asymmetric disposition between the two halves of the palindromic 147 bp DNA. As we had observed previously with two distinct stretching modes occurring in opposing halves of an isomorphous 145 bp NCP crystal, the differential crystal packing environment around the 1.5-turn location apparently promotes the asymmetry (18). In this sense, interaction of the H2B αC-helix from a neighboring particle with the DNA may help to stabilize the base flip-out distortion.
Aggressive Co2+/Ni2+-purine coordination
In a more recent study of Mn2+-NCP, in which data were collected at the Mn absorption edge, we identified a total of 45 Mn2+ binding sites (14). This is the same number of sites as found on average for Co2+ or Ni2+, which allows a comparison of localization preference between the two different metal types. This can shed light on differences in relative site affinities between the hard Mn2+ and Co2+/Ni2+, which are classified as having intermediate-softness (9).
While the distribution and site-specific occupancies of Co2+ compared to Ni2+ are very similar, considering only the DNA-associated ions, a substantially greater fraction of Co2+/Ni2+ (73%), relative to Mn2+ (42%), are observed to be engaged in direct coordination with the major groove (Figure 3 and Tables 2 and 3). Since the ionic radii of Co2+ (0.70 Å) and Ni2+ (0.66 Å) are both smaller than that of Mn2+ (0.75 Å), the former are in fact more difficult to dehydrate than Mn2+ (9,12). In addition, the preferred coordination geometry for the three metals is the same—octahedral. Thus, the elevated tendency of Co2+/Ni2+ to coordinate to major groove sites is a consequence of the increased affinity for the soft purine ring N7 atoms.
Figure 3.
Co2+/Ni2+-DNA binding site selectivity. (A and B) Ni2+ (A) and Co2+ (B) binding to GG dinucleotide sites at locations 61 and −35/−34, respectively. Displacement of the 3′ guanine into the major groove at position −34 (arrow) allows N7/O6 cross-linking and creates a secondary, low occupancy, site for N7 coordination to the 3′ base. An anomalous difference electron density map, contoured at 8σ (A) and 4σ (B), is shown superimposed on the model. (C and D) The guanine base atoms at locations 8 (C) or 14 (D) from the Ni2+-NCP structure (gold carbon atoms) were least-squares superimposed onto those of the Mn2+-NCP structure (green) to illustrate how conformational alterations associated DNA stretching and histone–DNA register shifting can influence metal binding. In both instances, Co2+/Ni2+ binding occurs in the stretched particle half, whereas Mn2+ association is not observed at the respective locations. This appears to be largely a consequence of steric clearance provided by positive slide at the TG elements (arrow) associated with the change in DNA orientation. (A–D) Co2+/Ni2+ ions and coordinate (-covalent) bonds appear as magenta spheres and lines, respectively.
Table 3.
Major groove coordination sitesa
| Binding site | DNA location | Mn2+ occupancy | Co2+ dist. (Å) | Co2+ occupancy | Ni2+ dist. (Å) | Ni2+ occupancy |
|---|---|---|---|---|---|---|
| TGGA | −35/−34 | (0.71) 0.47 | 2.2/2.3 | (0.74) 0.56 | 2.6/2.5 | (0.83) 0.55 |
| TGGA | −34 | 0.24 | 1.6 | 0.18 | 2.0 | 0.28 |
| AGGT | 61 | 0.73 | 2.4 | 0.63 | 2.3 | 0.66 |
| TGGA | −3* | 0.36 | 2.5 | 0.62 | 2.3 | 0.57 |
| TGGT | 48 | 0.68 | 2.1 | 0.59 | 2.2 | 0.75 |
| AGCA | 27 | 0.53 | 2.1 | 0.59 | 2.1 | 0.63 |
| AGAT | −56 | 0.16 | 2.3 | 0.47 | 2.1 | 0.68 |
| AGCT | −6* | − | 2.4 | 0.35 | 2.7 | 0.35 |
| CAGT | 29 | – | 2.7 | 0.34 | 2.5 | 0.49 |
| TGAT | 71 | – | 1.7 | 0.34 | 2.2 | 0.23 |
| TGGA | 24/25* | – | 2.5/2.4 | (0.28) 0.13 | 2.5/2.5 | (0.43) 0.21 |
| TGGA | 24* | – | 2.2 | 0.10 | – | – |
| TGGA | 25* | – | 2.8 | 0.05 | 2.6 | 0.22 |
| TGGA | 64/65 | – | – | (0.28) – | 2.3/2.8 | 0.20 |
| TGGA | 64 | – | 2.8 | 0.17 | – | – |
| TGGA | 65 | 0.15 | 2.6 | 0.11 | – | – |
| TGAA | 8* | – | 2.9 | 0.24 | 2.5 | 0.19 |
| AGCT | 5* | 0.43 (O6) | 2.4 | 0.14 | 2.5 | 0.23 |
| TGCC | 14* | – | 1.3 | 0.13 | 2.7 | 0.12 |
| CAGC | 4* | 0.14 | – | – | – | – |
| TGGA | −2* | 0.12 | – | – | – | – |
| AGGC | −16/−15* | – | – | – | – | – |
| TGCA | 58 | – | – | – | – | – |
| TGCA | −59 | – | – | – | – | – |
aThe sequence context for dinucleotide binding sites (bold) is given, with the coordinating purine bases underlined.
DNA location refers to the nucleotide distance from the central bp (0) at the particle pseudo two-fold axis.
The palindromic DNA runs from −73 to 73 in the 5′-3′ direction for either chain. Values shown are averages between the two symmetry-related particle halves, and sites are ordered from highest to lowest occupancy relative to Co2+ association. Distances are metal to purine N7 atom, with the exception of N7/O6 cross-linking at GG and an AGCT site, for which Mn2+ coordinates to the O6 atom of base 5 (14). The histone–DNA register for Co2+/Ni2+ differs from that of Mn2+ over 2.5 double helical turns flanking the nucleosome center (*).
All GG and GC dinucleotide elements are shown and others only if occupied by metal.
For comparison of relative affinities between different dinucleotide types, values in parentheses correspond to the summed occupancy for two different modes of coordination observed at a single GG site.
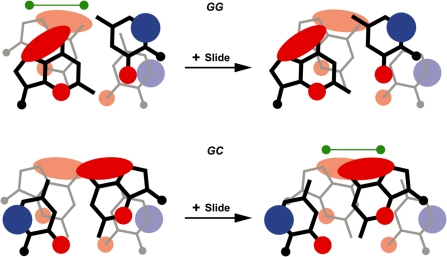
We found previously that Mn2+ binding in the NCP is governed largely by electrostatics, consistent with first principles (14). In this sense, the highest affinity major groove coordination sites coincide with GG and GC dinucleotides, in which the extensive electronegative zones of guanine are in close proximity (Figure 4). Specifically, Mn2+ binding is not observed at locations for which the value of the translational bp step parameter slide causes insufficient overlap of guanine bases (13). In general, these same principles appear to influence Co2+/Ni2+ coordination, whereby the highest metal occupancies are observed for GG dinucleotide sites and a similar discrimination towards GG and GC site conformation occurs (Table 3). However, in contrast to Mn2+, Co2+/Ni2+ also show medium to high occupancy association with guanine, as well as adenine, at GA or AG dinucleotides. The proximity of electropositive amine hydrogen atoms from adenine would generally disfavor ionic metal interactions. Moreover, whereas Mn2+ has been observed to coordinate solely to a guanine O6 atom of a GC site (13,14), Co2+/Ni2+ coordination sites are seen to always involve a purine N7 atom (Table 3). This exclusive nitrogenous ligand coordination behavior of Co2+/Ni2+ also involving high-affinity binding at less electrostatically favorable, adenine-rich locations arises from the increased contribution of polarizability relative to ionic forces in site discrimination for these softer metals.
Figure 4.
Guanine–guanine overlap in GG=CC and GC dinucleotides influences divalent metal association (13,14). Simplified representation of the electrostatic potential around bases in GG and GC steps, adapted from ref. 42. The extensive electronegative zone (red) associated with the N7 and O6 atoms of guanine creates a general hotspot for cation association to the major groove. Bp step slide in the positive direction has opposite effects in GG versus GC steps, whereby guanine-guanine overlap is decreased and increased, respectively. The greater charge density for GG steps lacking large positive slide and GC steps having positive slide appears to promote divalent metal binding (green dumbbells).
Although the presence of Co2+ or Ni2+ induces a conformational change over the central DNA component, metal association to the non-central elements appears to follow similar conformation-specific patterns as characterized for Mn2+ binding (Table 3 and Figure 4; 13,14). This can be seen by the strong correlation of metal occupancies, including several key vacant locations, between Mn2+ and Co2+/Ni2+ for GG (position −16/−15, + slide) and GC (position 58/−59, − slide) dinucleotide sites. Moreover, Co2+ and Ni2+ also display N7/O6 cross-linking at GG sites, in which the 3′ guanine is shifted into the major groove (Figure 3B). This allows simultaneous coordination of both guanine bases and creates an additional, lower occupancy N7 coordination site at the 3′ base.
The nucleosomal double helix displays a pronounced degree of mobility, which is exemplified by the fact that one can observe distinct stretching configurations for the same DNA sequence in different NCP crystal structures (17,18,25). Moreover, the DNA B-factors are systematically high in the NCP, suggesting that the double helix can also ‘breathe’ in the crystalline state. The shift in DNA register from the presence of Co2+/Ni2+ may be a consequence of the metals binding to and stabilizing an alternate conformation, or metal association may help to promote rearrangement.
Inspection of Co2+/Ni2+ occupancies over the central DNA region suggests that the register change, which breaks the symmetric alignment of the palindromic DNA, creates additional, high-affinity sites for metal association. In fact, a single bp shift in register or stretching of the double helix can dramatically alter the local DNA conformation (18). We had previously found that the proximity of the 5′ thymine methyl group at TG elements can modulate Mn2+ binding, which is favored by positive slide associated with minor groove bending that reduces potential steric interference with the coordinated metal hydrate (13). From the stretch-induced alterations in Co2+/Ni2+-NCP, increased positive slide at TG elements at positions 8 and 14 creates new metal binding sites in one particle half (Figure 3C and D). This emphasizes that substantial conformational alterations can come about from both stretch-specific structure as well as general DNA orientational changes, which can modulate major versus minor groove bending and associated structural parameters.
By forming strong coordinate-covalent bonds with purine N7 groups while maintaining conformational discrimination, Co2+ and Ni2+ can thus induce or select for distinct DNA structures. Moreover, although not clear due to the disorder of this region, the base flip-out is likely directly promoted by metal binding. We do observe metal coordination within the helical stack at this location (base 14 of TGCCT), and the flipped out guanine, as well as cytosine, bases could support metal coordination, which may not be observed because of high mobility (Figure 2). Considering that the conformational attributes for the GC and GG steps at this location in the Mn2+-NCP structure apparently preclude metal binding (low slide at TG and high positive slide for GG; 13,14), the base flip-out may also be directly promoted by creating new binding sites for Co2+/Ni2+ (see Figures 2 and 4).
Primary Co2+/Ni2+-histone coordination at histidine sites
The largest anomalous difference map peaks (Co2+, 29σ and Ni2+, 24σ) coincide with the major interparticle interface, in which divalent metal binding stabilizes interaction of electronegative elements from the H2A–H2B dimer with those of H3 and the H4 N-terminal tail (17). Co2+ and Ni2+ are observed to support the same coordinate bonds as Mn2+, namely with the carboxylate group of H3 Asp77 and the peptide carbonyl oxygen atom of H2B Val45. The preference for the same, octahedral, coordination geometry is likely key to facile metal substitution at this ionically driven binding site.
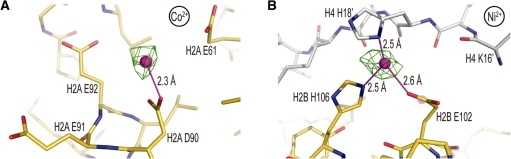
In addition to the above, ionic histone coordination, Co2+ and Ni2+ are also observed to coordinate exclusively with carboxylate groups at several other locations, including H3 Asp77, H2A Asp90 and H4 Asp24 (Figure 5A). On the other hand, the majority of observed histone association entails coordination to histidine—a favored protein ligand for intermediate-soft metals (see Table 2; 9). This mode of binding involves H2B residues His46 and His106 on the surface of the particle, as well as H2B His79 situated at the H2A–H2B dimer–dimer interfaces. Interestingly, one of the H2B His106 sites encompasses additional coordination with His18 of the H4 N-terminal tail from a neighboring particle (Figure 5B). Such internucleosomal histone–histone cross-linking could have functional implications for heavy metal toxicity.
Figure 5.
Co2+/Ni2+-histone binding site selectivity. (A) Co2+-aspartate coordination within an electronegative patch of H2A. (B) Ni2+ coordination to histidine, glutamate and an additional histidine ligand from the H4 tail of a neighboring particle (gray carbon atoms). (A and B) Co2+/Ni2+ ions and coordinate (-covalent) bonds appear as magenta spheres and lines, respectively. An anomalous difference electron density map, contoured at 4σ (A) and 8σ (B), is shown superimposed on the model.
Heavy monovalent cation association
Compared to multivalent cations, monovalent species are easily dehydrated and have reduced preference for a defined, regular coordination geometry (9,26). They therefore generally interact with macromolecules with lowered affinity and site selectivity. This is consistent with the monovalent metal binding sites found for Cs+-NCP and Rb+-NCP (see Table 2). In stark contrast to the much softer Ni2+/Co2+ ions, these hard alkali metals are observed coordinated to oxygen atoms at only a limited number of sites.
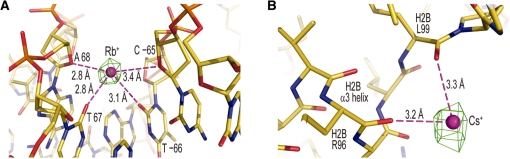
Cs+/Rb+ binding to the DNA involves the exocyclic groups of thymine and cytosine, as well as the sugar ring, in the minor groove. The apparent highest affinity site type, which occurs in both particle halves at the largest anomalous difference map values observed for Cs+ (7.2σ, 6.1σ) and Rb+ (5.4σ, 4.4σ), coincides with an extremely narrow AT dinucleotide minor groove element (Figure 6A). The narrowness of the groove and positioning of thymine O2 atoms in the AT steps allows simultaneous coordination to four different oxygen groups. This preferential mode of binding is identical to that characterized for monovalent cation association with oligonucleotide DNA and supports the premise that Na+ and K+ in the nucleus may stabilize minor groove narrowing over specific sequence elements (27).
Figure 6.
Cs+/Rb+ binding site selectivity. (A) Rb+ coordination in the minor groove at an AT dinucleotide element. (B) Cs+ coordination at an α-helix C-terminus. (A,B) Cs+/Rb+ ions and coordinate bonds appear as magenta spheres and broken lines, respectively. An anomalous difference electron density map, contoured at 4σ (A) and 5σ (B), is shown superimposed on the model.
With regard to histone binding, only one site is common to both Cs+ and Rb+, which occurs in both particle halves and involves coordination to multiple peptide carbonyl oxygen atoms around the C-terminus of the H2B α3 helix (Figure 6B). Cation binding is promoted by the juxtaposed carbonyl groups and the negative dipole component of the α-helix.
DISCUSSION
By virtue of their ability to form bonds with partial covalent character, soft metal cations can have a dominant effect on double helix structure. The tendency to elicit pronounced conformational changes upon binding can give rise to both local deformations or global form transitions. While this capacity makes soft metals generally toxic, it also underlies the therapeutic potential of platinum-based drugs (28). Although the exact structural nature of platinum lesions formed in the nucleosome is not clear (15,29–31), naked DNA with intra- or inter-strand cross-links displays pronounced kink distortions, which apparently induce apoptosis by interfering with transcription and other genomic transactions (28). In contrast to the purine N7-purine N7 cross-linking activity of Pt2+, we do not observe this mode of binding for Co2+ or Ni2+, which nonetheless display an absolute preference for N7 coordination in the major groove. Moreover, with the region of histone register shift notwithstanding, Co2+ and Ni2+ binding does not induce significant structural changes and is apparently largely governed by the preexisting DNA conformation in the nucleosome, as we have observed for association of Mn2+ (13,14). Therefore, relative to the very soft Pt2+, the much-reduced DNA-distorting activity of Co2+ and Ni2+ is consistent with the lower softness and binding affinity of these divalent metals.
In an earlier study where centered and off-centered NCP were treated with platinum anticancer agents, we found that cisplatin– and oxaliplatin–DNA adducts did not cause significant alteration of nucleosome positioning (32). However, nucleosomal platinum drug adducts do produce conformational perturbations in the double helix (30,31), and they may also be capable of histone–DNA register modulation in an analogous manner as we have found here for Co2+ and Ni2+ treatment. Since the occurrence of DNA stretching in the nucleosome is highly prevalent and some sequences display a mixture of stretching states (17,18,25,33–35), it is not surprising that a small molecule factor can influence this property (36). On the other hand, the incidence of base extrusion at the 1.5-turn position accompanying stretching induced by Co2+/Ni2+ binding would, a priori, appear to require a substantial energetic input. However, this is inconsistent with the general inability of Co2+/Ni2+ association to cause conformational changes in the DNA. Thus, it implies that the double helix at the 1.5-turn location is predisposed to conformational change, which is consistent with this site having the greatest DNA distorting potential and consequently the strongest influence on nucleosome positioning (18,37–39). G|C-rich sequences, in particular, appear prone to deformation as required for minor groove narrowing to fit the special histone-binding site at this location (39). Moreover, a 145-bp NCP construct of nearly identical DNA sequence as the NCP147 used in the present study displays extreme kinking into the minor groove over the same TGCCT/AGGCA element distorted by Co2+/Ni2+ association (18). Taken together, this indicates that extreme double helix deformations for certain sequence elements at the 1.5-turn location can be induced with relatively low energetic input by at least several factors. It is interesting to speculate that such conformational properties may have specific functions in natural genomic processes.
Alteration of chromatin structure has been implicated as one of the contributing factors in heavy metal-mediated carcinogenesis (6,7), and from what we observe here, Co2+ and Ni2+ may contribute to pathogenesis by altering nucleosome structure. Changes in histone–DNA register and double helix distortions could have a number of different effects by modifying compaction, dynamics or interactions with nuclear factors. For instance, Ni2+ appears to act as a cocarcinogen in part by inhibiting DNA repair (5–7), and alteration of nucleosome structure or preferential association at sites of DNA damage, akin to the deformed configuration we see at the 1.5-turn location, could contribute to the inhibitory activity. Moreover, high-affinity coordination of soft metal cations at DNA or histone sites would alter chromatin compaction attributes—a carcinogenic mechanism also proposed for Ni2+-mediated DNA hypermethylation (6,7,40). In addition to its ability to silence genes via DNA methylation, Ni2+ is known to modulate a number of histone post-translational modifications (6,7). In particular, Ni2+ binding to His18 of the H4 tail was previously proposed to explain loss of acetylation at Lys12 and Lys16 (41). Our findings lend support to this idea and further suggest that internucleosomal histidine–histidine cross-linking could contribute severely to pathogenesis.
Although more recent findings have indicated that the actual mutagenic potential of most heavy metals may not play the major role in cancer production (5–8), studies have shown that the DNA damaging activity of Ni2+ is enhanced by the presence of histone proteins (1,3). Moreover, the overall capacity of Co2+ and Ni2+ for generating reactive oxygen species is facilitated by metal association with peptides, in particular, histidine-containing (1,3,4). As such, the Co2+/Ni2+ coordination we observe in the nucleosome core suggests that multiple histone sites could provide an active source of reactive species generated immediately proximal to the DNA.
ACCESSION NUMBERS
RCSB Protein Data Bank accession codes: 3MGP (Co2+-NCP), 3MGQ (Ni2+-NCP), 3MGR (Rb+-NCP) and 3MGS (Cs+-NCP).
FUNDING
Academic Research Council grants 10/05 and 19/08 from the Ministry of Education (Singapore). Funding for open access charge: Ministry of Education (Academic Research Council grant 19/08).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to C. Schulze-Briese, M. Wang and V. Olieric for all of their help at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). We thank G.E. Davey for input on the manuscript.
REFERENCES
- 1.Kasprzak KS. Oxidative DNA and protein damage in metal-induced toxicity and carcinogenesis. Free Radic. Biol. Med. 2002;32:958–967. doi: 10.1016/s0891-5849(02)00809-2. [DOI] [PubMed] [Google Scholar]
- 2.Kawanishi S, Hiraku Y, Murata M, Oikawa S. The role of metals in site-specific DNA damage with reference to carcinogenesis. Free Radic. Biol. Med. 2002;32:822–832. doi: 10.1016/s0891-5849(02)00779-7. [DOI] [PubMed] [Google Scholar]
- 3.Bal W, Kasprzak KS. Induction of oxidative DNA damage by carcinogenic metals. Toxicol. Lett. 2002;127:55–62. doi: 10.1016/s0378-4274(01)00483-0. [DOI] [PubMed] [Google Scholar]
- 4.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 5.Beyersmann D, Hartwig A. Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Arch. Toxicol. 2008;82:493–512. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- 6.Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem. Res. Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arita A, Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. 2009;1:222–228. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galanis A, Karapetsas A, Sandaltzopoulos R. Metal-induced carcinogenesis, oxidative stress and hypoxia signalling. Mutat. Res. 2009;674:31–35. doi: 10.1016/j.mrgentox.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Glusker JP. Structural aspects of metal liganding to functional groups in proteins. Adv. Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 10.van de Sande JH, McIntosh LP, Jovin TM. Mn2+ and other transition metals at low concentration induce the right-to-left helical transformation of poly[d(G-C)] EMBO J. 1982;1:777–782. doi: 10.1002/j.1460-2075.1982.tb01247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duguid JG, Bloomfield VA, Benevides JM, Thomas GJ., Jr Raman spectroscopy of DNA-metal complexes. II. The thermal denaturation of DNA in the presence of Sr2+, Ba2+, Mg2+, Ca2+, Mn2+, Co2+, Ni2+, and Cd2+ Biophys. J. 1995;69:2623–2641. doi: 10.1016/S0006-3495(95)80133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bock CW, Katz AK, Markham GD, Glusker JP. Manganese as a replacement for magnesium and zinc: functional comparison of divalent ions. J. Am. Chem. Soc. 1999;121:7360–7372. [Google Scholar]
- 13.Davey CA, Richmond TJ. DNA-dependent divalent cation binding in the nucleosome core particle. Proc. Natl Acad. Sci. USA. 2002;99:11169–11174. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu B, Davey CA. Using soft X-rays for a detailed picture of divalent metal binding in the nucleosome. J. Mol. Biol. 2010;398:633–640. doi: 10.1016/j.jmb.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 15.Wu B, Droge P, Davey CA. Site selectivity of platinum anticancer therapeutics. Nat. Chem. Biol. 2008;4:110–112. doi: 10.1038/nchembio.2007.58. [DOI] [PubMed] [Google Scholar]
- 16.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 17.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 18.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J. Mol. Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 19.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 20.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 22.Hendrickson WA, Ogata CM. Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 1997;276:494–523. doi: 10.1016/S0076-6879(97)76074-9. [DOI] [PubMed] [Google Scholar]
- 23.Djinovic Carugo K, Helliwell JR, Stuhrmann H, Weiss MS. Softer and soft X-rays in macromolecular crystallography. J Synchrotron Radiat. 2005;12:410–419. doi: 10.1107/S0909049504025762. [DOI] [PubMed] [Google Scholar]
- 24.Chiu TK, Dickerson RE. 1 A crystal structures of B-DNA reveal sequence-specific binding and groove-specific bending of DNA by magnesium and calcium. J. Mol. Biol. 2000;301:915–945. doi: 10.1006/jmbi.2000.4012. [DOI] [PubMed] [Google Scholar]
- 25.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Draper DE, Misra VK. RNA shows its metal. Nat. Struct. Biol. 1998;5:927–930. doi: 10.1038/2901. [DOI] [PubMed] [Google Scholar]
- 27.McFail-Isom L, Sines CC, Williams LD. DNA structure: cations in charge? Curr. Opin. Struct. Biol. 1999;9:298–304. doi: 10.1016/S0959-440X(99)80040-2. [DOI] [PubMed] [Google Scholar]
- 28.Todd RC, Lippard SJ. Inhibition of transcription by platinum antitumor compounds. Metallomics. 2009;1:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danford AJ, Wang D, Wang Q, Tullius TD, Lippard SJ. Platinum anticancer drug damage enforces a particular rotational setting of DNA in nucleosomes. Proc. Natl Acad. Sci. USA. 2005;102:12311–12316. doi: 10.1073/pnas.0506025102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ober M, Lippard SJ. Cisplatin damage overrides the predefined rotational setting of positioned nucleosomes. J. Am. Chem. Soc. 2007;129:6278–6286. doi: 10.1021/ja0706145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ober M, Lippard SJ. A 1,2-d(GpG) cisplatin intrastrand cross-link influences the rotational and translational setting of DNA in nucleosomes. J. Am. Chem. Soc. 2008;130:2851–2861. doi: 10.1021/ja710220x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu B, Davey CA. Platinum drug adduct formation in the nucleosome core alters nucleosome mobility but not positioning. Chem. Biol. 2008;15:1023–1028. doi: 10.1016/j.chembiol.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 34.Edayathumangalam RS, Weyermann P, Dervan PB, Gottesfeld JM, Luger K. Nucleosomes in solution exist as a mixture of twist-defect states. J. Mol. Biol. 2005;345:103–114. doi: 10.1016/j.jmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Davey GE, Wu B, Dong Y, Surana U, Davey CA. DNA stretching in the nucleosome facilitates alkylation by an intercalating antitumour agent. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 37.Fitzgerald DJ, Anderson JN. DNA distortion as a factor in nucleosome positioning. J. Mol. Biol. 1999;293:477–491. doi: 10.1006/jmbi.1999.3171. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez AG, Anderson JN. Nucleosome positioning determinants. J. Mol. Biol. 2007;371:649–668. doi: 10.1016/j.jmb.2007.05.090. [DOI] [PubMed] [Google Scholar]
- 39.Wu B, Mohideen K, Vasudevan D, Davey CA. Structural insight into the sequence dependence of nucleosome positioning. Structure. 2010;18:528–536. doi: 10.1016/j.str.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Lee YW, Klein CB, Kargacin B, Salnikow K, Kitahara J, Dowjat K, Zhitkovich A, Christie NT, Costa M. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol. Cell. Biol. 1995;15:2547–2557. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoroddu MA, Schinocca L, Kowalik-Jankowska T, Kozlowski H, Salnikow K, Costa M. Molecular mechanisms in nickel carcinogenesis: modeling Ni(II) binding site in histone H4. Environ. Health Perspect. 2002;110(Suppl. 5):719–723. doi: 10.1289/ehp.02110s5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: dinucleotide conformational maps. J. Mol. Biol. 2000;295:71–83. doi: 10.1006/jmbi.1999.3236. [DOI] [PubMed] [Google Scholar]