Abstract
HU is one of the most abundant proteins in bacterial chromosomes and participates in nucleoid compaction and gene regulation. We report experiments using DNA stretching that study the dependence of DNA condensation by HU on force, salt and HU concentration. Previous experiments at sub-physiological salt levels revealed that low concentrations of HU could compact DNA, whereas larger HU concentrations formed a DNA-stiffening complex. Here we report that this bimodal binding behavior depends sensitively on salt concentration. Only the compaction mode was observed for 150 mM and higher NaCl levels, i.e. for physiological salt concentrations. Similar results were obtained for the more physiological salt K-glutamate. Real-time studies of dissociation kinetics revealed that HU unbound slowly (minutes to hours under the conditions studied) but completely for salt concentrations at or above 100 mM NaCl; the lifetime of HU complexes was observed to increase with the HU concentration at which the complexes were formed, and to decrease with salt concentration. Higher salt levels of 300 mM NaCl completely eliminated observable HU binding to DNA. Finally, we observed that the dissociation kinetics depend on force applied to the DNA: increased applied force in the sub-piconewton range accelerates dissociation, suggesting a mechanism for DNA tension to regulate chromosome structure and gene expression.
INTRODUCTION
HU protein (histone-like protein from Escherichia coli strain U93) is one of the most abundant proteins in bacterial nucleoids and plays an important architectural role (1–3). Two 9.5-kDa variants of HU exist in E. coli, α and β, which can constitute homodimers (HU-α2 and HU-β2) or heterodimers (HU-αβ). The heterodimer is the major form of HU in E. coli (4). In vivo HU concentrations vary between a high level of 30 000 copies per cell in exponential phase and a low level of 7500 in stationary phase (5,6). HU binds to DNA in a largely sequence-independent manner generating dynamically bent DNA. Co-crystal structures of HU–DNA showed that binding and DNA distortion involve formation of two sharp kinks in the DNA spaced by 9 bp, generated by insertions of prolines located at the ends of two β-ribbon arms of HU that reside in the DNA minor groove (7,8). The bends in DNA induced by HU binding have been measured to be from 53° to more than 140° in the DNA as assayed by X-ray crystallography, gel electrophoresis, DNA cyclization, atomic force microscopy and Fluorescence Resonance Energy Transfer (FRET), and are thought to be relatively flexible (7,9–15).
The high levels of HU in vivo combined with its sequence-independent binding and bending activities are consistent with HU having an important architectural role in chromosome compaction. In support of this, 4′,6′-diamidino-2-phenylindole (DAPI)-stained nucleoids of hupAB mutant strains of E. coli and Salmonella are unfolded, and DNA supercoiling density is reduced (16–22). Numerous studies have also demonstrated HU to be an important accessory factor in DNA replication, transcription, and recombination processes (1,6,17,23,24).
Estimates of the equilibrium constant (Kd) for non-specific binding of HU to DNA vary widely, having been estimated to range up to 29 µM, depending on the dimeric variety of HU and experimental conditions (25–27). HU has also been observed to bind with much higher affinity to certain types of structured DNA molecules, such as nicked, bent, gapped, three- or four-way junction, or AT-rich DNA. The Kd of specific binding to these DNA structures were found to be between 10 and >105 times higher than non-specific binding (8,10,26,28–30).
Three groups have used single-DNA stretching experiments to analyze HU–DNA interactions (13,31,32). In such experiments, tethered 10–50 kb DNAs are put under controlled forces and their mechanical response is studied. These types of experiments are sensitive to binding of proteins to the double helix if those proteins distort DNA, and are capable of observing kinetics of binding in response to changes in bulk solution conditions. Single-DNA stretching experiments all observed that low concentrations (below 100 nM) of HU caused a compaction effect, i.e. a reduction in extension of the DNA–protein complex at a given force, relative to the extension of naked DNA (13,32,33). However, at higher HU concentrations, a second ‘mode’ of binding was observed, namely the abrogation of bending (12) and a stiffening (‘rigid filament’) behavior (13,32). The stiffening behavior is thought to be a result of tandem binding of HU molecules along the double helix, and it is intriguing since it is contradictory to the presumed chromosome-compaction function of HU (34). However, it is important to note that the HU to DNA ratio for the stiffening binding mode was roughly one dimer per tens of bp, higher than the cellular level of one dimer per 300 bp (5,9,35).
In order to reconcile the wide variations of observed structure and affinity of HU for DNA, distinct modes of binding have been proposed. At least three modes classified by binding constants and binding enthalpies for a moderate concentration of salt (150 mM Na+) and relatively low temperature (15°C) were observed in a recent study, using a combination of isothermal calorimetry and FRET techniques. It was suggested that at low HU/DNA molar ratio a non-cooperative 34 bp mode involves DNA bending, whereas higher HU/DNA drives binding from a 34- to a 10- to a 6-bp mode, leading to stiffening (25).
We became interested in the role played by salt concentration in selecting binding modes and controlling affinity of HU for DNA, since single-DNA studies had observed the stiffening of DNA by HU at salt concentrations appreciably below the 100–200 mM salt levels considered physiologically relevant to E. coli (36). More specifically, experiments of van Noort et al. were done at 60 mM KCl (13), while experiments of Skoko et al. were done in 40 mM NaCl (32). Intriguingly, experiments of Schnurr et al. on Bacillus stearothermophilus (Bst) HU (BstHU) in 100 mM NaCl suggest a less dominant stiffening function, although it is clear from those studies that BstHU has a much higher affinity for DNA than does E. coli HU (31). Evidence for strong dependence of the strength of non-specific HU–DNA interactions (Kd values) on salt concentration also can be found in the biochemical literature (1,37,38). Studies on binding sites ranging from 9 to 42 bp in length have been done under a variety of salt conditions with a wide range of results (14,27).
Intertwined with questions of the role of salt is that of the stability of HU–DNA complexes. The HU in exponentially growing cells is sufficient to bind ∼8% of the E. coli chromosome, which can divide every 20 min, suggesting that kinetic repartitioning of HU on DNA may occur. The only previous report on unbinding kinetics for molecules in bulk solution was based on a plasmid-linking-number assay for studying the competitive partition of HU between linear and supercoiled DNA molecules (16), a process potentially distinct from simple dissociation of HU from DNA. Previous single-DNA studies have noted that dissociation of HU–DNA complexes is very slow (32), and indeed that ‘exchange’ of protein to bulk DNA fragments occurs much more rapidly (32); however, systematic studies of dissociation kinetics have not been done.
Dissociation of protein from DNA in vivo potentially can be influenced by other physical factors including force (39,40). Force has been observed to alter dissociation rates of ligands from receptors (41–43), and DNA unwinding activity of hRPA protein (44). Whether or not force can drive bound HU or IHF from DNA has been discussed (13,45), but no experiment has clearly demonstrated force-dependent affinity of a DNA-binding protein.
In this report, we use single-DNA pulling by magnetic tweezers as a tool to study the dependence of HU–DNA interactions on force, salt and HU concentration. Our primary method is the use of real-time monitoring of DNA condensation by HU to assay binding of the protein. We used this to track dissociation of HU–DNA complexes, and to ascertain the dependence of unbinding kinetics on HU concentration, applied tension and univalent salt level.
MATERIALS AND METHODS
Protein expression and purification
Heterodimeric HU was purified from RJ5814 (ihfB::cat fis::kan-767 endA::Tn10 his ilv λcI857 N+ containing pPL-hupAB from R. McMacken) by a protocol involving cation exchange chromatography on SP-Sepharose and FPLC mono S followed by FPLC gel filtration through Superose 75 (GE Healthcare LifeSciences) (32).
Manipulation of HU–DNA complexes with magnetic tweezers
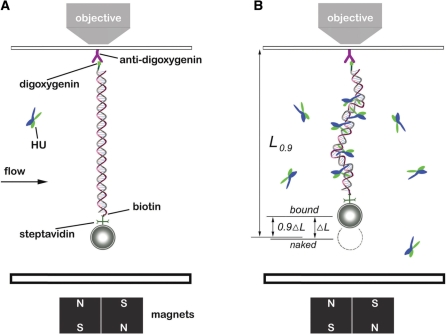
A 48.5-kb λ DNA (Promega, Madison, WI) was end-labeled with biotin and digoxygenin as previously described (46). Labeled λ DNA was incubated with 2.8-µm diameter streptavidin-coated paramagnetic beads (Dynabeads M-280, Invitrogen, Grand Island, NY) in Phosphate-buffered saline (PBS) for 15 min. Then the bead-DNA constructs were injected into a flow cell and tethered to a piece of anti-digoxygenin (Roche Diagnostics, Indianapolis, IN) coated cover glass (Figure 1A).
Figure 1.
Experimental configuration. (A) Schematic of magnetic tweezers setup. DNAs end-labeled with biotin and digoxygenin were attached to paramagnetic microbeads through biotin-streptavidin cross-linkers. Then the bead-DNA constructs were adhered to a piece glass slide on a flow cell by digoxygenin-antibody linkage. The images were magnified and tracked by a piezo controlled 100× objective, and captured by a CCD camera. The bead was manipulated by motor-driven permanent magnets near the flow cell. The solution inside the flow cell could be changed, allowing HU–DNA association and dissociation to be controlled. (B) DNA after HU binding. The extension of the HU–DNA complex (Lbound) was shorter than the original naked DNA length (Lnaked) due to bending of DNA by the proteins. The difference of Lnaked and Lbound was denoted by ΔL and taken as an indicator of amount of protein bound. During protein dissociation experiments, the time t0.9 for the extension to return 90% of the way back to Lnaked (to the level L0.9), was used to measure the complex dissociation lifetimes.
Vertical magnetic tweezers (MT) were constructed based on previous systems with slight modification (32,47,48). A 100× 1.4 NA microscope objective was used to image the magnetic beads. Four cubic NdFeB magnets (0.5′ × 0.5′ × 0.5′, K&J Magnetics Inc., Jamison, PA) were mounted on a stepper-motor-driven translator, producing a variable magnetic field gradient near the microscope stage. A computer program written in Labview (National Instruments, Austin, TX) controlled the movements of the magnets, tracked transverse motions of the bead, and measured vertical bead position using a focusing algorithm. The transverse motion was captured by a digital camera (PL-741, Pixelink, Ottawa, ON) and real time images were analyzed to calculate the force according to a fluctuation technique (49). Briefly, the paramagnetic bead tethered to a single DNA (Figure 1A) can be considered to be a pendulum of extension z. For extensions greater than half of the DNA contour length, the applied force is determined by  where kB is Boltzmann’s constant, and T is the absolute temperature, and (x2 + y2) is the mean square displacement of the bead in the x–y plane.
where kB is Boltzmann’s constant, and T is the absolute temperature, and (x2 + y2) is the mean square displacement of the bead in the x–y plane.
The force between the bead and permanent magnets stretched the DNA whose extension was measured using a piezoelectric objective positioner (NV40/1, PiezoSystem Jena, Hopedale, MA) and automated focus software written in Labview. The force (f) and extension (L) relationship was calibrated and compared with the established mechanical properties of single DNA  . Here, L0 is the contour length of DNA, A = ∼50 nm is the persistence length of a single DNA (50). All experiments were carried out at a temperature of 25°C (298 K), with temperature held at that value using an objective ring heater (OHXX, 20/20 Technologies, Wilmington, NC).
. Here, L0 is the contour length of DNA, A = ∼50 nm is the persistence length of a single DNA (50). All experiments were carried out at a temperature of 25°C (298 K), with temperature held at that value using an objective ring heater (OHXX, 20/20 Technologies, Wilmington, NC).
Force-extension measurements
Once a single DNA tether was confirmed, the PBS in the flow cell was substituted with working buffers which contained 20 mM HEPES, 0.5 mg/ml BSA, plus from 40 mM to 300 mM NaCl, pH 7.5. During solution exchanges, the force applied to the DNA tether was held at ∼2 pN to prevent it from sticking to the cover glass. For experiments with HU, force-extension curves were measured at successively higher concentrations, with 30 min incubation done before each measurement. Extension of the DNA at a given force (from 0.05 to 6.0 pN) was measured in the working buffer and HU solutions with attention paid to making sure the DNA–protein complex length had stabilized before each extension measurement.
HU dissociation to protein-free solution
After the force-extension measurements, the HU solution was washed away in about 1 min by 300 µl working buffer, which was 5× the volume of the flow cell (∼60 µl). Immediately following the wash, the extension was continuously monitored with the force held at either 0.1 or 0.3 pN. If the HU concentration was more than 500 nM, the flow cell was washed again with 200 µl buffer after 1 hour. As dissociation occurred, the extension of the HU–DNA complex was observed to gradually return to its naked DNA level, by smooth but non-exponential dynamics.
For 150 and 200 mM NaCl buffers, the average extensions of HU–DNA complexes were shorter than those of the naked DNAs. To define a dissociation time we used the extension difference between the bound state extension and the original extension of the naked DNA (ΔL= Lnaked – Lbound). The time t0.9, at which the extension returned 90% of the way back to Lnaked, i.e. where the extension returned to  , was taken to be the dissociation time (Figure 1B). We used t0.9 instead of t0.5 (50% length recovery) because t0.5 had a smaller signal to noise ratio, due to the length differences being 1–4 µm while thermal fluctuation was ∼1 µm.
, was taken to be the dissociation time (Figure 1B). We used t0.9 instead of t0.5 (50% length recovery) because t0.5 had a smaller signal to noise ratio, due to the length differences being 1–4 µm while thermal fluctuation was ∼1 µm.
For HU concentration greater than 150 nM in the 100 mM NaCl buffer, when rigid filaments formed, Lbound was defined to be the minimum extension of the HU–DNA complex that appeared in the dissociation process.
RESULTS
HU displays ‘bimodal’ DNA compaction and stiffening at or below 100 mM NaCl
Force-extension measurements (see Materials and methods section) for naked λ DNA were first performed in buffer containing a given amount of NaCl. No hysteresis (different extensions as force was increased or decreased) was observed in the extension as a function of force for any NaCl concentration. Then, buffer containing a given HU concentration was added and force-extension curves were again obtained. This procedure was repeated in separate experiments for each combination of HU (25–1000 nM) and NaCl (40–300 mM) concentrations studied.
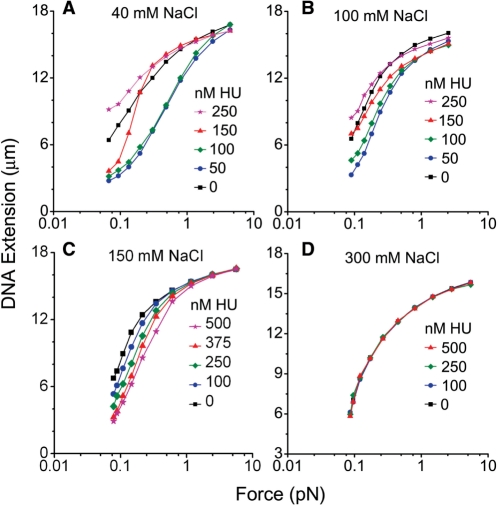
Figure 2A and B shows the end-to-end extension of the DNA–protein complexes as a function of HU concentration, for a series of fixed forces indicated to the right of the panels. For each force, the extension of naked DNA is indicated by a dashed line. For solutions in which the NaCl concentration was less than or equal to 100 mM, both compaction and elongation were observed as indicated by the non-monotonic behavior of DNA extension versus HU concentration, in accord with previous reports (13,32). Extensions of HU–DNA were shorter than that of naked DNA if the HU concentrations were less than or equal to 100 nM, indicating the formation of flexible hinges (51). The maximum compaction occurred for ∼50 nM HU, close to previous reports (13,32). The difference between HU–DNA and naked DNA shrank (Figure 2A and B, dotted lines) as force was increased. At 0.1 pN force, the maximum compaction ratio was 53% for 40 mM, and 49% for 100 mM NaCl. On the other hand, the extension became longer than that of naked DNA if the HU concentration was increased to 250 nM or more, indicating the presence of rigid filaments.
Figure 2.
Dependence of DNA extension on force, salt and HU concentration. Dashed lines show extension of naked DNA under corresponding forces. Each experimental data point, presented as the mean value, was obtained from 6–10 measurements. (A) Bimodal binding of HU and single λ-DNA in 40 mM NaCl buffer. Compaction was observed for HU concentrations < 100 nM and was maximum at ∼50 nM HU. From 50 nM to higher concentrations, the extension increased gradually and could become longer than 0 nM HU, indicating the formation of rigid filaments. (B) Bimodal binding as in A, for 100 mM NaCl. Essentially the same dependence of binding mode on HU concentration was observed as for 40 mM NaCl. (C) Unimodal binding in 150 mM NaCl buffer. As the HU concentration was increased, the extension decreased with concentration to the maximum concentration studied of ∼1000 nM HU. No rigid filament stiffening effect was observed. (D) No binding was detectable through extension changes in 300 mM NaCl buffer. Extension remained unchanged relative to that of naked DNA for all HU concentrations and applied forces studied.
The DNA extensions for 40 and 100 mM NaCl can also be plotted as a function of force as shown in Figure 3A and B, and show the behavior observed previously (13,32); low concentrations of HU (100 nM and below) shift the force extension curve to higher forces (formation of flexible bends), while larger concentrations (250 nM or above) lead to a shift of the force-extension curve back to lower forces (formation of rigid filaments). Overall the bimodal binding behavior we observe is in good accord with previous reports (13,32).
Figure 3.
Force-extension curves of HU–DNA complexes. Each data point represents the average of 6–10 measurements. (A) HU-dependence of force-extension curve of a λ-DNA in 40 mM NaCl buffer. Force-extension curve shifted to higher force if the added HU concentration was <250 nM, signifying the formation of flexible hinges. The curve moved to lower force if HU was >250 nM, exhibiting the formation of rigid filaments. (B) HU-dependence of force-extension curve as in A, for 100 mM NaCl. The same ‘bimodal’ behavior was observed as in 40 mM NaCl. (C) Extension in 150 mM NaCl buffer. Force-extension curve continuously shifted to lower force as the HU concentration increased. (D) Force-extension remained unchanged for all HU concentrations in 300 mM NaCl.
HU only compacts DNA for NaCl between 150 and 200 mM
We next carried out experiments at a higher, more physiological NaCl concentration of 150 mM (26,36), with the surprising result that the shifts of extension of λ DNA became monotonic with HU concentration (Figure 2C), showing a progressive reduction of extension as HU concentration was raised. No stiffening effect (non-monotonic behavior of extension versus concentration as in Figure 2A and B) was observed. Similar behavior was observed for 200 mM NaCl. The maximum compaction ratio at 0.1 pN in 150 mM NaCl was 49%, similar to those for 100 and 40 mM. Correspondingly, force-extension curves always shifted to lower force for HU concentration ranging from 25 to 1000 nM (Figure 3C, compare concentration ordering of curves to that of Figure 3A and B).
In repeated experiments we verified the surprising result that the small change in salt concentration from 100 to 150 mM drastically changed the character of HU–DNA binding, particularly at higher HU concentrations.
DNA compaction is eliminated for 300 mM or greater NaCl
In experiments with 300 mM NaCl concentration, no change in the extension versus HU concentration was observed (Figure 2D), indicating that binding of HU to DNA no longer occurred, or possibly that HU was binding but no longer bending DNA. Force-extension curves at each HU concentration matched those of naked DNA, indicating that neither flexible hinge bends nor the rigid filaments could form (Figure 3D). To sum up the observations to this point, we observed that at low salt (40 and 100 mM NaCl) both flexible hinge bending and stiffening binding modes occur; at physiological salt levels (150–200 mM NaCl) only compaction by bending appeared to occur, and at high salt (300 mM NaCl) no evidence of binding at all was observed.
Higher HU concentration during binding or lower salt increase stability of HU–DNA complexes
To more precisely analyze the kinetics of dissociation of HU from DNA we carried out further kinetic experiments. After allowing HU to bind to DNA at given HU and NaCl concentrations, the HU–DNA complex was held by ∼2 pN force and washed with protein-free buffer to permit dissociation. HU–DNA extension was taken as an indicator of the dissociation process which was continuously monitored, and the point at which the extension returned 90% of the way back to the initial naked DNA level (t0.9, see ‘Materials and Methods’ section) was used to measure the complex lifetime. Dissociation experiments were carried out at low forces (∼0.1 pN) both to maximize the extension change to facilitate quantitative measurements, and to avoid perturbing the kinetics using applied force (see below).
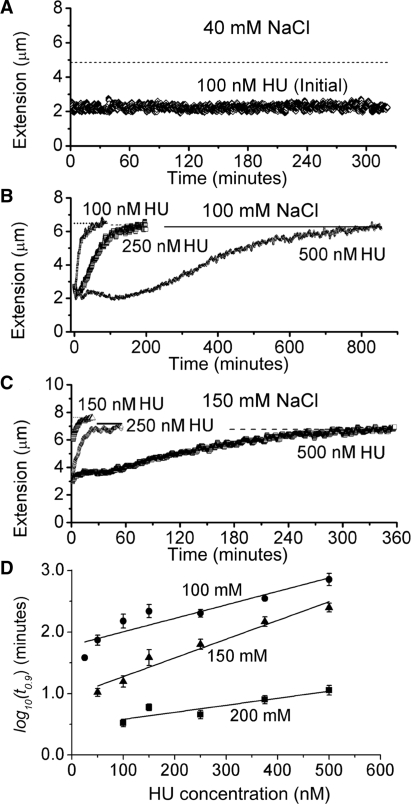
In 40 mM NaCl buffer, with <0.1 pN holding force, the extension of HU–DNA complexes which formed in high HU concentration did not return to that of naked λ DNA after washing. Figure 4A shows an experiment for a complex formed at 100 nM HU which remained stable at 0.1 pN after 320 min. In general, we found that the 40 mM NaCl HU–DNA complexes were essentially stable for many hours and we were not able to measure t0.9 values for them.
Figure 4.
Dissociation monitored via change in extension and dissociation lifetimes (t0.9). Straight lines in (A), (B), (C) indicate the 90% extension point L0.9 used to determine t0.9. (A) Incomplete dissociation in 40 mM NaCl buffer after replacement of HU solution with protein-free buffer. For example, a tether formed at 100 nM HU (i.e. initial concentration) remained stable at 0.1 pN after 320 min. (B) Slow dissociation in 100 mM NaCl buffer. t0.9 was 200 min for the HU–DNA complexes formed at 250 nM and 850 min for 500 nM. In the first 12 min, the decrease in the extension curve of 250 nM indicated the dissolution of rigid filaments. HU–DNA complex lifetimes became longer as the initial HU concentration increased. (C) Fast dissociation in 150 mM NaCl buffer. The t0.9 was 50 min for the HU–DNA complexes formed at 250 nM HU and 330 min for 500 nM, which were both shorter than the corresponding t0.9 values in 100 mM NaCl. (D) t0.9 determined from time courses, plotted for reversible binding in 100, 150 and 200 mM NaCl buffers. t0.9 values increase as HU concentration during binding is increased and as salt concentration is decreased. Each data point shows an average of three to five separate experiments. Taking 375 nM HU for example, t0.9 was 8 ± 1 (SEM) min for 200 mM NaCl, 150 ± 30 min for 150 mM and 350 ± 30 min for 100 mM. t0.9 data for different salt concentrations showed a roughly exponential dependence on HU concentration during binding; straight lines show exponential fits.
By contrast, in similar experiments carried out with 100 mM NaCl, the extensions increased gradually towards the length of naked λ DNA after washing (Figure 4B). For complexes formed with more than 250 nM HU, we observed that the extension first decreased to a minimum value before increasing towards the naked DNA extension, indicating that the rigid filament behavior was lost first, followed by removal of flexible hinge bends. This non-monotonic kinetic behavior recapitulated the behavior seen in the equilibrium binding curves of Figure 2B, where flexible hinges and rigid filaments formed subsequently as HU concentration was increased.
For similar experiments on HU–DNA complexes formed in 150 and 200 mM NaCl, we observed that extension always increased monotonically to the naked DNA length (Figure 4C). This behavior was consistent with the observation of flexible hinge behavior only in HU solution (Figure 2C) at these salt concentrations.
Larger HU concentration during binding or lower salt concentration increase dissociation lifetimes
From the dissociation time courses at 0.1 pN discussed above we measured 90% lifetimes of HU dissociation to protein-free buffer (t0.9). We note that in general the extension versus time showed quite complex (non-monotonic) dynamics, so fitting the decays to exponentials or extraction of first-order lifetimes was not possible. On the other hand the measurement of the 90% point was well past the level of HU binding where the transition from stiffening to bending mode binding occurred, providing a robust measure of the time required for the dissociation process.
The t0.9 values increased with the concentration of HU that was used to form the complexes. As mentioned above, lifetime values could not be measured for 40 mM NaCl as the complexes were stable. By contrast, in 100 mM NaCl, dissociation occurred and we were able to measure lifetimes as a function of concentration; for example t0.9 was 200 min for the HU–λ DNA complex formed at 250 nM, while t0.9 was 850 min for 500 nM HU (see time courses in Figure 4B). As HU concentration during binding was increased, the time required for dissociation to protein-free buffer increased.
As salt concentration was increased, dissociation occurred more rapidly. For 150 mM example time courses shown in Figure 4C the t0.9 was 50 min for complexes formed at 250 nM HU, and 330 min for 500 nM HU complexes.
A series of experiments of the type shown in Figure 4B and C were carried out for 100, 150 and 200 mM NaCl buffers to obtain three to six measurements per HU concentration used. The resulting t0.9 values were averaged; the results are shown in Figure 4D. The trends of longer lifetimes for higher HU concentration during binding, and longer lifetimes for lower salt concentrations mentioned above are strong; note that a logarithmic t0.9 scale is used in Figure 4D, and that there is roughly an exponential dependence of the complex lifetimes on initial HU concentration (regression coefficients of the exponential fits in Figure 4D were 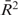 = 0.83, 0.94, 0.75, respectively). There is a similar strong dependence of lifetime on decreasing salt concentration. We note that the 100 mM NaCl data points for more than 500 nM HU correspond to many hour-long complex dissociation times. We also note that for each salt concentration there was an HU concentration below which t0.9 became too short to measure reliably because the dissociation was nearly completed during the ∼60 s exchange of buffer.
= 0.83, 0.94, 0.75, respectively). There is a similar strong dependence of lifetime on decreasing salt concentration. We note that the 100 mM NaCl data points for more than 500 nM HU correspond to many hour-long complex dissociation times. We also note that for each salt concentration there was an HU concentration below which t0.9 became too short to measure reliably because the dissociation was nearly completed during the ∼60 s exchange of buffer.
Binding mode selection in K-glutamate buffer is similar to NaCl buffer
Some proteins are known to bind differently to DNA in buffers based on the more physiological salt K-glutamate (52) than in buffers containing NaCl or KCl. To examine this possibility for HU, we performed association and dissociation experiments in K-glutamate buffer. Bimodal binding was observed in 100 mM K-glutamate (Supplementary Figure S1A), similar to 100 mM NaCl buffer, with the small difference that the maximum compaction is nearer to 100 nM HU instead of 50 nM. Monotonic binding was obtained in 150 mM K-glutamate (Supplementary Figure S1B). In dissociation kinetics experiments, the t0.9 for 500 nM HU in 150 mM K-glutamate buffer was observed to be very similar to 500 nM HU in 150 NaCl buffer (Supplementary Figure S2).
Increased DNA tension accelerates dissociation
Given the results for t0.9 (Figure 4D) shown for 0.1 pN applied force, we wanted to determine whether higher forces would significantly change the dissociation kinetics; this is theoretically expected since applied tension should increase the free energy penalty of binding a DNA-bending protein due to the force-extension work associated with reduction of overall complex extension (39,40). We carried out experiments similar to those shown in Figure 4D, but at a higher force of 0.3 pN. Example time courses for corresponding experiments done with 150 nM and 250 nM HU during binding, but with 0.1 and 0.3 pN tension during dissociation are shown in Figure 5A (all for 150 mM NaCl). As can be seen, the dissociation time course is faster for the higher force.
Figure 5.
Force effects on t0.9 in 150 mM NaCl buffer. (A) Example dissociation time courses under different forces. As exemplified by 250 nM initial HU, the t0.9 was 51 min at 0.1 pN, but 6.8 min at 0.3 pN. Straight lines are L0.9 values. (B) Dependence of t0.9 values on the force and the initial HU concentration. For instance, the average t0.9 for 250 nM HU was 68 ± 13 min at 0.1 pN, 6.5 ± 0.2 min at 0.3 pN. Each data point represents three to six experimental measurements. Linear fits were performed for the logarithmic values of t0.9 values.
A series of such experiments were carried out for 150 mM NaCl to obtain averaged results as shown in Figure 5B. Similar to Figure 4D, dissociation lifetimes were spread over a wide range as a function of HU concentration during binding (the exponential fits shown in Figure 5B for 0.1 and 0.3 pN have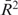 = 0.95, 0.92, respectively). Complexes held at a force of 0.3 pN shared a lifetime t0.9 less than one-third of that observed for 0.1 pN, over the entire concentration range studied (shift between different force data in Figure 5B).
= 0.95, 0.92, respectively). Complexes held at a force of 0.3 pN shared a lifetime t0.9 less than one-third of that observed for 0.1 pN, over the entire concentration range studied (shift between different force data in Figure 5B).
DISCUSSION
DNA stiffening and then DNA bending are sequentially eliminated by increasing salt
We have used single-DNA stretching methods to monitor HU–DNA interactions, and have shown that varying salt concentration over a relatively narrow range has a dramatic effect on the qualitative nature of HU–DNA interactions. At low salt (40 mM and 100 mM NaCl and 100 mM K-glutamate) we recover previous observations (13,32) of HU concentration-dependent binding modes: at low HU concentration we observe DNA compaction by flexible bends formed along DNA, while at higher HU concentrations we observe stiffening of the double helix.
However, increasing salt concentration just 50 mM further to 150 mM eliminates the stiffening-mode binding; in this new ‘unimodal’ regime, only DNA-bending-driven compaction is observed (Figures 2 and 3, and Supplementary Figure S1B). Unimodal DNA-bending HU binding is seen also for 200 mM NaCl, and then at 300 mM NaCl, no evidence of binding at all is observed, consistent with HU being a basic protein which interacts with DNA in part via electrostatic interactions which are weakened (screened) by higher salt concentrations (53). We therefore find a dramatic effect of increasing salt concentration on mode and stability of HU–DNA interactions. Noting that cytoplasmic K+ in E. coli is thought to be relatively variable, around 200 mM for ambient growth but can extend up to 800 mM under conditions of osmotic stress (38), our results are potentially important for understanding how HU binds DNA in vivo. On the face of it, our results suggest that in vivo, HU is likely only to bend DNA. However, we note that in vivo other factors including macromolecular crowding may well play a role in controlling HU binding to DNA, so one must be cautious when drawing conclusions concerning in vivo binding from in vitro experiments.
HU–DNA complex dissociation kinetics depend sensitively on salt concentration
Given that we could observe systematic shifts in force-extension curves for HU–DNA complexes as a function of HU concentration, we used those shifts to monitor unbinding of HU from DNA in experiments where the initial HU solution was washed away and replaced with protein-free buffer (Figure 4). For the low-salt 40 mM NaCl case we reproduced previous reports (12,13,31,32) of slow or stalled dissociation kinetics. However, for larger salt concentrations (100 mM and higher) we observed gradual dissociation of the HU–DNA complexes, eventually observing the return of their elasticity back to that of naked DNA. The dissociation kinetics became more rapid with increasing salt concentration (Figure 4D), again attributable to the electrostatic component of HU binding to DNA. We have observed similar strong effects of salt concentration on dissociation kinetics in experiments with the N-terminal DNA-binding domain of λ-integrase (Int-DBD), and we expect this to be a general feature of DNA-binding proteins. Previous studies have shown that addition of competitor DNA in the washing buffer greatly accelerates the dissociation of HU from DNA (12,31), suggesting that exchange kinetics is faster than dissociation kinetics.
We find that for the 100 mM NaCl case, the dissociation kinetics of the DNA-stiffened complexes are non-monotonic, proceeding by compaction followed by decompaction. This most likely reflects that HU initially bound in ‘DNA-stiffening-decompacting’ mode converts to ‘DNA-bending-compacting’ mode as dissociation proceeds.
On rates versus off-rates for non-specific binding of proteins to large DNA molecules
We note that the off-kinetics for the large DNA molecules that we have studied are most likely not indicative of the microscopic off-rates for individual proteins from short DNA, but instead reflect the lifetime of arrays of proteins bound along a DNA molecule. This can be seen by considering the apparent Kd for non-specific binding that one would infer from the halfway point of the shift of the force-extension curves between the naked and saturated binding limits. As an example consider the 150 mM NaCl case where unimodal binding is observed (Figures 2C and 3C); the force curve is halfway shifted near 250 nM HU, providing an estimate for the non-specific Kd (roughly analogous to a gel-shift estimate). The complex dissociation time for this case (Figure 4D, point for 150 mM NaCl and 250 nM HU) is ∼60 min. In our experiments we see no delay in formation of complexes when we flow in HU; the ‘on-time’ for complex formation is appreciably less than one minute.
This reveals a paradox: on and off rates should be balanced for this case, since according to the titration data (Figures 2C and 3C) it is at the apparent non-specific Kd. A possible resolution of this paradox is that there is quite strong binding cooperativity of clusters of HU on long DNAs, allowing rapid formation followed by strong stabilization of the bound complexes. Evidence for cooperativity of HU binding has been reported using FRET techniques even at HU:DNA molar ratios near 1:1 (12). We are developing single-DNA-based tools for determination of binding cooperativity of DNA-bending proteins to test this hypothesis. On top of this there may be appreciable sequence-dependence of affinities and kinetics of HU–DNA binding and unbinding. We are studying other proteins to determine the generality of the ‘on-off paradox’; we have already reported similar effects for HMG box proteins (32) as well as for the E. coli nucleoid protein Fis (54).
Low-salt conditions ‘freeze’ HU on DNA
The strong increase of complex lifetimes with decreasing NaCl concentrations (Figure 4D) leads to the phenomenon that at moderately low NaCl concentration (40 mM), dissociation of HU from DNA to protein-free solution can no longer be observed on experimentally observable time scales (Figure 4A), in accord with previous reports (32). This ‘frozen’ low-salt regime is consistent with the formation of discrete complexes in native gels only under very low salt concentrations (26,37). This illustrates an important feature of the single-DNA approach: protein–DNA interactions can be monitored under conditions where complexes are in dynamic chemical equilibrium on relatively short time scales.
HU–DNA complexes display ‘memory’ of the HU concentration at which they formed
Figure 4D shows that the lifetimes of the HU–DNA complexes depend strongly on the HU concentration at the time the complexes were formed. While this seems intuitively reasonable, it also makes clear that the dissociation process is not simply related to a microscopic off-rate: if the proteins in an HU–DNA complex were to independently dissociate, the complex lifetimes would all be nearly the same (and comparable to the inverse of that off-rate). Instead, the lifetimes strongly depend on the HU–DNA structures established at the time of binding. In effect, the complexes have a ‘memory’ of the initial HU solution conditions.
Force destabilizes HU–DNA complexes
We have demonstrated that force can drive unbinding of HU from DNA, and therefore affects the binding affinity. This has been theoretically suggested (40,55) on the grounds that applied force causes a free energy cost due to the compaction of DNA by DNA-bending proteins, but this effect was not previously demonstrated (note that a number of experiments have demonstrated destabilization of loops formed by DNA-binding proteins, but not actual unbinding of protein from DNA). Our data (Figure 5B) indicate that the dissociation rates of HU–DNA complexes in physiological 150 mM NaCl buffer depend rather sensitively on force: a shift of force from 0.1 to 0.3 pN generates a roughly three-fold reduction in HU–DNA complex lifetime. The dependence of dissociation rate on force can most simply be understood in terms of the reduction in stability of the bound state relative to the transition state for protein release, i.e. a reduction in the free energy barrier for the dissociation process (41).
Given that this level of force is significantly below forces generated in DNA during its transcription (56) and replication (57), forces generated in DNA in vivo can modify patterns of binding of HU; similar effects are likely for other DNA-bending proteins. Since HU is both a structural and gene-regulatory protein, forces along DNA in the cell can remodel the bacterial chromosome and alter gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JFM lab was supported by the National Science Foundation [DMR-0715099, PHY-0852130] to JFM lab; National Institutes of Health [U54CA143869-01 (NU-PS-OC)]; Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust; National Institutes of Health [GM038509] to R.C.J. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr John Graham, Dr Dunja Skoko and Professor Jie Yan for technical assistance and helpful discussions, Jenna McCracken for help with HU purification and Professor M. Thomas Record for helpful discussions.
REFERENCES
- 1.Johnson R.C., Johnson L.M., Schmidt J.W., Gardner J.F. Major nucleoid proteins in the structure and function of the Escherichia coli chromosome. In: Higgins N.P., editor. The Bacterial Chromosome. Washington, DC: ASM Press; 2005. pp. 65–132. [Google Scholar]
- 2.Rouviere-Yaniv J., Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc. Natl Acad. Sci. USA. 1975;72:3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swinger K.K., Rice P.A. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Rouviere-Yaniv J., Kjeldgaard N.O. Native Escherichia coli HU protein is a heterotypic dimer. FEBS Lett. 1979;106:297–300. doi: 10.1016/0014-5793(79)80518-9. [DOI] [PubMed] [Google Scholar]
- 5.Azam T.A., Iwata A., Nishimura A., Ueda S., Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon N.E., Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc. Natl Acad. Sci. USA. 1984;81:424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swinger K.K., Lemberg K.M., Zhang Y., Rice P.A. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swinger K.K., Rice P.A. Structure-based analysis of HU-DNA binding. J. Mol. Biol. 2007;365:1005–1016. doi: 10.1016/j.jmb.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodges-Garcia Y., Hagerman P.J., Pettijohn D.E. DNA ring closure mediated by protein HU. J. Biol. Chem. 1989;264:14621–14623. [PubMed] [Google Scholar]
- 10.Kamashev D., Balandina A., Rouviere-Yaniv J. The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 1999;18:5434–5444. doi: 10.1093/emboj/18.19.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavoie B.D., Shaw G.S., Millner A., Chaconas G. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 12.Sagi D., Friedman N., Vorgias C., Oppenheim A.B., Stavans J. Modulation of DNA conformations through the formation of alternative high-order HU-DNA complexes. J. Mol. Biol. 2004;341:419–428. doi: 10.1016/j.jmb.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 13.van Noort J., Verbrugge S., Goosen N., Dekker C., Dame R.T. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wojtuszewski K., Mukerji I. HU binding to bent DNA: a fluorescence resonance energy transfer and anisotropy study. Biochemistry. 2003;42:3096–3104. doi: 10.1021/bi0264014. [DOI] [PubMed] [Google Scholar]
- 15.Paull T.T., Haykinson M.J., Johnson R.C. HU and functional analogs in eukaryotes promote Hin invertasome assembly. Biochimie. 1994;76:992–1004. doi: 10.1016/0300-9084(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 16.Broyles S.S., Pettijohn D.E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 1986;187:47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- 17.Hillyard D.R., Edlund M., Hughes K.T., Marsh M., Higgins N.P. Subunit-specific phenotypes of Salmonella typhimurium HU mutants. J. Bacteriol. 1990;172:5402–5407. doi: 10.1128/jb.172.9.5402-5407.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh L.S., Rouviere-Yaniv J., Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: changes associated with salt shock. J. Bacteriol. 1991;173:3914–3917. doi: 10.1128/jb.173.12.3914-3917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rouviere-Yaniv J., Yaniv M., Germond J.E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 20.Dri A.M., Rouviere-Yaniv J., Moreau P.L. Inhibition of cell division in hupA hupB mutant bacteria lacking HU protein. J. Bacteriol. 1991;173:2852–2863. doi: 10.1128/jb.173.9.2852-2863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huisman O., Faelen M., Girard D., Jaffe A., Toussaint A., Rouviere-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J. Bacteriol. 1989;171:3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paull T.T., Johnson R.C. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 23.Oberto J., Nabti S., Jooste V., Mignot H., Rouviere-Yaniv J. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One. 2009;4:e4367. doi: 10.1371/journal.pone.0004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy S., Dimitriadis E.K., Kar S., Geanacopoulos M., Lewis M.S., Adhya S. Gal repressor-operator-HU ternary complex: pathway of repressosome formation. Biochemistry. 2005;44:5373–5380. doi: 10.1021/bi047720t. [DOI] [PubMed] [Google Scholar]
- 25.Koh J., Saecker R.M., Record M.T., Jr DNA binding mode transitions of Escherichia coli HU (alphabeta): evidence for formation of a bent DNA – protein complex on intact, linear duplex DNA. J. Mol. Biol. 2008;383:324–346. doi: 10.1016/j.jmb.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinson V., Takahashi M., Rouviere-Yaniv J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol. 1999;287:485–497. doi: 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 27.Wojtuszewski K., Hawkins M.E., Cole J.L., Mukerji I. HU binding to DNA: evidence for multiple complex formation and DNA bending. Biochemistry. 2001;40:2588–2598. doi: 10.1021/bi002382r. [DOI] [PubMed] [Google Scholar]
- 28.Castaing B., Zelwer C., Laval J., Boiteux S. HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks or gaps. J. Biol. Chem. 1995;270:10291–10296. doi: 10.1074/jbc.270.17.10291. [DOI] [PubMed] [Google Scholar]
- 29.Kamashev D., Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pontiggia A., Negri A., Beltrame M., Bianchi M.E. Protein HU binds specifically to kinked DNA. Mol. Microbiol. 1993;7:343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- 31.Schnurr B., Vorgias C., Stavans J. Compaction and supercoiling of single, long DNA molecules by HU protein. Biophys. Rev. Lett. 2006;1:29–44. [Google Scholar]
- 32.Skoko D., Wong B., Johnson R.C., Marko J.F. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 33.Lia G., Bensimon D., Croquette V., Allemand J.F., Dunlap D., Lewis D.E., Adhya S., Finzi L. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl Acad. Sci. USA. 2003;100:11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dame R.T., Goosen N. HU: promoting or counteracting DNA compaction? FEBS Lett. 2002;529:151–156. doi: 10.1016/s0014-5793(02)03363-x. [DOI] [PubMed] [Google Scholar]
- 35.Drlica K., Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konopka M.C., Weisshaar J.C., Record M.T., Jr Methods of changing biopolymer volume fraction and cytoplasmic solute concentrations for in vivo biophysical studies. Methods Enzymol. 2007;428:487–504. doi: 10.1016/S0076-6879(07)28027-9. [DOI] [PubMed] [Google Scholar]
- 37.Balandina A., Kamashev D., Rouviere-Yaniv J. The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 2002;277:27622–27628. doi: 10.1074/jbc.M201978200. [DOI] [PubMed] [Google Scholar]
- 38.Bonnefoy E., Takahashi M., Rouviere-Yaniv J. DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J. Mol. Biol. 1994;242:116–129. doi: 10.1006/jmbi.1994.1563. [DOI] [PubMed] [Google Scholar]
- 39.Marko J.F., Siggia E.D. Driving proteins off DNA using applied tension. Biophys. J. 1997;73:2173–2178. doi: 10.1016/S0006-3495(97)78248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan J., Marko J.F. Effects of DNA-distorting proteins on DNA elastic response. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2003;68:011905. doi: 10.1103/PhysRevE.68.011905. [DOI] [PubMed] [Google Scholar]
- 41.Marshall B.T., Long M., Piper J.W., Yago T., McEver R.P., Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 42.Wu L., Xiao B., Jia X., Zhang Y., Lu S., Chen J., Long M. Impact of carrier stiffness and microtopology on two-dimensional kinetics of P-selectin and P-selectin glycoprotein ligand-1 (PSGL-1) interactions. J. Biol. Chem. 2007;282:9846–9854. doi: 10.1074/jbc.M609219200. [DOI] [PubMed] [Google Scholar]
- 43.Zhu C., Yago T., Lou J., Zarnitsyna V.I., McEver R.P. Mechanisms for flow-enhanced cell adhesion. Ann. Biomed. Eng. 2008;36:604–621. doi: 10.1007/s10439-008-9464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Vlaminck I., Vidic I., van Loenhout M.T., Kanaar R., Lebbink J.H., Dekker C. Torsional regulation of hRPA-induced unwinding of double-stranded DNA. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq067. Advance Access Published on 2 March 2010; doi:10.1093/nar/gkq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali B.M., Amit R., Braslavsky I., Oppenheim A.B., Gileadi O., Stavans J. Compaction of single DNA molecules induced by binding of integration host factor (IHF) Proc. Natl Acad. Sci. USA. 2001;98:10658–10663. doi: 10.1073/pnas.181029198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith S.B., Finzi L., Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 47.Skoko D., Li M., Huang Y., Mizuuchi M., Cai M., Bradley C.M., Pease P.J., Xiao B., Marko J.F., Craigie R., et al. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc. Natl Acad. Sci. USA. 2009;106:16610–16615. doi: 10.1073/pnas.0909077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan J., Maresca T.J., Skoko D., Adams C.D., Xiao B., Christensen M.O., Heald R., Marko J.F. Micromanipulation studies of chromatin fibers in Xenopus egg extracts reveal ATP-dependent chromatin assembly dynamics. Mol. Biol. Cell. 2007;18:464–474. doi: 10.1091/mbc.E06-09-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strick T.R., Allemand J.F., Bensimon D., Bensimon A., Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 50.Marko J.F., Siggia E.D. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 51.Yan J., Marko J.F. Localized single-stranded bubble mechanism for cyclization of short double helix DNA. Phys. Rev. Lett. 2004;93:108108. doi: 10.1103/PhysRevLett.93.108108. [DOI] [PubMed] [Google Scholar]
- 52.Leirmo S., Harrison C., Cayley D.S., Burgess R.R., Record M.T., Jr Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry. 1987;26:2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- 53.Jones S., Shanahan H.P., Berman H.M., Thornton J.M. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003;31:7189–7198. doi: 10.1093/nar/gkg922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skoko D., Yoo D., Bai H., Schnurr B., Yan J., McLeod S.M., Marko J.F., Johnson R.C. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J. Mol. Biol. 2006;364:777–798. doi: 10.1016/j.jmb.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H., Marko J.F. Maxwell relations for single-DNA experiments: Monitoring protein binding and double-helix torque with force-extension measurements. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2008;77:031916. doi: 10.1103/PhysRevE.77.031916. [DOI] [PubMed] [Google Scholar]
- 56.Wang M.D., Schnitzer M.J., Yin H., Landick R., Gelles J., Block S.M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 57.Maier B., Bensimon D., Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc. Natl Acad. Sci. USA. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.