Abstract
The Mcm2-7 complex is the eukaryotic replicative helicase, a toroidal AAA+ molecular motor that uses adenosine triphosphate (ATP) binding and hydrolysis to separate duplex DNA strands during replication. This heterohexameric helicase contains six different and essential subunits (Mcm2 through Mcm7), with the corresponding dimer interfaces forming ATPase active sites from conserved motifs of adjacent subunits. As all other known hexameric helicases are formed from six identical subunits, the function of the unique heterohexameric organization of Mcm2-7 is of particular interest. Indeed, prior work using mutations in the conserved Walker A box ATPase structural motif strongly suggests that individual ATPase active sites contribute differentially to Mcm2-7 activity. Although only a specific subset of active sites is required for helicase activity, another ATPase active site (Mcm2/5) may serve as a reversible ATP-dependent discontinuity (‘gate’) within the hexameric ring structure. This study analyzes the contribution that two other structural motifs, the Walker B box and arginine finger, make to each Mcm2-7 ATPase active site. Mutational analysis of these motifs not only confirms that Mcm ATPase active sites contribute unequally to activity but implicates the involvement of at least two additional active sites (Mcm5/3 and 6/2) in modulating the activity of the putative Mcm2/5 gate.
INTRODUCTION
In eukaryotes, the replicative helicase is the minichromosome maintenance (Mcm) complex, a heterohexamer of six individually essential proteins numbered two through seven [Mcm2-7; (1)]. Each Mcm subunit belongs to the AAA+ family of adenosine triphosphatases (ATPases) that typically form active sites at dimer interfaces (2–4). One subunit of an ATPase active site contributes both Walker A and Walker B motifs, while the adjoining subunit contributes the arginine finger motif (2). In addition to catalytic roles in ATP binding and hydrolysis, these motifs likely make variable contributions to the inter-site communication needed to enable coordinated DNA unwinding (2). Mcm2-7 forms a toroidal complex with an open central channel that putatively constitutes the substrate (i.e. DNA) binding site flanked by six ATPase active sites (3,5–7).
The heterohexameric organization of Mcm2-7 is unique among known helicases (7) and appears to be functionally important because each of the six Mcm subunits are completely conserved in all eukaryotes examined to date (1,8,9). As hexameric helicases are typically formed from six copies of an identical subunit, the assumed equal functional involvement of each subunit forms the cornerstone of all mechanistic models of hexameric helicase function (7,10,11). In contrast, because Mcm2-7 is formed from six distinct subunits, it contains six unique ATPase active sites. Five of the six ATPase active sites (Mcm5/3, 3/7, 7/4, 4/6 and 6/2) have been directly demonstrated by subunit association experiments (Figure 1A), and mutational analysis of the Mcm3/7, 7/4 and 6/2 dimers has identified the active site motifs contributed by each subunit to the active sites (12,13). The sixth site (Mcm2/5) is too unstable to be observed by these means but is inferred to exist by (i) considerations of the toroidal nature of the complex; and (ii) the similar effect that mutations in this putative active site have on the association rate of the complex with ssDNA and its relative ability to bind circular single-strand (ss)DNA (13–16).
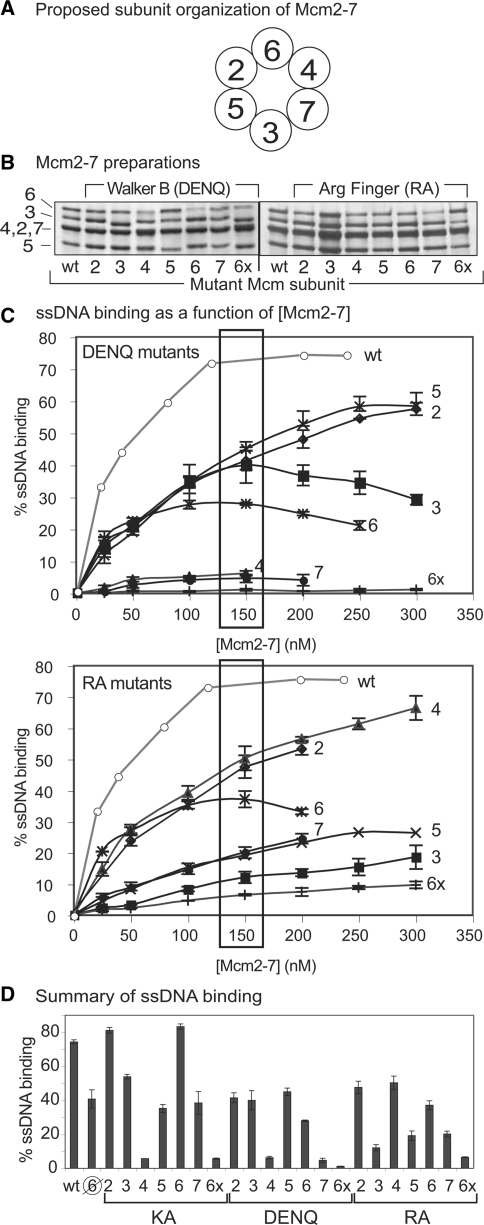
Figure 1.
DNA binding of mutant Mcm2-7 complexes. (A) Proposed subunit organization of Mcm2-7 (12,13). (B) Silver-stained SDS-PAGE gel of wild-type and indicated mutant Mcm2-7 hexamers. (C) ATPγS-dependent ssDNA binding as a function of (Mcm2-7) for DENQ (top) or RA (bottom) mutant complexes. The number to the right of each curve denotes the mutant subunit; 6x refers to Mcm2-7 complexes in which all six subunits contain the indicated mutation. Boxes around the 150 nM points indicate the data used to create Figure 1D. (D) Summary of Mcm2-7 ssDNA binding by the pentameric preparation ( ) and the single and sextuple (6x) KA (left), DENQ (middle) and RA (right) mutant hexamers (at 150 nM). The data for the Mcm2-7 complexes containing the indicated KA mutants are from ref. 5 and are shown for comparison. Assays for all three sets of mutant complexes were performed under identical conditions. Although the Mcm2-7 complex containing Mcm7KA appears to bind ssDNA, this binding is largely independent of ATP (5), and as such, we consider this mutant to be defective in ssDNA binding.
) and the single and sextuple (6x) KA (left), DENQ (middle) and RA (right) mutant hexamers (at 150 nM). The data for the Mcm2-7 complexes containing the indicated KA mutants are from ref. 5 and are shown for comparison. Assays for all three sets of mutant complexes were performed under identical conditions. Although the Mcm2-7 complex containing Mcm7KA appears to bind ssDNA, this binding is largely independent of ATP (5), and as such, we consider this mutant to be defective in ssDNA binding.
Biochemical analysis of Mcm subcomplexes (14,15,17) allows the basic classification of the subunits into two functional groups: one that contains the Mcm4, 6 and 7 subunits, which form hexameric subcomplexes [Mcm467 (18) and Mcm7/4 (17)] with in vitro helicase activity (helicase subunits), while the remaining subunits (regulatory subunits) appear to have a minimal role in DNA unwinding and, in fact, inhibit the in vitro activity of the helicase subunits under some conditions (14,16,19). Although the function of the Mcm regulatory subunits is unknown, analysis of the ssDNA association rates of mutant Mcm2-7 complexes and the ability of these complexes to bind circular ssDNA suggests that the Mcm2/5 ATPase active site functions as an ATP-dependent discontinuity or ‘gate’ in the toroidal Mcm structure (5,12,14).
Although an in vivo function for the putative Mcm2/5 gate is currently unknown, various data suggest that the topological state of the gate may either assist in initially loading Mcm2-7 onto replication origins or, alternatively, regulate the helicase activity of the complex (5,12,14). If true, the Mcm2/5 gate must communicate with the active site(s) required for helicase function. Because adjacent subunits communicate primarily through nucleotide occupancy and resultant conformational changes in their shared ATPase active sites, mutations in the active sites that link the gate to the helicase subunits (i.e. the Mcm5/3 and 6/2 ATPase active sites) might mimic gate defects by interfering with this communication.
This study focuses on the DNA binding and helicase properties of Mcm2-7 complexes containing Walker B mutations and reexamines and extends the properties of the previously published arginine finger mutant complexes. Our results not only confirm that the Mcm7/4 and 4/6 sites are essential for in vitro helicase activity but additionally indicate that the Mcm ATPase active sites that flank Mcm2/5 (i.e. Mcm5/3 and 6/2) modulate the biochemical activities of the putative Mcm2/5 gate.
MATERIALS AND METHODS
Reagents, Mcm mutations, computer images
Buffers and other reagents are detailed in the Supplementary Data. The Saccharomyces cerevisiae Mcm mutations used in this study are amino acid substitutions in the Walker B (DE→NQ, DENQ) and arginine finger (R→A, RA) motifs, respectively (Table 1). All constructs have been presented previously and were completely sequenced to verify that they contain only the indicated mutation (5,12,15). The acquisition and editing of computer images used in Figures 1 and 2 are described in Supplementary Data.
Table 1.
Mcm mutants used in this study
| Mutations | Walker A box (KA) | Walker B box (DENQ) | Arg finger (RA) |
|---|---|---|---|
| MCM2 | 542–GDPGTAKSQ | 604–LIDEFDK | 673–LSRFD |
| MCM3 | 408–GDPSTAKSQ | 470–CIDEFDK | 539–LSRFD |
| MCM4 | 567–GDPSTSKSQ | 629–CIDEFDK | 698–LSRFD |
| MCM5 | 415–GDPGTAKSQ | 477–CIDEFDK | 546–LSRFD |
| MCM6 | 574–GDPSTSKSQ | 636–CIDEFDK | 705–MSRFD |
Figure 2.
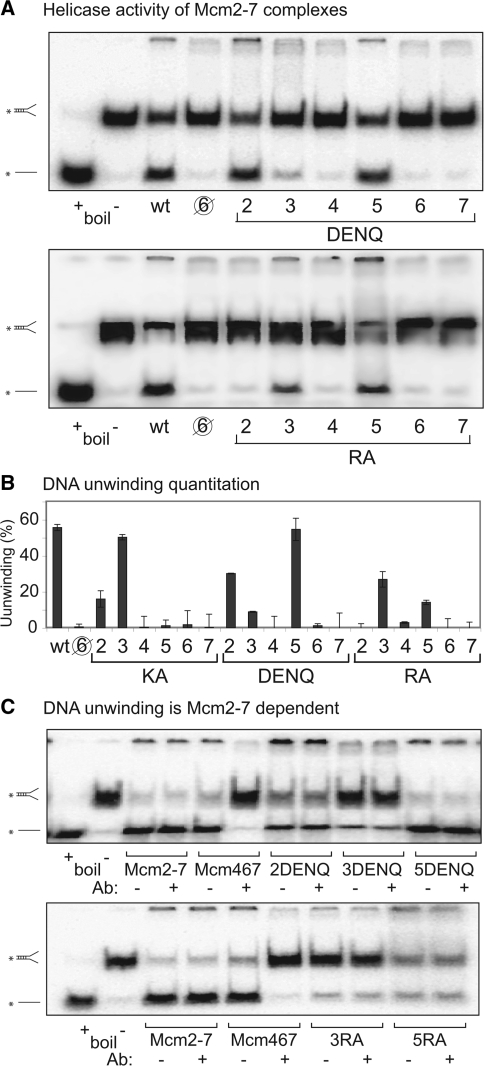
(A) Helicase activity of wild type (wt) Mcm2-7, the pentameric preparation ( ), and single DENQ (top) and RA (bottom) mutant Mcm2-7 complexes. Electrophoretic positions of the fork substrate and dissociated single strand are shown at left. The lane marked ‘boiled (+)’ serves as a positive control and shows the position of the radioactive single strand after unwinding of the fork; the lane marked ‘boiled (−)’ shows the position of the radioactive fork in the absence of dissociation. The identity of the mutant subunit is listed below the gel. (B) Quantitation of unwinding activities shown in (A). Previous results with the KA mutant complexes (14) are listed for comparison. (C) Confirmation that DNA unwinding of the DENQ and RA mutant preparations is Mcm2-7-dependent. Pair-wise helicase assays of the indicated Mcm2-7 and Mcm467 preparations in the presence or absence of an antibody that specifically blocks Mcm467 DNA unwinding (14) are shown.
), and single DENQ (top) and RA (bottom) mutant Mcm2-7 complexes. Electrophoretic positions of the fork substrate and dissociated single strand are shown at left. The lane marked ‘boiled (+)’ serves as a positive control and shows the position of the radioactive single strand after unwinding of the fork; the lane marked ‘boiled (−)’ shows the position of the radioactive fork in the absence of dissociation. The identity of the mutant subunit is listed below the gel. (B) Quantitation of unwinding activities shown in (A). Previous results with the KA mutant complexes (14) are listed for comparison. (C) Confirmation that DNA unwinding of the DENQ and RA mutant preparations is Mcm2-7-dependent. Pair-wise helicase assays of the indicated Mcm2-7 and Mcm467 preparations in the presence or absence of an antibody that specifically blocks Mcm467 DNA unwinding (14) are shown.
Proteins and purification
Wild-type and mutant Mcm2-7 complexes were expressed in baculovirus-infected insect cells and purified through multiple chromatography steps as previously described [see Supplementary Data, (12) and (15)]. Although a gel filtration step was used to enrich for hexamers, molecular weight resolution is limited by this technique, and we cannot reliably separate hexamers (∼600 kDa) from pentamers (∼500 kDa). The protein preparations used in this study were characterized (5,12,14) by both quantitative western blotting using subunit-specific antibodies and known amounts of individually purified Mcm subunits as standards to establish the stoichiometry of co-migrating subunits (i.e. Mcm2 4, and 7; Supplementary Table S1), and by gel filtration or co-immunoprecipitation to confirm that the final preparations maintained a hexameric oligomerization (Supplementary Figure S1). By these criteria, although most of our preparations are limited for Mcm6, at least 37–76% of the protein in these preparations are heterohexamers [except those containing the Mcm7RA (23% heterohexamers) and Mcm6DENQ (13% heterohexamers) subunits]. In addition, this study utilizes a previously described pentameric Mcm complex that contains all subunits except Mcm6 (subsequently called the Mcm pentameric preparation) (5,14). Protein concentrations were quantified as described previously (5,12,14) and, unless otherwise noted, refer to moles of hexamers.
DNA binding assays
Double filter binding assays for ssDNA (including the association rate enhancement and circularization studies) were carried out as described [Supplementary Methods, (5,14)], using an oligonucleotide probe (oligo 826, TGTCTAATCCCGAAAGGCCCTGCCACTGAAATCAACACCTAAAGCATTGA) that was 5′-radiolabeled using T4 polynucleotide kinase and [γ32P]ATP. Unless otherwise noted, the data points for the ssDNA binding, association and circularization assays represent the averages of ≥3 repeats of the same experiment, and the error bars correspond to the standard deviations.
Association assay
This assay has been described previously (14). For preincubation experiments, Mcm complexes were incubated with 5 mM ATPγS for 30 min at 30°C prior to addition of radiolabeled oligo 826 and the remaining reaction components at t = 0. At the indicated times, 5-µl samples were withdrawn and analyzed by filter binding. The association kinetics were plotted as described (20,21) using Equation (1):
where (R) and (O) are the total concentrations of Mcm complex and ssDNA, respectively, and (RO) is the concentration of the Mcm-ssDNA complex at time t. The slope of the line represents the apparent association rate constant (ka).
Circularization assay
This assay has been described previously (14). Briefly, 5 mM sodium fluoride and competitor DNA [either 10 µg/µl circular or linear M13mp18 ssDNA or 1× TE (no competitor control)] in buffer B2 was added to the standard reactions but varied in the order of addition. The radiolabeled probe used was oligo 826. Reactions labeled ‘no comp’ contained all of the reagents (except M13 competitor ssDNA) added simultaneously and incubated at 30°C. In the reactions labeled ‘cir 1st’, all reagents except ATPγS were present at t = 0 and incubated at 30°C; nucleotide was added at t = 60 min, and incubation continued at 30°C. In the reactions labeled ‘cir 2nd’, all reagents other than β-glycerophosphate, sodium fluoride, oligo 826 and M13 ssDNA were present at t = 0 and incubated at 30°C; the remaining reagents were added at t = 60 min, and incubation continued at 30°C. Mcm/radiolabeled probe complexes were measured using the double filter binding assay at t = 120 min.
Helicase assay
Helicase assays were performed essentially as described [see Supplementary Data and (5,14)]. To distinguish the helicase activity of Mcm2-7 from that of any possible contaminating Mcm467 subcomplexes in our mutant preparations, helicase assays in Figure 2C were treated with a specific neutralizing antibody (anti-Mcm7, sc-6688, Santa Cruz) that inhibits the helicase activity of Mcm467 but not of Mcm2-7 (14). The experiments were conducted ≥2 times, and the gel images shown depict typical results.
RESULTS
Experimental rationale
To date, helicase activity from recombinant Mcm2-7 has only been observed following co-expression of all six subunits in an insect cell system (14,22). Due to the inherent variability of this system and the difficulty of purifying hexameric complexes away from Mcm oligomers of similar size, our analysis suggests that, on average, only about half of the protein in our preparations is in the form of Mcm2-7 heterohexamers, with the balance of the preparation being a mixture of Mcm subcomplexes (see ‘Materials and Methods’ section, Supplementary Table S1 and Supplementary Figure S1). This mixture of Mcm species potentially makes it difficult to specifically establish the biochemical activities of the intact Mcm2-7 complex. However, this issue can be largely mitigated by knowing the biochemical activities of Mcm subcomplexes and using this information to discount their contribution to assays of heterohexameric Mcm2-7 activity.
Toward this end, we have generated and assayed a representative Mcm subcomplex that is likely the major non-hexameric species in our preparations. As the limiting subunit in our current Mcm preparations is Mcm6, the bulk of non-heterohexameric Mcm species in the preparations are likely deficient for Mcm6. Thus, to model these subcomplexes, we purified and characterized a pentameric Mcm preparation lacking Mcm6 (henceforth referred to as the pentameric preparation). In the experiments that follow, the pentameric preparation serves as a metric for Mcm subcomplexes. Thus, biochemical activities that are present in our Mcm2-7 preparations but lacking in the pentameric preparations are considered to reflect a unique activity of the heterohexamer.
Mcm2-7 requires only a subset of ATPase active sites for ssDNA binding
Mcm2-7 binds ssDNA in an ATP-dependent manner (5,16). To examine the role(s) that the six Mcm Walker B and arginine finger motifs make to ssDNA binding, mutant Mcm2-7 complexes containing either Walker B or arginine finger mutations were expressed, purified, and characterized (Figure 1B, Supplementary Figure S1, Supplementary Table S1). These heterohexameric complexes contain a single subunit with a mutation in either the Walker B [substitution of the conserved acidic residues (DE) to their amide counterparts (NQ), i.e. DENQ mutants] or arginine finger (substitution of the conserved arginine to alanine, i.e. RA mutants) motif in the company of the remaining five wild-type Mcm subunits, as indicated [(12,15); Table 1]. We previously characterized the ATPase activities of these 12 mutant complexes, and both DENQ and RA substitutions individually eliminate ATP hydrolysis from their resident active site, as demonstrated in mutational studies of the dimeric Mcm7/4 ATPase active site (12,13).
Using a DNA filter-binding assay (5), ATPγS-dependent ssDNA binding of mutant Mcm2-7 complexes to a radiolabeled oligonucleotide was assayed and compared to wild type Mcm2-7 binding. Control complexes that contain the indicated mutation in all six of the Mcm subunits either eliminate (6xDENQ) or greatly reduce (6xRA) ATPγS-dependent ssDNA binding (Figure 1C), confirming the global involvement of the six Mcm ATPase active sites in ssDNA binding. However, complete integrity of the Mcm toroidal structure is not required for this activity because the pentameric preparation demonstrates substantial ATPγS-dependent ssDNA binding (Figure 1D).
In contrast to expectations of equal active site involvement, analogous mutations in specific subunits differentially affect ssDNA binding as a function of Mcm2-7 concentration (Figure 1C). Supplementary Table 2 contains a list of the apparent ssDNA dissociation constant (Kd) for each preparation, as well as those previously obtained for the Walker A mutant (lysine to alanine substitution, i.e. KA mutant) complexes for comparison (5). Several mutant preparations (i.e. Mcm3DENQ, 6DENQ and 6RA) demonstrate decreases in ssDNA binding at higher protein concentrations, perhaps suggesting that these particular mutations promote the formation of non-productive aggregates. These results are summarized at a single protein concentration in Figure 1D and compared with the ssDNA binding results of the previously published KA mutant complexes (5).
The mutant Mcm2-7 complexes can be separated into two classes. Most of the mutations increase the dissociation constant for ssDNA binding 3- to 6-fold relative to wild-type Mcm2-7. Because this reduction in affinity does not occur with the Mcm pentameric preparation (Supplementary Table S2), the reduction in ssDNA affinity of the mutant complexes could be mechanistically significant, suggesting that these active sites are involved in the propagation of conformational changes within the complex that optimize ssDNA binding. In sharp contrast, the 4DENQ and 7DENQ mutations essentially ablate ssDNA binding (i.e. mutants bind similarly to the 6xDENQ complex); while the 3RA, 5RA and 7RA mutations severely reduce ssDNA binding comparable to the analogous 6xRA complex (7- to 8-fold, Figure 1C). This reduction strongly indicates a direct involvement of the component ATPase active sites in ssDNA binding. With the exception of the Mcm7KA complex (5), mutant Mcm2-7 complexes that still bind ssDNA do so in an ATPγS-dependent manner (data not shown). These data confirm that Mcm2-7 active sites participate unequally in ssDNA binding and most strongly implicate the primary involvement of the Mcm7/4 (4KA, 4DENQ, and 7RA), 3/7 (7KA, 7DENQ, and 3RA) and (to a lesser extent) the 5/3 (5RA) ATPase active sites in this activity.
Mcm2-7 active sites contribute differentially to DNA unwinding
We next examined the ability of the mutant Mcm2-7 complexes to unwind a short DNA fork substrate (Figure 2A). These results are quantified (Figure 2B) and compared to the results for the KA mutant complexes (14). Although most of the mutations severely reduce or eliminate DNA unwinding, several mutant complexes retain demonstrable helicase activity, which in some cases approach wild-type levels. Although most Mcm subcomplexes lack helicase activity (e.g. the pentameric preparation, Figure 2A), it is well known that the hexameric Mcm467 subcomplex efficiently unwinds DNA (18). To verify that the helicase activity of our mutant Mcm2-7 preparations was not caused by Mcm467 contamination, helicase assays were repeated with addition of a neutralizing antibody that specifically inhibits unwinding by Mcm467 but not wild-type Mcm2-7 (14). As shown in Figure 2C, addition of the antibody has no effect on the DNA unwinding activity of the mutant Mcm2-7 preparations, confirming that their helicase activity does not result from contaminating Mcm467.
If one hypothesizes that loss of helicase activity is due to functional loss of the indicated mutant active site, the unwinding potential of each mutation can be correlated with a particular ATPase active site within the Mcm2-7 complex. Three different classes of sites exist: (i) all mutations in the Mcm7/4 (4KA, 4DENQ and 7RA) and 4/6 (6KA, 6DENQ and 4RA) active sites eliminate DNA unwinding; (ii) none of the mutations within the Mcm5/3 (3KA, 3DENQ and 5RA) site eliminate unwinding; and (iii) mutations within the remaining active sites (Mcm2/5, 3/7 and 6/2) have mixed effects, with some mutations eliminating activity and others having little effect. These results directly demonstrate that Mcm ATPase active sites contribute unequally to helicase activity.
Mutations in the Mcm6/2 and Mcm4/6 ATPase active sites alter the ability of Mcm2-7 to bind circular ssDNA
In contrast to analogous mutations in the other Mcm ATPase active sites, defects in the Mcm2/5 site have one of two specific biochemical effects that both depend upon the order in which ATP enters the reaction. The first involves the ability of Mcm2-7 to bind circular ssDNA (i.e. the circularization assay). This activity is measured using an ssDNA binding assay in which an unlabelled circular ssDNA substrate competes for the binding of Mcm2-7 to a radiolabeled linear substrate (14). Although this assay measures circular ssDNA binding via a reduction in linear ssDNA binding, we will subsequently refer to this activity as circular ssDNA binding for simplicity.
Using this assay, wild-type Mcm2-7 was previously shown to bind much better to circular ssDNA if added to the reaction before ATPγS (Figure 3A, cir 1st) rather than after ATPγS preincubation [Figure 3A, cir 2nd; (14)]. This ATPγS preincubation effect depends upon both the Mcm2/5 ATPase active site and the oligomerization state of the complex. Mcm2-7 containing the 5KA mutant subunit (5) or the Mcm pentameric preparation [shown in Figure 3A for comparison; (5,14)] both bind circular ssDNA well, regardless of ATP preincubation. In contrast, the Mcm2-7 complex containing the 2RA mutant subunit or the Mcm467 subcomplex both bind circular ssDNA poorly regardless of ATP preincubation, with little effect on linear ssDNA binding (Figure 3A, Supplementary Table S3) (5). Because the closely related toroidal helicase SV-40 large T-antigen (TAg) binds ssDNA within its central channel (23), the ability of TAg to bind a DNA molecule lacking an end (i.e. circular DNA) would likely require remodeling of the complex to allow DNA access to the central channel. In contrast, the free end of a linear DNA substrate might bind the helicase by simply threading into the central channel without toroid opening. Thus, the ability of Mcm2-7 to bind circular DNA has been interpreted as changes in the topology of the Mcm toroidal structure that favor either opening (e.g. Mcm5KA complex) or closing (e.g. Mcm2RA complex) of the toroidal structure to facilitate or block access of circular ssDNA to the central channel, respectively (14).
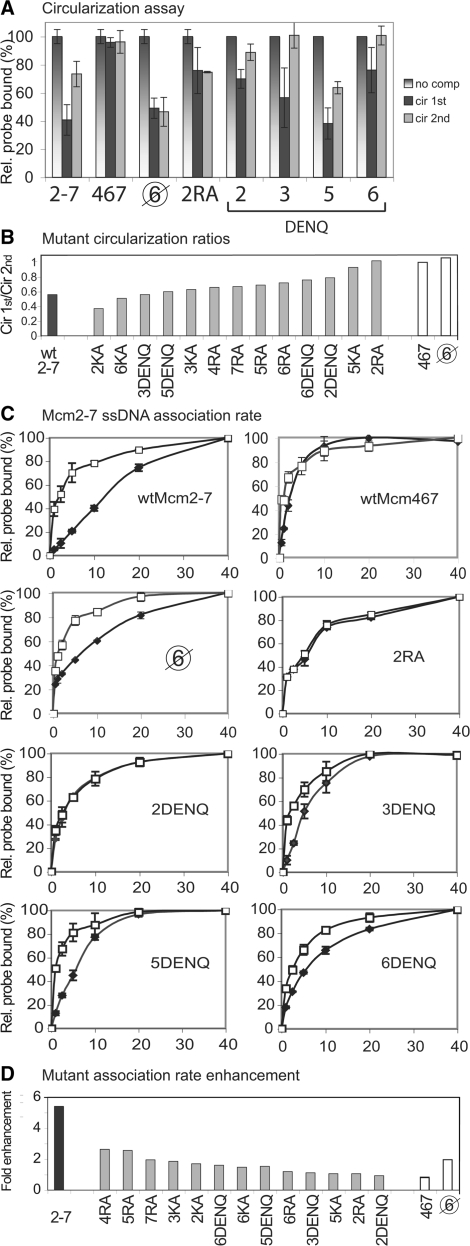
Figure 3.
Effects of ATP preincubation on Mcm2-7 circular ssDNA binding and ssDNA association by the wild-type (wt) and mutant complexes. (A) Circularization assay. This assay measures the ability of a circular ssDNA substrate (i.e. cir) to compete with the standard radiolabeled oligonucleotide substrate for Mcm2-7 binding as a function of ATPγS preincubation. The three vertical bars in the graph for each protein represent (i) normalized relative binding in the absence of circular competitor DNA; (ii) competition experiments in which both the cold circular competitor and radiolabeled oligonucleotide are added to the Mcm complex for 30 min prior to ATPγS addition (cir 1st); and (iii) conditions in which the Mcm preparation was incubated with ATPγS prior to addition of both cold circular ssDNA competitor and radiolabeled oligonucleotide (cir 2nd). Ring closure results for wild type Mcm2-7, Mcm467, the pentameric preparation ( ), and 2RA complexes have been previously published (14) and are shown here for comparison. (B) Summary of circularization. The ratio of cir1st/cir 2nd for each mutant Mcm2-7 preparation (Supplementary Table 3) is plotted in ranked order. (C) Mcm2-7/ssDNA association as a function of time. The Mcm preparations were either preincubated (open squares) or not (filled diamonds) with ATPγS for 30 min prior to addition of the ssDNA substrate, as described in the ‘Materials and methods’ section. The filter binding method and the radiolabeled ssDNA substrate used are the same as in Figure 1C. Note: the association rate data for the wild-type Mcm2-7, Mcm467, pentamer (
), and 2RA complexes have been previously published (14) and are shown here for comparison. (B) Summary of circularization. The ratio of cir1st/cir 2nd for each mutant Mcm2-7 preparation (Supplementary Table 3) is plotted in ranked order. (C) Mcm2-7/ssDNA association as a function of time. The Mcm preparations were either preincubated (open squares) or not (filled diamonds) with ATPγS for 30 min prior to addition of the ssDNA substrate, as described in the ‘Materials and methods’ section. The filter binding method and the radiolabeled ssDNA substrate used are the same as in Figure 1C. Note: the association rate data for the wild-type Mcm2-7, Mcm467, pentamer ( ), and Mcm2RA have been previously published [(5) and Supplementary Data in ref. 14), and are shown here for comparison. (D) Summary of association rate enhancement of the mutant Mcm2-7 preparations with ssDNA. The ratio of the association rate in the absence of ATPγS preincubation to the association rate in the presence of ATPγS preincubation for each mutant Mcm2-7 preparation (Supplementary Table 4) is plotted in ranked order.
), and Mcm2RA have been previously published [(5) and Supplementary Data in ref. 14), and are shown here for comparison. (D) Summary of association rate enhancement of the mutant Mcm2-7 preparations with ssDNA. The ratio of the association rate in the absence of ATPγS preincubation to the association rate in the presence of ATPγS preincubation for each mutant Mcm2-7 preparation (Supplementary Table 4) is plotted in ranked order.
Although the Walker A and arginine finger mutants have been previously studied with the circularization assay (5,14), the Walker B mutants have not been similarly examined (Figure 3A). In contrast to the control complexes, Mcm2-7 preparations containing the Walker B mutation in either the Mcm6/2 (i.e. Mcm2DENQ) or the Mcm4/6 (i.e. Mcm6DENQ) ATPase active site bind circular ssDNA poorly regardless of ATPγS preincubation. This result suggests that these mutations cause the corresponding complexes to preferentially exist in a topologically closed form. Note that this result is the opposite that is observed for the topologically open pentameric preparation (Figure 3A). Moreover, the results shown in Figure 2C strongly suggest that the poor ability of these mutant preparations to bind circular ssDNA is not due to the presence of contaminating Mcm467.
Because prior analysis of the Walker A and arginine finger mutations failed to identify defects in the Mcm6/2 or 4/6 active sites that affect circular ssDNA binding, we reexamined our data to identify subtle defects that may have been previously missed. To facilitate comparison between the various mutant complexes, the ratio of circular DNA binding in the absence of ATPγS preincubation to that in the presence of ATPγS preincubation was calculated (cir 1st/cir 2nd, Supplementary Table S3) and plotted in ranked order from lowest to highest (Figure 3B). Using this criterion, the wild-type Mcm2-7 complex demonstrates a ratio of 0.56, while complexes that demonstrate no change in circular ssDNA binding following ATP preincubation [i.e. Mcm467 and the 5KA and 2RA complexes (5,14)] have ratios of 0.92–1.01. In addition to both Mcm2DENQ (0.79) and Mcm6DENQ (0.76), the next most defective Mcm2-7 complex contains the Mcm6RA (0.72) mutation. Because both the 2DENQ and 6RA mutations are believed to ablate the Mcm6/2 ATPase active site (12,13), the similar defect in circular ssDNA binding provides additional support that the Mcm6/2 active site helps regulate the toroidal topology of Mcm2-7. In contrast, while the Mcm4RA and 6DENQ mutations both ablate the Mcm4/6 active site, the Mcm4RA mutation demonstrates only a moderate circular ssDNA binding defect similar to a variety of other Mcm ATPase mutations (Figure 3B), results that neither support nor dispute the involvement of the 4/6 active site in circular ssDNA binding.
Mutations in the Mcm6/2 and Mcm5/3 ATPase sites alter the binding kinetics of Mcm2-7 with ssDNA
The second biochemical defect specifically associated with the Mcm2/5 active site concerns the association kinetics of the complex for linear ssDNA. Wild-type Mcm2-7 binds ssDNA slowly, but the association rate is enhanced (>5-fold) by preincubation of the complex with ATP or ATPγS prior to the addition of linear ssDNA [Figure 3C, (5)]. Reminiscent of circular ssDNA binding, both the Mcm467 subcomplex and Mcm2-7 complexes containing the Mcm5KA or 2RA mutation obviate the ATP preincubation effect on ssDNA association (i.e. they load onto ssDNA at a rate similar to the wild-type complex in the presence of ATP preincubation). In contrast to the circular ssDNA binding assay results, the pentameric preparation resembles the wild-type Mcm2-7 complex in its ssDNA association kinetics, indicating that although this effect is strongly influenced by the Mcm2/5 active site, it may not be directly related to the topological closure of the Mcm2-7 toroid [Figure 3C; (14)].
Among the Walker B mutant Mcm2-7 complexes that still appreciably bind ssDNA, we measured their association rates both with and without ATPγS preincubation (Figure 3C, Supplementary Table 4). The Mcm2DENQ complex uniquely demonstrates no rate enhancement upon ATPγS preincubation, a result identical to that observed for the Mcm5KA and 2RA mutant complexes. Because the pentameric Mcm preparation acts like the wild-type Mcm2-7 complex in this assay (Figure 3C), the association rate enhancement in the absence of ATP preincubation cannot be explained by a difference in the level of non-heterohexameric complexes in the Mcm2DENQ preparations.
Our prior analysis of Mcm mutants failed to identify a defect in the Mcm6/2 ATPase active site that affects the Mcm2-7 association rate, so we reexamined our data to identify subtle defects that may have been previously overlooked. To provide a ranked order of the effects of different Mcm mutations on ssDNA association kinetics, the magnitude of the ATPγS-dependent association rate enhancement for each mutant complex was calculated (ka with ATPγS preincubation/ka without ATPγS preincubation). By this criterion, the wild-type Mcm2-7 complex demonstrates an ∼5.4-fold rate enhancement, while the 5KA, 2RA and 2DENQ complexes demonstrate essentially no enhancement (0.93- to 1.06-fold, Figure 3D, Supplementary Table 4). In addition to these three mutants, the 3DENQ and the 6RA mutations are the next most defective (1.11- and 1.19-fold, respectively) (5,12,14). Because both the 2DENQ and 6RA mutations are believed to ablate the Mcm6/2 ATPase active site (12,13), the partial defect caused by 6RA provides additional support that the Mcm6/2 active site helps to regulate the ssDNA association rate of Mcm2-7. In contrast to the Mcm6/2 active site, however, only the Walker B mutation in the Mcm3/5 active site (i.e. 3DENQ) is strongly defective for ssDNA association rate enhancement, while the arginine finger allele for this active site (Mcm5RA, Figure 3D) has a much less severe defect. Although differences between mutants in the same active site may be expected (see below), the contrasting results with the 3DENQ and 5RA alleles neither support nor discount the involvement of the Mcm5/3 ATPase active site in regulating the Mcm2-7 ssDNA association rate.
DISCUSSION
We show that inclusion of either the Mcm2DENQ or 6RA mutation in Mcm2-7, similar to the previously analyzed 5KA and 2RA mutations, causes defects in both ATP-dependent ssDNA association rate enhancement and circular ssDNA binding. Moreover, complexes containing the Mcm3DENQ mutation are unusually defective in the ATP-dependent ssDNA association rate enhancement effect, while the Mcm6DENQ mutation causes a partial defect in circular ssDNA binding. Taken together, our results indicate that the Mcm6/2 and 5/3 ATPase active sites (and perhaps the Mcm4/6 site) are involved in the putative Mcm2/5 gate activity (14), as well as confirm and extend prior observations that the six Mcm ATPase active sites contribute differentially to Mcm2-7 function.
Concerns regarding Mcm oligomerization
One premise of this study is that the results reflect the activity of structurally intact Mcm2-7 hexamers, but several experimental factors may limit the validity of this assumption. The structural stability of the Mcm complex is one such potential issue, but evidence indicates that Mcm oligomerization is stable under our experimental conditions. Unlike other helicases, such as TAg (24), Mcm oligomerization does not require ATP (13). Moreover, Mcm2-7 is stable to the extent of dilution used in our assays. For example, analytical gel filtration of purified hexamers in the absence of ATP results in enormous protein dilution, but complexes largely retain their hexameric size [Supplementary Figure 1C (5); and Supplementary Figures 1B and 5B (14)]. In addition, Mcm2-7 complexes diluted during immunoprecipitation retain their initial oligomeric state (Supplementary Figure 1). Although these data do not exclude the possibility of a rapid and reversible opening and closing of the Mcm toroidal structure, we observe robust and reproducible differences between various mutant Mcm2-7 preparations, which strongly argue that the propensity to open and close the Mcm ring depends upon the state of individual ATPase active sites.
In addition, the presence of Mcm subcomplexes in our preparations might similarly obscure our results. To address this possibility, we directly evaluated a defined non-hexameric Mcm complex (pentameric preparation). Comparison between the properties of the mutant Mcm2-7 preparations with those of the pentameric preparation allowed us to separate the contribution that Mcm hexamerization and mutant ATPase active sites make toward activity. For example, the pentameric preparation binds circular ssDNA much better than the Mcm2-7 heterohexamer, suggesting that most Mcm subcomplexes that are competent to bind ssDNA will bind circular ssDNA ‘better’ than the heterohexameric Mcm2-7. In contrast, several of our mutant Mcm2-7 preparations bind circular ssDNA binding ‘worse’ than Mcm2-7 and the pentameric preparation (e.g. Mcm2DENQ), an observation difficult to explain by the participation of topologically open Mcm subcomplexes. Because the Mcm467 subcomplex poorly binds circular ssDNA (5), this observation could in principle be explained by contaminating Mcm467 subcomplexes in the mutant preparations. However, through immunoneutralization of Mcm467 (14), we show that Mcm467 activity does not account for the DNA helicase activity of our mutant Mcm2-7 preparations (Figure 2C), indicating that Mcm467 contamination in our preparations is negligible.
The differential contribution of Mcm ATPase active sites to ssDNA binding and helicase activity
Although prior studies of the Mcm Walker A mutants demonstrated functional differences among the Mcm2-7 ATPase active sites (5,14), additional analysis indicated that they not only disrupt the ATP hydrolysis of the active site in which they reside but also downregulate the ATPase activity of the entire Mcm2-7 complex (15). The dual roles of the Walker A motifs in both ATP hydrolysis and hexamer regulation potentially confuse the interpretation that Mcm active sites are differentially required for ssDNA binding and DNA unwinding. In contrast, the Mcm Walker B and arginine finger mutants lack a dominant inhibitory effect on ATP hydrolysis (12), making them better candidates for studying the contribution of individual actives to DNA binding and unwinding. Although both motifs are predominately involved with ATP hydrolysis rather than binding [the Walker B motif positions the nucleophilic water, while the arginine finger motif assists ATP hydrolysis through its interactions with the γ-phosphate of ATP (2)], these motifs likely differ from one another in their ability to transmit information concerning ATPase active site occupancy among subunits within the complex. Thus, mutations in the Walker B and arginine finger motif might well be expected to have different and informative biochemical defects even if they reside in the same ATPase active site.
Our results with the DENQ and RA mutant complexes confirm the differential involvement of Mcm ATPase active sites in ssDNA binding and unwinding. The Mcm7/4 and 4/6 (and possibly 3/7) sites are key to DNA unwinding, while the Mcm3/7 and 7/4 (and possibly 5/3) sites are required for ssDNA binding. Although we cannot preclude the possibility that the activity changes caused by our mutations are due to subtle structural changes within the complex (rather than to alterations in ATP binding or hydrolysis per se), the fact that independent mutations in the same active site often have similar molecular phenotypes supplies strong support to our hypothesis that individual ATPase active sites contribute differentially to Mcm2-7 activity. These results also support previous studies indicating that subcomplexes containing only the Mcm4, 6 and 7 (18) or just the Mcm4 and 7 (17) subunits are completely competent for in vitro DNA unwinding in the absence of the remaining Mcm subunits, suggesting that the Mcm7/4 active site is the main motor that drives DNA unwinding. In addition, these results are in agreement with recent experiments using the archaeal homohexameric Mcm complex that indicate that only two of the six ATPase actives sites are needed for DNA unwinding (25).
Involvement of the Mcm regulatory subunits in Mcm2-7 topology and association rate
Mutations in the Mcm2/5 active site affect the ability of the complex to bind circular ssDNA (14) and increase its association rate with linear ssDNA (5,14). Together, these observations form the basis of the Mcm2-7 gate model (14). Our analysis demonstrates that certain Walker B and arginine finger mutations in the two ATPase active sites that flank Mcm2/5 (Mcm6/2 and Mcm5/3) have similar, although weaker, effects on both the ability to bind circular ssDNA and on the association rate. These new data suggest that the Mcm6/2 and 5/3 sites are involved in the putative Mcm gate activity, possibly by communicating the status of gate closure to the active sites involved in helicase activity, acting allosterically to control gate function (see below), or serving as ‘minor gates’ for DNA entry into the central channel of the complex (see below).
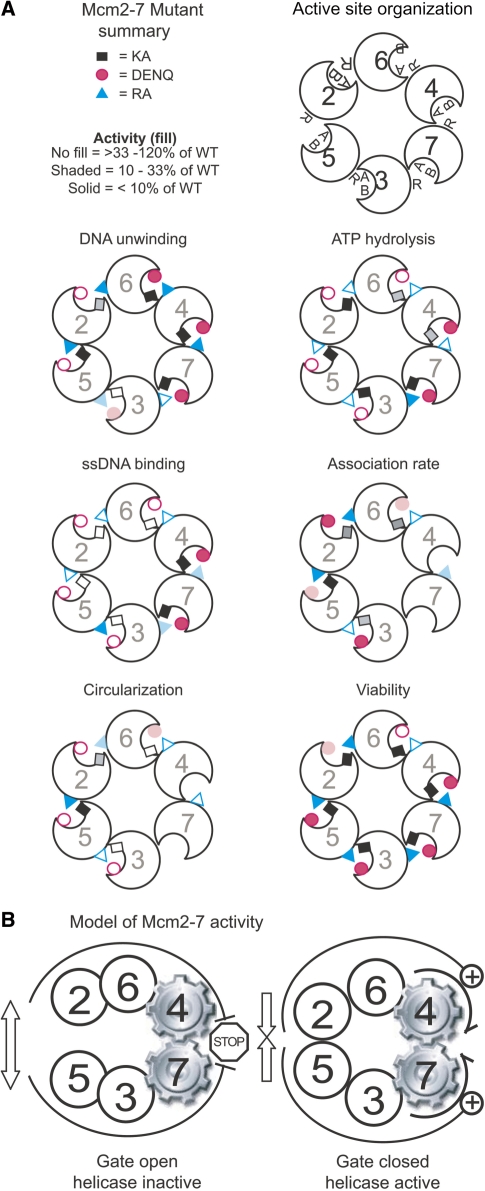
In summary, different Mcm2-7 ATPase active sites appear to have become specialized to perform the various component biochemical activities (ATP hydrolysis, ssDNA binding, DNA unwinding and, possibly, gate function) that together enable Mcm2-7 to fulfill its role in DNA replication. Active sites involved in particular functions tend to be adjacent within the complex (Figure 4A), supporting the notion that different regions of the complex have become specialized for different aspects of Mcm2-7 activity.
Figure 4.
(A) Correlation between various biochemical and genetic defects of individual Mcm ATPase alleles and their location within the Mcm2-7 ring. Walker A (KA, denoted by black or gray rectangle), Walker B (DENQ, denoted by red circle) and arginine finger (RA, denoted by cyan triangle) mutations of each subunit are mapped onto the putative circular organization of Mcm2-7, as indicated. To illustrate the trends in these data in a simple manner, the activity of each mutation on the indicated property is categorized into one of three groups and labeled as follows: approximately wild-type (120–33% the levels of wild type Mcm2-7, no fill), partially defective (33–10%, shaded), or completely defective (<10% the activity of wild type, solid fill). These categories are for illustration only and do not imply, for example, that a mutant placed in the wild-type category for one activity globally functions as a true wild type Mcm2-7 complex. The summary of association rates was derived from Supplementary Table S4, and missing alleles correspond to those that bind ssDNA too poorly to reliably quantify an association rate. Trends in the relative ATPγS preincubation stimulation of association rates were quantified as follows: unfilled symbols correspond to a rate stimulation between 5.4- (wt) and 2.4-fold; shaded fill corresponds to a rate stimulation of <2.4- to 1.4-fold; and solid fill corresponds to a rate stimulation of <1.4-fold. Trends in the circularization activity were plotted as the ratio of cir1st/cir2nd: unfilled corresponds to a ratio of 0.4–0.7 (∼wt); shaded fill 0.7–0.8; and solid fill 0.9–1.0 (completely defective). Data concerning the KA mutations are summarized from (5,14,15), the ATP hydrolysis and viability data of the DENQ and RA alleles are from (12), and the ssDNA association rate and circularization data for the RA mutants are from (14). (B) Model of Mcm2-7 activity. The Mcm5/3 and 6/2 active sites communicate the status (open versus closed) of the Mcm2/5 gate to the Mcm7/4 motor. When the gate is open (left), helicase activity is inhibited; when the gate is closed (right), helicase activity is stimulated.
Model for the activity of the Mcm regulatory subunits in Mcm2/5 gate function
At least two issues must be addressed in any model of how ATPase active sites might regulate Mcm2-7 function: (i) the relationship between topological closure of the Mcm2-7 toroid and helicase activity; and (ii) the relationship between nucleotide occupancy of the various ATPase active sites.
Evidence that helicase activity requires topological closure of Mcm2-7
Several observations indicate that DNA unwinding by Mcm2-7 requires a closed hexameric complex. First, while several different Mcm oligomers have been shown to unwind DNA [Mcm2-7, 467 and 7/4 (14,17,18)], all three species appear to form hexameric toroids, suggesting a functional requirement for topological closure in DNA unwinding. Conversely, addition of the Mcm5/3 dimer to the Mcm467 subcomplex inhibits its helicase activity, likely through disruption of the Mcm467 hexameric ring (16,26). Moreover, several observations demonstrate an inverse relationship between results from the circular ssDNA binding assay (a metric for topological closure of the complex) and helicase activation. For instance, even though the hexameric Mcm7/4 complex is capable on its own to unwind DNA, the open Mcm pentameric preparation (containing Mcm2-5 and 7, but lacking Mcm6) binds circular ssDNA but lacks helicase activity (14). Further, solution conditions that alter the topology of the complex also alter its ability to unwind DNA. Chloride-containing buffers favor an open conformation of Mcm2-7 (i.e. favor circular ssDNA binding) and concomitantly inhibit DNA unwinding; glutamate-containing buffers favor a closed Mcm topology (i.e. block circular ssDNA binding) and stimulate DNA unwinding (14).
Evidence that the Mcm regulatory ATPase active sites are allosteric rather than catalytic
Prior studies indicate that the Mcm2-7 ATPase active sites turn over ATP at very different rates. The Mcm5/3, 6/2 and 4/6 dimers have a very low rate of ATP hydrolysis relative to the Mcm3/7 and 7/4 dimers (12,13,15,17). Moreover, analysis of ATP hydrolysis in Mcm2-7 complexes containing DENQ or RA mutations generally supports the observations of dimer ATP hydrolysis levels (12), suggesting that the regulatory subunits are not part of an ordered hydrolysis cycle that promotes DNA unwinding. Furthermore, the S. cerevisiae Mcm2DENQ (12) and Mcm2KR (16) mutations decrease ATP hydrolysis at the Mcm6/2 active site but are viable, indicating that ATP hydrolysis at this site is largely unnecessary for DNA replication. Finally, our analysis indicates that many of the mutations among the regulatory active sites affect the ability of the complex to bind circular ssDNA, strongly suggesting that the binding of ATP to these sites affects the topology of the Mcm2-7 complex.
Figure 4B summarizes the above observations and suggests a working model that supposes that helicase activity from Mcm7/4 and an open Mcm2/5 gate are normally mutually exclusive, and the status of the gate (i.e. open versus closed) is transmitted to the helicase motor by allosteric changes propagated by the two flanking ATPase active sites (Mcm5/3 and 6/2). This arrangement may allow helicase activity to be controlled by cellular factors that regulate nucleotide occupancy or turnover of the Mcm2/5, 5/3 and/or 6/2 active sites.
Alternatively, as stated above, the Mcm5/3, 6/2 and (possibly) 4/6 active sites may function as ‘minor gates’ in addition to the putative ‘major’ Mcm2/5 gate. Such structural features are worth considering if the Mcm2-7 complex does indeed bind dsDNA in its central channel at origins of replication [as indicated by recent structural work (27)] but ultimately functions as a classical helicase by encircling ssDNA while unwinding (7). For example, the Mcm2/5 gate may allow initial dsDNA entry into the central channel of the complex, while the minor gate(s) could facilitate the subsequent expulsion of one of the two single strands from the central channel upon initial dsDNA melting.
Comparison of in vivo and in vitro Mcm activity
The ability of the MCM alleles studied here to support viability in yeast is summarized in Figure 4A (12,15). First, although many of these alleles have demonstrable in vitro DNA unwinding, they are lethal in vivo, suggesting that these mutations block an essential function not directly related to helicase activity. Second, while the S. cerevisiae Mcm4RA and 6DENQ mutants are both completely viable, these mutations inhibit the in vitro helicase activity of Mcm2-7 (this study) and Mcm467 (12). While there is currently no explanation for these observations, it may suggest the interesting possibility that some ATPase active sites have currently unknown but essential functions, or that additional factors in vivo modulate the activity of these sites (see CMG complex below).
Possible modulation of Mcm function – the CMG complex
Several higher-order complexes that contain the six Mcm subunits in the company of additional DNA replication factors have recently been identified (28,29). The contribution of the Walker A mutants to DNA unwinding has recently been studied in each of the six Mcm subunits of the higher order CMG (Cdc45/Mcm2-7/GINS) complex from Drosophila (22). Comparison between the results of the Drosophila CMG complex and the S. cerevisiae Mcm2-7 complex indicates that, in both cases, individual Mcm ATPase active sites contribute unequally to DNA unwinding. Intriguingly, however, the subunits whose Walker A mutations result in the most deleterious effect are almost completely reversed between the model systems, with the Mcm3 and 5 Walker A mutations in the CMG complex causing the largest decreases in DNA unwinding activity (22). Although the effects of these mutations on ATP hydrolysis, ssDNA binding and viability in the Drosophila CMG complex are not yet known, the differences in their DNA unwinding contributions suggest several possible explanations: (i) that the Mcm2-7 complexes in these two organisms function differently; (ii) that Cdc45 and the GINS complex modulate Mcm2-7 functions in the CMG complex; or (iii) that the apparent differences between the two complexes correspond to relatively minor differences in allosteric communication between the active sites rather than major catalytic differences between them. In support of the last point, it should be noted that even in S. cerevisiae, among the three motifs studied to date (Walker A, Walker B and arginine finger), mutations exist in each active site that have a range of defects—some eliminate DNA winding, while others have relatively little effect. Future work will be required to sort out the basis for these differences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The American Cancer Society (RSG-05-113-01-CCG) and the National Institutes of Health (1RO1GM83985-1A1) to A.S. Funding for open access charge: National Institutes of Health (1RO1GM83985-1A1).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Julia van Kessel, Debra Irvine, and members of the Schwacha and Zakian labs for thoughtful comments on the manuscript.
REFERENCES
- 1.Bochman M, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 3.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochman ML, Schwacha A. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J. Biol. Chem. 2007;282:33795–33804. doi: 10.1074/jbc.M703824200. [DOI] [PubMed] [Google Scholar]
- 6.Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 8.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Richards TA, Aves SJ. Ancient diversification of eukaryotic MCM DNA replication proteins. BMC Evol. Biol. 2009;9:60. doi: 10.1186/1471-2148-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr. Opin. Struct. Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem. Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol. Cell. Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey MJ, Indiani C, O'Donnell M. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 2003;278:4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 14.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Schwacha A, Bell SP. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell. 2001;8:1093–1104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- 16.Stead BE, Sorbara CD, Brandl CJ, Davey MJ. ATP binding and hydrolysis by Mcm2 regulate DNA binding by Mcm complexes. J. Mol. Biol. 2009;391:301–313. doi: 10.1016/j.jmb.2009.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanter DM, Bruck I, Kaplan DL. Mcm subunits can assemble into two different active unwinding complexes. J. Biol. Chem. 2008;283:31172–31182. doi: 10.1074/jbc.M804686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 19.Sato M, Gotow T, You Z, Komamura-Kohno Y, Uchiyama Y, Yabuta N, Nojima H, Ishimi Y. Electron microscopic observation and single-stranded DNA binding activity of the Mcm4,6,7 complex. J. Mol. Biol. 2000;300:421–431. doi: 10.1006/jmbi.2000.3865. [DOI] [PubMed] [Google Scholar]
- 20.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J. Mol. Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 21.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 22.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 25.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Yabuta N, Kajimura N, Mayanagi K, Sato M, Gotow T, Uchiyama Y, Ishimi Y, Nojima H. Mammalian Mcm2/4/6/7 complex forms a toroidal structure. Genes Cells. 2003;8:413–421. doi: 10.1046/j.1365-2443.2003.00645.x. [DOI] [PubMed] [Google Scholar]
- 27.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.