Abstract
Here we investigated the regulation of NF-κB activity by post-translational modifications upon reconstitution of NF-κB p65-deficient cells with the wild-type protein or phosphorylation-defect mutants. Analysis of NF-κB target gene expression showed that p65 phosphorylations alone or in combination function to direct transcription in a highly target gene-specific fashion, a finding discussed here as the NF-κB barcode hypothesis. High-resolution microscopy and surface rendering revealed serine 536 phosphorylated p65 predominantly in the cytosol, while serine 468 phosphorylated p65 mainly localized in nuclear speckles. TNF stimulation resulted in the translocation of the cytosolic p65 kinase IKKε to the nucleus and also to promyelocytic leukemia (PML) nuclear bodies. This inducible IKKε translocation was dependent on p65 phosphorylation and was prevented by the oncogenic PML-RARα fusion protein. Chromatin immunoprecipitation experiments revealed the inducible association of IKKε to the control regions of several NF-κB target genes. In the nucleus, the kinase contributes to the expression of a subset of NF-κB-regulated genes, thus revealing a novel role of IKKε for the control of nuclear NF-κB activity.
INTRODUCTION
Cells recognize intruding microorganisms with the help of specific membrane-bound or intracellular receptors and respond with the synthesis of proinflammatory mediators such as tumor necrosis factor (TNF) and interleukin 1 (IL-1). Once secreted, these cytokines in turn trigger their cognate receptors and thus help to rapidly amplify the inflammatory response (1). The NF-κB transcription factor is a key component for the production of many cytokines and also acts as a central mediator of cytokine-triggered effects (2). The five members of the NF-κB family of transcription factors can form different dimer combinations, but a heterodimer between p50 and the strongly transactivating p65 subunit is the most frequently detected form (3). All inducers of the canonical NF-κB activation pathway lead to the proteasomal elimination of inhibitory IκB proteins and thus release the DNA-binding subunits (4). IκB degradation depends on its prior phosphorylation by the IκB kinase complex that consists of the IκB kinases (IKK) IKKα, IKKβ and the regulatory subunit IKKγ/NEMO (5).
After release from IκB and nuclear translocation, the dimeric DNA-binding subunits can bind to their cognate DNA sequences and trigger expression of hundreds of target genes (6). Some NF-κB-dependent genes are important for the immune response, while others regulate cell survival and proliferation.
It is currently unclear how the free NF-κB dimers control key parameters of the target gene-specific response. Each individual NF-κB activating stimulus leads to the induction of a specific overlapping and distinct subset of genes (7). All parameters (induction, kinetics, cofactor recruitment, amplitude and termination) are specifically tailored for each gene in order to suit the specific requirements of the inducing stimulus (3). The individual contribution of the respective NF-κB subunits to cytokine-induced gene expression patterns was revealed by gene array experiments (8). In addition, a systematic analysis of binding sites for the p65 subunit by chromatin immunoprecipitation (ChIP) assays revealed thousands of binding sites in the genome of monocytes (9). Although ‘-omics’ approaches identified many genes containing NF-κB binding sites in their regulatory regions (9,10), each inflammatory gene must be expressed and turned off with peculiar kinetics that fit to its specific function. The mechanisms that specify these transcriptional programs are still not well understood and include dimer exchange, differential chromatin organization and modification of the DNA-binding subunits by post-translational modifications (7).
These modifications occur for all NF-κB DNA-binding subunits, but are most extensively characterized for p65 which can be regulated by ubiquitination, nitrosylation, acetylation, prolyl isomerization, monomethylation and phosphorylation (11–16). The functional consequences of these modifications are quite distinct, as exemplified by regulatory acetylation. Acetylation of p65 at lysines 122 and 123 impairs p65 transactivation, while acetylation at lysines 218 and 221 inhibits IκBα binding and increases p65-dependent transcription (17,18). Phosphorylation of p65 is found at many sites, but the most extensively studied are serine 276 (19) as well as serine 468 and 536 which are both contained in the C-terminal transactivation domain (20,21). The physiological relevance of p65 phosphorylation was revealed in a knock-in mouse model expressing a p65 protein with a serine 276 to alanine mutation. These animals showed aberrant gene expression which caused embryonic lethality at different time points (22). Mechanistic work has shown that phosphorylation can control several parameters such as protein/protein interactions and p65 ubiquitination. Phosphorylation at serine 276 results in conformational changes that allow binding of CREB-binding protein (CBP)/p300 (19), while phosphorylation of serine 536 favors binding of TATA-binding protein-associated factor II31, a component of TFIID (23). TNF-induced p65 ser 468 phosphorylation is a prerequisite for its association with a protein complex consisting of the acetyltransferase GCN5 and the ubiquitin E3 ligase components COMMD1 and Cullin-2, which lead to target gene specific p65 ubiquitination and degradation (24,25). Another ubiquitin E3 ligase regulating p65 stability is PDLIM2, which mediates LPS-induced p65 polyubiquitination and sequesters the transcription factor in promyelocytic leukemia nuclear bodies (PML-NBs) (26). PML-NBs are subnuclear domains which critically depend on the presence of the PML tumor suppressor protein and exert a variety of biological functions ranging from control of apoptosis to hematopoietic differentiation and gene transcription (27).
Among the kinases that phosphorylate serine 468 and 536 is IKKε (also called IKKi), a non-canonical IKK (28,29). IKKε is also an important mediator of the interferon response, as it phosphorylates the transcription factors IRF3 and IRF7, which form a complex with NF-κB to establish a multi-protein enhanceosome mediating the production of type I interferons (30). Recent evidence showed several additional functions for IKKε. The kinase also participates in the clearance of the nuclear receptor corepressor (NCoR) from distinct promoters (31). Mouse models also revealed the importance of IKKε for low-grade chronic inflammation as it occurs in obesity (32). Integrative genomic approaches identified IKKε as a breast cancer oncogene amplified and overexpressed in >30% of breast tumors and derived cell lines (28,33). Overexpression of IKKε is sufficient to transform cells, whereas siRNA-mediated knockdown of the kinase decreases the survival of breast cancer cells (33).
Here we have investigated the physiological role of regulatory p65 phosphorylation by reconstitution experiments. The impact of p65 phosphorylation on gene expression was highly gene specific and accordingly p65 phosphorylation at serine 468 and 536 occurred at distinct intracellular locations. TNF stimulation resulted in the inducible association of IKKε to p65 and the p65-dependent transport of the kinase to the nucleus and PML-NBs. IKKε was required for the expression of specific genes and ChIP experiments revealed the recruitment of IKKε to the control regions of many inflammatory target genes, thus identifying a novel role for IKKε in the nucleus.
MATERIALS AND METHODS
Cells, reagents, plasmids and antibodies
Mouse embryonic fibroblasts (MEFs) deficient for p65 were a gift of Dr Hiroyasu Nakano (Tokyo, Japan) and IKKε−/− MEFs were kindly provided by Dr Shizuo Akira (Osaka, Japan). MEFs that were stably reconstituted with p65 wild-type or point mutants were produced by infection with lentiviral vectors and subsequent selection. Primary lung macrophages were isolated from euthanized female and male C57/BL6 mice. The trachea was exposed and macrophages were isolated essentially as described (34). Cells were seeded in 24-well plates (300 000 cells/well) and further analyzed for gene expression. TNF was purchased from ImmunoTools and always used in a standard concentration of 20 ng/ml. Protein A/G sepharose was from Santa Cruz Biotechnology, Dynabeads® protein G was from Invitrogen and the IKKε inhibitor BX795 was purchased from Axon Medchem. Antibodies recognizing IKKε (H-116), p65 (C-20), p65 (A), PML (H-238) and (PG-M3) were purchased from Santa Cruz Biotechnology. The antibodies recognizing Flag (M2), tubulin (tub 2.1) (Sigma), IKKε (12142), β-actin (8227) (Abcam) and HA (3F10) (Roche) were from the indicated suppliers. The anti-IgG and the two phospho-specific antibodies recognizing p65 phosphorylated at serine 468 (3039S) or serine 536 (3031S) were from Cell Signaling Technology. Secondary horseradish peroxidase coupled anti-rabbit or anti-mouse antibodies were purchased from Dianova. Secondary Cy3-coupled anti-rabbit and anti-mouse antibodies were purchased from Jackson ImmunoResearch Laboratories, the Alexa Fluor®488-conjugated anti-rabbit antibody was from Invitrogen. Plasmids encoding HA-p65, HA-p65 S468A, HA-p65 S536A, HA-p65 S536A/S468A, GFP-p65 (25), Flag-IKKε, Flag-IKKε-K38A (35), PML-IV, PML-III, PML-RARα (36) and shPML (37) were described as published.
RNA extraction and real-time PCR
Cells were lysed and total RNA was extracted using the RNeasy kit (Qiagen). The quality of RNA was controlled by agarose gel electrophoresis and ethidium bromide staining. Synthesis of cDNA was done with 1 μg of RNA from oligo (dT)-20 primers using the Superscript II first strand synthesis system (Invitrogen). Real-time PCR was performed using specific primers and the SYBR Green ROX Mix (Thermo Scientific). The sequence of the primers is given in the Supplementary Data. The PCR reactions were analyzed using an Applied Biosystems 7300 real-time PCR system. All experiments were performed in triplicate, data were normalized to the housekeeping gene β-actin and the relative abundance of transcripts was calculated by the comparative ΔΔCT method.
ChIP and re-ChIP assays
Cells were cross-linked with 1% (v/v) formaldehyde, collected and then lysed in RIPA buffer [10 mM Tris pH 7.5, 150 mM NaCl, 1% (v/v) NP-40, 1% (w/v) Na-Desoxycholat, 0.1% (w/v) SDS, 1 mM EDTA and 10 µg/ml aprotinin]. DNA was sheared by sonification using a Branson sonifier 250. After removal of cellular debris by centrifugation, equal amounts of DNA were incubated with 2 μg of anti-p65, anti-IKKε or control IgG antibodies previously bound to protein G-coupled Dynabeads®. After rotating for 2 h at 4°C on a spinning wheel, the magnetic beads were washed five times using the following buffers: RIPA buffer (twice), RIPA high-salt buffer [2 M NaCl, 10 mM Tris pH 7.5, 1% (v/v) NP-40, 0.5% (v/v) Na-Desoxycholate and 1 mM EDTA], RIPA and TE buffer (10 mM Tris pH 7.5 and 1 mM EDTA). The precipitated fragments were eluted with TE buffer containing 1% SDS. For re-ChIP assays, the eluted material was diluted 1:10 with RIPA buffer lacking SDS and precipitated again with the monoclonal anti-IKKε antibody. The eluted DNA was quantified by real-time PCR using specific primer sets flanking the expected p65 binding site. The specificity of the IKKε antibodies was further ensured by controls that revealed inducible IKKε binding to the Icam1 promoter only in IKKε containing cells, but not in Ikbke-deficient MEFs (Supplementary Figure S1). Sequences of the primers used for ChIP experiments is given in the Supplementary Data.
Immunofluorescence and three-dimensional surface rendering
Cells were grown on cover slips in 12-well dishes. Stimulation was performed with 20 ng/ml TNF for the indicated periods. Cells were fixed in a cold 1:1 solution methanol:acetone for 1 min, rehydrated in phosphate-buffered saline (PBS) and subsequently blocked with 10% goat serum in PBS for 1 h. The cells were incubated with primary antibodies (diluted in PBS containing 1% goat serum) at 4°C overnight. The next day, cells were washed three times in PBS during 10 min, then incubated with the Cy3-coupled anti-rabbit or with the Cy3-coupled anti-mouse alone or combined with Alexa Fluor®488 anti-rabbit for the colocalization experiments (all diluted in PBS containing 1% goat serum). Nuclear DNA was stained with Hoechst 33324. The cover slips were mounted on microscope slides with Kaiser’s glycerol gelatine and further analyzed using an inverted Nikon Eclipse 2000E microscope. Three-dimensional confocal analysis was done by analysis of fixed cells using a laser scanning microscope (Leica TCS SP2 AOBS) equipped with differential interference contrast optics. Confocal pictures were obtained using a 40× Leica oil immersion objective (HCX PL APO CS, numerical aperture: 1.25). The detector gain and amplifier offset was set to obtain pixel intensities within the linear range. Optical reconstruction of confocal Z-stacks (step size: 0.15 µm), three-dimensional volume renderings and animated videos were generated using the Imaris software package (Bitplane AG). The Photoshop CS4 (Adobe Systems) program was used to optimize contrast and brightness. All immunofluorescence data are representative for >80% of interphase cells. Dying or mitotic cells and also cells expressing aberrantly high levels of the proteins were not analyzed.
Immunoprecipitation and Western blotting
Cells were washed in PBS and collected by centrifugation. The cell pellet was resuspended in NP-40 lysis buffer [20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM phenylmethylsulfonylfluoride, 10 mM NaF, 0.5 mM sodium vanadate, leupeptine (10 µg/ml), aprotinin (10 µg/ml) and 1% (v/v) NP-40] and incubated on ice for 20 min. After removal of cellular debris by centrifugation for 10 min, equal amounts of proteins contained in the supernatant were further analyzed. Immunoprecipitations were performed upon addition of 2 μg of the indicated antibodies together with 25 µl of protein A/G sepharose. After rotating for 4 h at 4°C, the supernatant was collected and the beads were washed for five times in lysis buffer. The precipitated proteins were eluted by boiling in 1.5× SDS sample buffer for 5 min. Western blotting was done by SDS–PAGE and semi-dry transfer to polyvinylidene difluoride membranes. After incubation with primary and secondary peroxidase-coupled antibodies, protein were detected using the enhanced chemoluminiscence system (GE Healthcare) according to the instructions given by the manufacturer.
RESULTS
NF-κB p65 phosphorylation regulates gene expression in a target gene specific manner
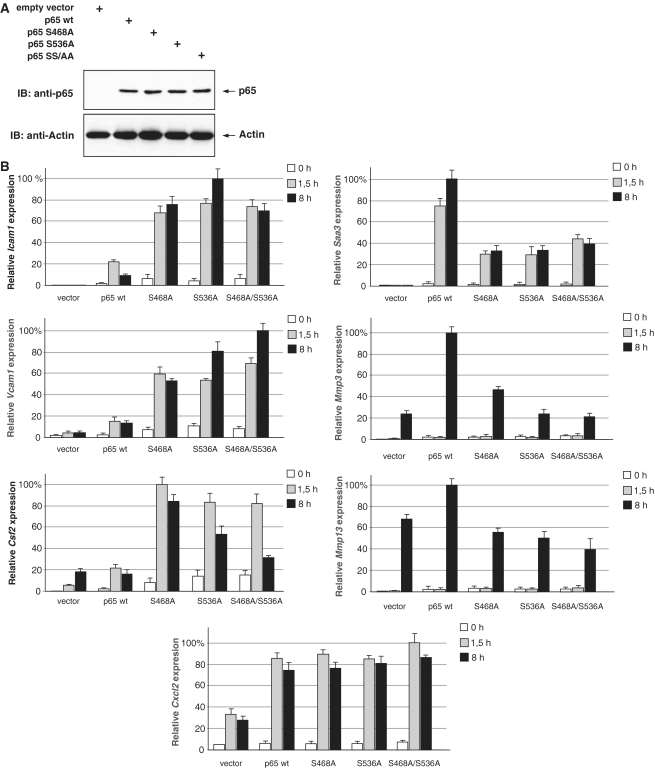
The functional relevance of p65 phosphorylation for its ability to trigger gene expression has been widely investigated by reporter gene assays. To analyze the role of p65 phosphorylation in a more physiological setting, p65-deficient MEFs were retransfected to stably express physiological levels of p65 wild-type or point mutants where serine 468 or 536 were changed individually or together to alanine (Figure 1A). Cells were left untreated or stimulated with TNF for different periods and gene expression of several known p65 target genes identified in a previous study (38) was quantified by real-time PCR (Figure 1B). TNF-induced expression of one gene group is down-regulated by p65 phosphorylation, as judged by the higher transcriptional activity of the non-phosphorylatable mutants. This group comprises the Icam1, Vcam1 and Csf2 genes. In contrast, expression of the Saa3, Mmp3 and Mmp13 genes requires intact p65 phosphorylation for full gene activation. Effects of p65 phosphorylation were not detected for the Cxcl2 gene, showing that the regulatory function of these phosphorylations is strictly dependent on individual regulated gene.
Figure 1.
Analysis of phosphorylation-dependent NF-κB p65 activity. (A) p65-deficient MEFs were stably reconstituted to express physiological levels of HA-p65 and its point mutated versions that were changed in serine 468 (p65 S468A), serine 536 (p65 S536A) or both sites (p65 SS/AA). One fraction of the cells was lysed and equal amounts of protein were tested by immunoblotting (IB) for comparable expression of p65. (B) The indicated cells were treated with TNF for 1, 5 or 8 h. The expression of selected NF-κB target genes by real-time PCR. In order to facilitate comparison, maximal gene activation was arbitrarily set as 100%. Experiments were performed in triplicates, error bars display standard deviations.
Is the differential transcriptional activity of the mutants attributable to differences in p65 promoter occupancy? To address this question, ChIP assays were performed with MEFs stably expressing p65 wild-type or the point mutated p65 proteins. Cells were left untreated or stimulated for various periods with TNF, followed by ChIP experiments using p65 specific antibodies. Quantification of genomic fragments encompassing NF-κB binding sites controlling the various NF-κB target genes allowed to detect inducible p65 binding to all regions with the exception of the Mmp3 gene for which no functional binding site was found. For the group of genes that are negatively regulated by p65 phosphorylation, ChIP experiments showed an increased and prolonged occurrence of the serine 468 mutant at the NF-κB binding sites (Supplementary Figure S2A). This finding is fully compatible with the recently reported relevance of serine 468 phosphorylation for promoter-specific proteasomal elimination of p65 (24,25). On the other hand, the recruitment of the p65 serine 536 alanine mutant was comparable to that of the wild-type protein at the NF-κB sites contained in the Vcam1 and Csf2 promoters, suggesting that its increased transcriptional activity must rely on promoter occupancy-independent mechanisms. The partial correlation between transcriptional activity and promoter recruitment was also seen for the group genes that depend on p65 phosphorylation (Supplementary Figure S2B), while p65 recruitment to the Cxcl2 control region was not regulated by phosphorylation (Supplementary Figure S2C).
Distinct intracellular distribution of NF-κB p65 phosphorylations
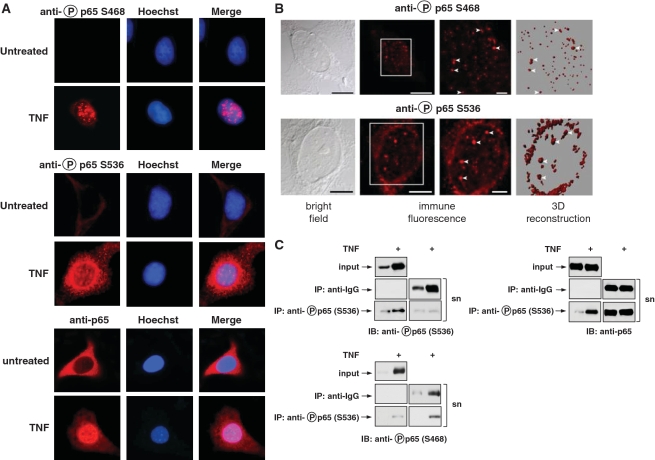
To reveal the intracellular localization of the phosphorylated p65 proteins, immunofluorescence studies with phospho-specific antibodies were performed. Analysis of TNF-induced cells showed the accumulation of serine 468 phosphorylated endogenous p65 mainly in nuclear microspeckles (Figure 2A). In contrast, the majority of serine 536 phosphorylated p65 was cytosolic and accumulated around the nucleus. To investigate the intracellular distribution of phosphorylated p65 in more detail, confocal image stacks were processed to reveal their three-dimensional distribution pattern. Both phosphorylated p65 forms showed a prominent organization into microspeckles of heterogeneous sizes (Figure 2B), animated versions of these structures are displayed in the Supplementary Data. TNF stimulation of cells leads to the nuclear translocation of the vast majority of p65 to the nucleus, while serine 536 phosphorylated p65 is mainly found in the cytosol (Figure 2A). This differential distribution is only explainable by a non-quantitative phosphorylation of p65, a possibility that was tested experimentally. Cells were stimulated with TNF and cell extracts were used for immunoprecipitation with an antibody recognizing serine 536 phosphorylated p65. Subsequent western blotting using the supernatant allowed to detect trace amounts of p65 phosphorylated at serine 536 or 468, but a large fraction of unmodified p65 (Figure 2C). These data show that (I) only a fraction of p65 is phosphorylated and (II) phosphorylation can occur at serine 536 and 468 simultaneously.
Figure 2.
Intracellular distribution of p65 phosphorylation. (A) HeLa cells were left untreated or stimulated with TNF for 30 min. The subcellular localization of endogenous p65 or its phosphorylated forms was visualized by indirect immunofluorescence using anti-phospho-S468, anti-phospho-S536 or anti-p65 antibodies (red). Nuclear DNA was revealed by Hoechst staining. (B) HeLa cells were stimulated for 30 min with TNF and further analyzed by confocal microscopy. A bright field picture, two magnifications of immunofluorescence pictures and a three-dimensional reconstruction of the same cell are displayed. The Z-stack pictures were processed using the Imaris software package for surface rendering. To facilitate the orientation, white arrow heads point to speckles that are displayed by immunofluorescence and also by 3D reconstruction. (C) HeLa cells were stimulated with TNF for 30 min as shown. Lysates were subjected to immunoprecipitation (IP) with anti-IgG control or anti-phospho-p65-S536 antibodies. The input, supernatant (sn) and the immunoprecipitated proteins were further analyzed by western blotting using anti-p65 or phospho-specific antibodies recognizing serine 468 or 536.
IKKε is required only for a subset of NF-κB target genes
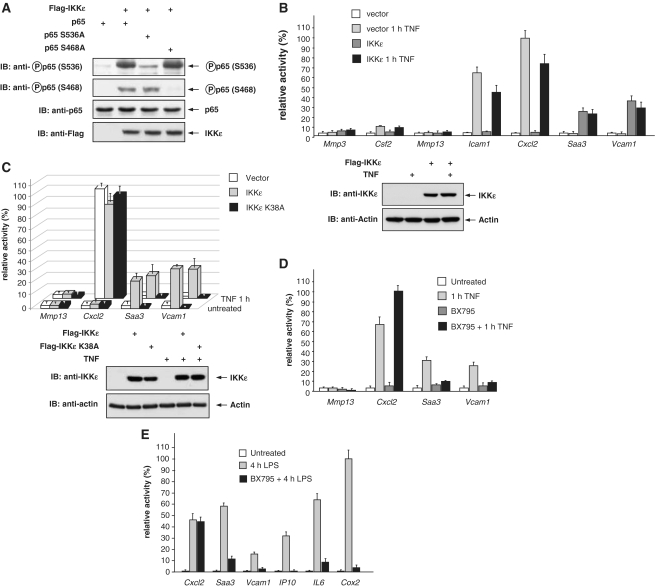
Given the differential contributions of p65 phosphorylations at serines 468 and 536 for gene expression, it was then interesting to investigate the relevance of IKKε, as this kinase has the ability to phosphorylate p65 at these two sites (Figure 3A). Loss-of-function experiments using shRNA-mediated IKKε knock-down showed that TNF-induced serine 536 phosphorylation was independent from IKKε, while serine 468 phosphorylation was largely impaired in the absence of this kinase (data not shown). MEFs lacking the Ikbke gene were reconstituted to express physiological levels of the kinase and then left untreated or stimulated with TNF. Real-time PCR experiments showed that from all the seven genes analyzed in Figure 1 only two (Saa3 and Vcam1) were IKKε dependent (Figure 3B). In both cases the expression of IKKε was sufficient to trigger transcription even in the absence of TNF stimulation. Also Mmp3 and Mmp13, two genes which were not induced 1 h after TNF stimulation, remained unaffected by IKKε even after prolonged TNF stimulation (data not shown). Consistent with Figure 1B, the expression of Cxcl2, Csf2 and Icam1 genes was triggered already 1 h after TNF stimulation, but this induction was IKKε independent. A similar experimental approach was taken in order to investigate the role of the IKKε kinase function for its ability to trigger gene expression. Cells were reconstituted to express the wild-type kinase or a kinase inactive IKKε lysine 38 to alanine mutant, followed by TNF stimulation and real-time PCR analysis of the two IKKε-regulated genes and two adequate controls. These experiments revealed induced Saa3 and Vcam1 expression only in the presence of the wild-type kinase (Figure 3C). To investigate the contribution of the kinase function by an independent setting, TNF-induced gene expression was determined in the presence of the specific small molecule IKKε inhibitor BX795 (39). These experiments revealed the selective inhibition of TNF-triggered Saa3 and Vcam1 expression (Figure 3D), supporting the relevance of the IKKε kinase function for the expression of a specific subset of genes. The role of IKKε for target gene specific gene expression was also seen in primary alveolar macrophages stimulated with LPS (Figure 3E). In this setting, several genes including Saa3, Vcam1, IP10, IL6 and Cox2 were largely dependent on IKKε kinase activity.
Figure 3.
IKKε-dependent expression of NF-κB target genes. (A) p65-deficient MEFs were transfected with the various expression vectors as shown. Thirty-six hours later they were analyzed by western blotting for expression and phosphorylation of p65 as shown. (B) Ikbke-deficient MEFs were reconstituted to express physiological levels of Flag-IKKε. After 24 h, the cells were treated with TNF for 1 h or left untreated. Gene expression was determined by real-time PCR and normalized to the level of β-actin. Adequate IKKε expression was determined by western blotting using an anti-IKKε antibody. (C) Ikbke−/− MEFs were reconstituted to express Flag-IKKε or the point mutated kinase-inactive version Flag-IKKε-K38A. Cells were further analyzed as in (B). (D) MEFs were treated for 1 h with BX795 (1 μM) or left untreated prior to stimulation with TNF as shown. Gene expression was further quantified by real-time PCR. (E) Primary mouse lung macrophages were preincubated for 1 h with 1 μM BX795 or left untreated prior to stimulation for 4 h with LPS. Gene expression was determined by real-time PCR and normalized to the level of β-actin. All real-time experiments were performed in triplicates, error bars display standard deviations. Maximal gene activation was arbitrarily set as 100%.
TNF-triggered nuclear translocation and chromatin association of IKKε depends on p65
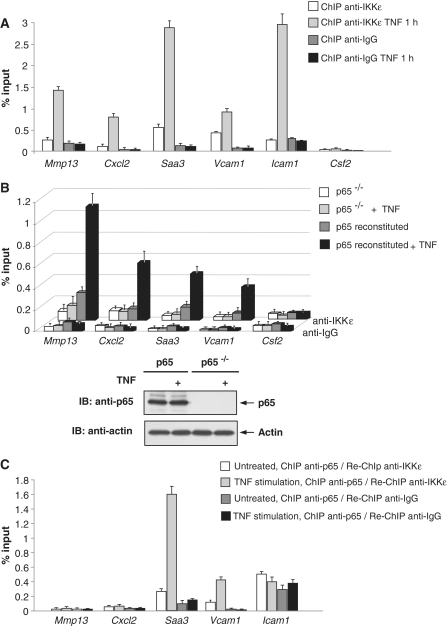
The recent years have mounted evidence that some mainly cytosolic components of the NF-κB signaling cascade such as IKKα or GSK3β can also be found at specific NF-κB target genes where they participate in the control of gene expression (40,41). Given the functional relevance of IKKε for the expression of individual genes, it was then interesting to investigate the potential association of IKKε to chromatin areas containing NF-κB binding sites. ChIP assays with untreated controls or TNF-stimulated cells revealed TNF-inducible recruitment of endogenous IKKε to genome fragments encompassing the NF-κB sites of a surprisingly high number of p65 target genes (Figure 4A). All promoters showed TNF-inducible IKKε recruitment or some basal binding of IKKε even in the unstimulated state, as seen for the NF-κB binding sites contained in the Saa3 and Vcam1 promoters. These experiments suggest that IKKε recruitment is not only seen for genes that actually depend on IKKε (such as Saa3 and Vcam1), but also for genes that do not rely on its activity. This finding is compatible with recent results describing a more general role of IKKε for gene expression upon its contribution to the clearance of the corepressor NCoR from selected promoters (31). To investigate the potential role of the IKKε substrate protein p65 for the chromatin recruitment of the kinase, further ChIP experiments were performed. TNF-inducible chromatin association of IKKε was determined in MEFs that were either p65-deficient or reconstituted to express the p65 protein. ChIP experiments revealed the absolute necessity of the p65 protein for IKKε promoter recruitment (Figure 4B). These data raise the possibility that IKKε is recruited to its target promoters upon interaction with p65, a scenario that was tested by sequential ChIP experiments. TNF-stimulated cells or adequate controls were used to immunoprecipitate chromatin with anti-p65 antibodies, followed by a second round of immunoprecipitation (re-ChIP) with anti-IKKε antibodies. These experiments showed p65-associated IKKε at the genomic fragments containing NF-κB sites controlling the Saa3 and Vcam1 promoters (Figure 4C). Remarkably, the expression of these two genes fully depends on IKKε. In contrast, Re-ChIP experiments failed to detect p65-associated IKKε at the other promoters, suggesting that chromatin association of IKKε can also employ further mechanisms.
Figure 4.
TNF-induced and NF-κB p65-dependent chromatin recruitment of IKKε. (A) MEFs were stimulated with TNF as shown, followed by ChIP analysis using the indicated specific and anti-IgG control antibodies. IKKε association with different promoter regions was detected by real-time PCR using specific primers. The amounts of p65-associated DNA are presented as the percentage recovered out of the total input DNA (percent input). Experiments were performed in triplicates, error bars display standard deviations. (B) p65-deficient MEFs were transfected with a vector encoding p65 or an adequate control. After 24 h, the cells were stimulated with TNF as shown and ChIP assays for IKKε were performed as in (A). The lower part shows a control western blot ensuring adequate p65 expression. (C) MEFs were stimulated with TNF as shown and used for the ChIP procedure using anti-p65 antibodies or controls, followed by elution and re-ChIP using anti-IKKε antibodies. Experiments were performed in triplicates, error bars display standard deviations.
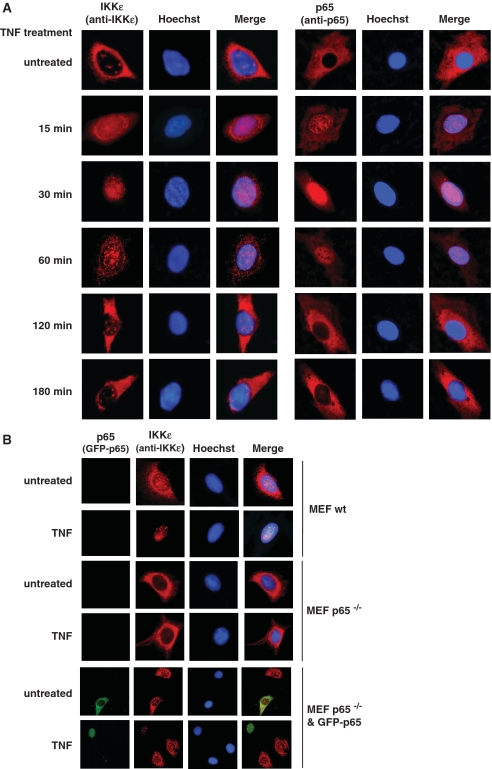
The TNF-induced recruitment of IKKε to selected promoters raises the question whether TNF also affects the intracellular localization of the kinase. Cells were stimulated for various periods with TNF and the localization of endogenous IKKε was analyzed by immunofluorescence. In unstimulated cells most of the kinase was found in the cytosol and a residual fraction in nuclear bodies. Already 15 min after stimulation of the TNF receptor, a substantial fraction of IKKε was found in the nucleus. Relocalization of the kinase to the cytosol started around 1 h after stimulation and was complete two hr after addition of TNF (Figure 5A). The kinetic behavior of IKKε nuclear import and export mirrored that of p65, which showed an oscillatory behavior with most p65 in the nucleus 30–60 min after TNF treatment, followed by nuclear export and a cytosolic localization of p65 at later time points. The similar kinetics of IKKε and p65 for nuclear import and the requirement of p65 for chromatin association of IKKε raise the question whether IKKε depends on p65 for its nuclear translocation. To address this issue, TNF-triggered nuclear import of IKKε was compared between wild-type MEFs and p65−/− MEFs. These experiments showed no nuclear translocation of IKKε in the absence of p65, while only p65−/− cells reconstituted to express p65 showed TNF-triggered IKKε accumulation in the nucleus (Figure 5B).
Figure 5.
TNF-induced and NF-κB p65-dependent nuclear translocation of IKKε. (A) HeLa cells were stimulated with TNF for the indicated periods. The intracellular localization of endogenous IKKε and p65 proteins was analyzed by indirect immunofluorescence using anti-p65 and anti-IKKε antibodies (red). Nuclear DNA was revealed by Hoechst staining. (B) Wild-type MEFs and p65-deficient MEFs transfected with empty vector or a plasmid encoding GFP-p65 were stimulated for 30 min with TNF as shown. The subcellular localization of GFP-p65 (green) and of endogenous IKKε (red) was analyzed by indirect immunofluorescence.
NF-κB p65 phosphorylation is required for TNF-inducible association with IKKε
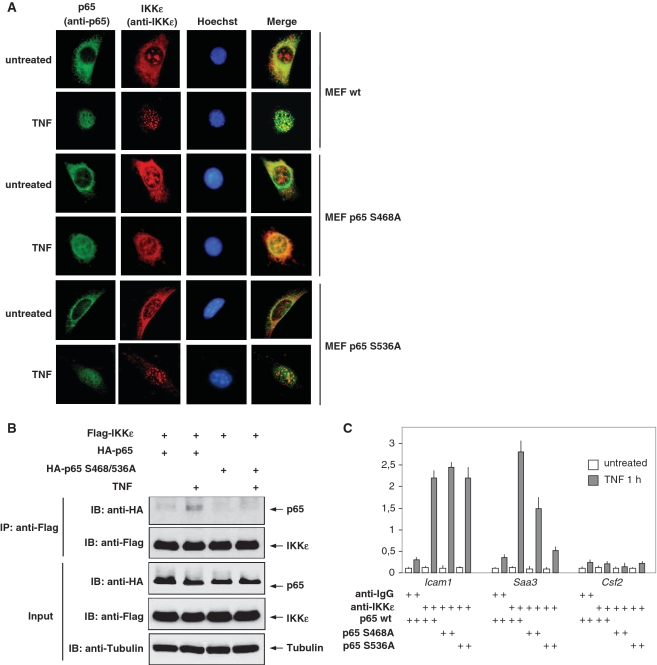
The necessity of p65 for nuclear import of IKKε raises the question on a possible contribution of p65 phosphorylation for this process. To address this question, MEFs expressing wild-type p65 or non-phosphorylatable mutants were analyzed for TNF-triggered nuclear import of IKKε (Figure 6A). These studies revealed an impaired nuclear translocation of IKKε in the presence of p65 S468A, thus revealing a contribution of p65 phosphorylation for nuclear import. The recently reported inducible association of IKKε and p65 in LPS stimulated cells (31) prompted us to investigate the occurrence of TNF-triggered complex formation between the kinase and its substrate by coimmunoprecipitation experiments. Cells were transfected to express IKKε, p65 or a non-phosphorylatable double point mutant thereof. Control cells or TNF-treated cells were lysed, followed by immunoprecipitation of IKKε. Western blotting allowed to detect TNF-inducible binding between IKKε and p65 wild-type but not p65 SS/AA (Figure 6B), indicating that the inducible association depends on p65 phosphorylation. To test the role of p65 phosphorylation for the recruitment of IKKε to target gene promoters, ChIP experiments were performed with cells stably expressing wild-type p65 or versions mutated at serines 468 or 536. TNF-triggered association of IKKε to the Icam1 promoter was not affected by p65 phosphorylation, while recruitment to the Saa3 promoter was largely dependent on serine 536 phosphorylation (Figure 6C).
Figure 6.
Phosphorylation-dependent association of IKKε and p65. (A) MEFs stably reconstituted with p65 wild-type or the point mutated versions p65 S468A and p65 S536A were stimulated with TNF and the subcellular localization of IKKε (red) and p65 (green) was visualized by indirect immunofluorescence. (B) HEK293 cells transfected to express Flag-IKKε together with HA-p65 or its point mutated version HA-p65 SS/AA were stimulated with TNF, lysed and cell extracts subjected to immunoprecipitation as shown. The input material and the immunoprecipitated proteins were further analyzed by western blotting using anti-HA and anti-Flag antibodies. (C) MEFs stably reconstituted with p65 wild-type or the point mutated versions p65 S468A and p65 S536A were stimulated with TNF for 1 h, followed by ChIP analysis using the indicated specific and anti-IgG control antibodies. IKKε association with the indicated promoter regions was detected by real-time PCR using specific primers, error bars show standard deviations from three experiments.
TNF-triggered nuclear translocation and chromatin association of IKKε depend on its kinase function
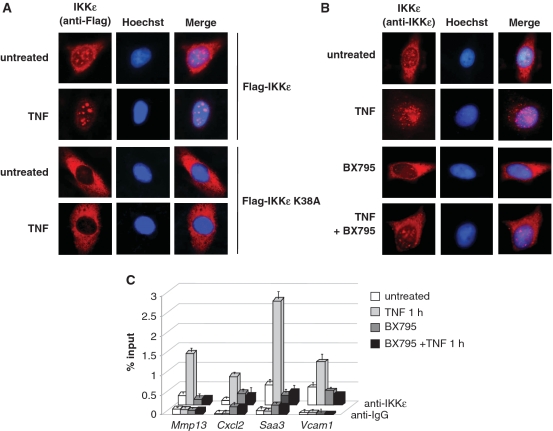
IKKε-mediated gene induction depends on its kinase activity, which may be explained by the necessity to phosphorylate nuclear substrates or alternatively by a kinase-dependent nuclear import of IKKε. To distinguish between these possibilities, Ikbke−/− cells were transfected to express the wild-type or kinase inactive IKKε. After stimulation with TNF, immunofluorescence studies showed a faithful recapitulation of TNF-triggered nuclear import of the wild-type kinase, while IKKε K38A was found exclusively in the cytosol (Figure 7A). In addition, these experiments showed completely absent IKKε localization to nuclear speckles in unstimulated cells, a finding that we consistently observed for a small fraction of the kinase. To reveal the importance of the kinase function for nuclear translocation by an independent experimental approach, TNF-triggered nuclear import of endogenous IKKε was studied in the presence of the IKKε inhibitor BX795. Inhibition of IKKε kinase function strongly interfered with TNF-triggered translocation of IKKε to subnuclear speckles (Figure 7B). Similarly, ChIP experiments showed that BX795 efficiently blocked TNF-inducible recruitment of IKKε to its cognate promoters (Figure 7C). Collectively, these results reveal the importance of the IKKε kinase function for TNF-induced nuclear import and all downstream events. Some proteins such as NF-κB p65 show a constant shuttling between cytosol and nucleus (42), while others depend on a distinct inducing signal to allow nuclear translocation (43). To investigate the relevance of these two possibilities for nuclear entry of IKKε, cells were treated with different combinations of TNF and leptomycin B, an inhibitor of nuclear export. Immunofluorescence analysis showed no effect of leptomycin B on the intracellular localization of IKKε (Supplementary Figure S3A), but confirmed the reported nuclear accumulation of p65 (Supplementary Figure S3B) even in the absence of TNF stimulation (42). In contrast, nuclear export of IKKε occurring after prolonged treatment with TNF was prevented by leptomycin B (data not shown), suggesting that nuclear translocation of IKKε depends on an active TNF-triggered import rather than a constant shuttling of the kinase between the nucleus and the cytosol.
Figure 7.
Nuclear import and chromatin association of IKKε depends on its kinase activity. (A) Ikbke-deficient MEFs were reconstituted to express Flag-IKKε or Flag-IKKε-K38A. Thirty-six hours after transfection, cells were left untreated or stimulated with TNF for 30 min. The subcellular localization of IKKε was visualized by indirect immunofluorescence using an anti-Flag antibody (red). (B) MEFs were incubated for 1 h with BX795 (1 μM) as shown and then further treated for 30 min with TNF. Subcellular localization of endogenous IKKε was visualized by indirect immunofluorescence using an anti-IKKε antibody. (C) MEFs were preincubated for 1 h with 1 μM BX795 or left untreated prior to stimulation for 1 h with TNF. ChIP assays for IKKε were performed using specific anti-IKKε and anti-IgG control antibodies and the precipitated DNA was quantified by real-time PCR using specific primers. Experiments were performed in triplicates, error bars display standard deviations.
IKKε can be recruited to PML-NBs
TNF stimulation of cells results in the accumulation of IKKε in the nucleoplasm, but also in distinct speckled structures. What type of nuclear speckles is containing the IKKε protein? As components of PML-NBs are known to regulate the transcription of some NF-κB target genes (44), we tested the possible localization of IKKε to these subnuclear structures. Expression of the PML splice variant PML-IV allowed full recruitment of IKKε to PML-NBs even in the absence of TNF stimulation. In contrast, the splice variant PML-III (which lacks exon 8) did not lead to the retention of IKKε to nuclear speckles, but still allowed TNF-induced nuclear entry of the kinase (Figure 8A). Fusion of the human Pml gene to the retinoic acid receptor α (Rarα) gene by the t (15,17) chromosomal translocation generates a dominant negative PML-RARα fusion protein which is causative for acute promyelocytic leukemia (45). Expression of PML-RARα largely prevented TNF-triggered recruitment of IKKε to PML-NBs (Figure 8A). Conversely, shRNA-mediated down-modulation of PML precluded TNF-induced recruitment of IKKε to PML-NBs (Figure 8B), supporting the relevance of PML-NBs also in a loss-of-function approach. The overlapping localization of IKKε with PML-NBs was also seen at the level of the endogenous proteins. TNF stimulation allowed the nuclear import of IKKε and the partial colocalization of the kinase with PML-NBs (Figure 8C). Knock-down of PML prevented the accumulation of IKKε in PML-NBs and also in the nucleoplasm, thus revealing the imporance of these subnuclear structures for the nuclear import of IKKε. Given the partial localization of IKKε to PML-NBs, it was then interesting to investigate whether also p65 serine 468 phosphorylation occurs in these subnuclear structures. Immunofluorescence studies showed only a minor colocalization between PML and serine 468 phosphorylated p65 (Figure 8D). On the other hand, the downmodulation of PML by shRNA resulted in a changed intranuclear distribution of phosphorylated p65 which occurred throughout the nucleus in a microspeckled fashion (Figure 8D), indicating the relevance of PML for the intranuclear distribution of serine 468 phosphorylated p65.
Figure 8.
TNF-induced recruitment of IKKε to PML-NBs. (A) HeLa cells were transfected to express the PML splicing forms PML-III, PML-IV or the PML-RARα fusion protein. Thirty-six hours after the transfection, cells were left untreated or stimulated with TNF for 30 min. The subcellular localization of PML and endogenous IKKε was visualized by indirect immunofluorescence using an anti-PML (red) and anti-IKKε antibody (green). (B) Upper: Hela cells transfected with a plasmid expressing a PML-specific shRNA were treated with TNF for 30 min as shown and analyzed for subcellular localization of endogenous IKKε using an anti-IKKε antibody. Lower: the efficient knock-down of PML was controlled by transfection of the vector directing the synthesis of the PML specific shRNA and detection of PML by immunofluorescence. The limited transfection efficiency allows to observe PML down-regulation only in transfected cells. (C) Cells were transfected with a vector for a PML specific shRNA or a control. After 36 h, cells were stimulated for 30 min with TNF as shown and the localization of the endogenous proteins was revealed by immunofluorescence. (D) Hela cells were treated as in (C) and analyzed for the occurrence and subcellular localization of endogenous PML and serine 468 phosphorylated p65 as shown. (E) Schematic model summarizing the mechanisms and functions of p65 phosphorylation and its phosphorylation by IKKε and other kinases.
DISCUSSION
Spatial organization of p65 phosphorylation
This study suggests that the functional relevance of a phosphorylation site is also influenced by its intracellular distribution. While the p65 protein is found in the entire nucleus except the nucleolus in TNF-stimulated cells, the phosphorylated p65 proteins accumulate in specific subcellular regions. Immunofluorescence experiments showed a strong enrichment of serine 536 phosphorylated p65 in the area surrounding the nucleus. This localization is compatible with published data suggesting a functional role of p65 serine 536 phosphorylation for controlling the kinetics of p65 nuclear import (29). In contrast, p65 serine 468 phosphorylation was predominantly nuclear and occurred in a speckled distribution which shows strong accumulation of serine 468 phosphorylated p65 in distinct nuclear regions. As several groups of NF-κB target genes are not randomly distributed in the genome and are enriched in distinct chromosomal clusters (9,10), it will be relevant to investigate whether these genomic regions are selectively enriched with phosphorylated p65. Our data show that the effects of p65 phosphorylation depend on the individual target gene, an observation which is supported by the microscopical data which revealed focal points of p65 phosphorylation in the nucleus. The accumulation of phosphorylated p65 in distinct regions raises the question for the molecular mechanisms underlying this inhomogeneous distribution. Systematic approaches such as ChIP to CHIP experiments will help to address these important questions in the future.
The NF-κB barcode hypothesis
Within a given cell type, stimulation with distinct NF-κB activating agents stereotypically allows the generation of DNA-binding dimers, while target gene expression displays clear differences in respect to the regulated genes (4). In addition, co-regulated genes often show stimulus-specific dynamic parameters of the transcriptional response (3,41). Our data support the idea that these key parameters are controlled by mechanisms that include post-translational modification of the DNA-binding subunits. This study shows that the impact of each individual phosphorylation site is highly gene specific and that reporter gene assays are therefore not adequate to investigate the functional consequences of NF-κB modifications. Our findings are in line with studies that extensively investigated the functional consequences of p65 serine 276 phosphorylation on gene expression (22,46–48). The functional outcome for target gene transcription is strictly dependent on the individual phosphorylation site. For example, mutation of serine 276 impairs Icam-1 transcription (22,49), while this study shows that prevention of serine 468 or 536 phosphorylation stimulates Icam-1 expression. While the Cxcl2 gene is not affected by serine 468 or 536 phosphorylation, a recent study revealed that expression of this gene is up-regulated by a phosphomimetic p65 threonine 435 aspartic acid mutant (50). These gene-specific effects are not confined to phosphorylation sites, but have also been reported for p65 modifications by acetylation or monomethylation (15,51). All these results are fully consistent with the NF-κB barcode hypothesis. According to this concept, post-translational modifications alone or in combination, generate distinct patterns that function to direct transcription in a target gene-specific fashion. Therefore it will be an important future task to identify the NF-κB modification patterns at different genomic NF-κB binding sites. These results may bear the seeds for a new generation of NF-κB inhibitors which do not preclude general NF-κB functions but rather interfere with distinct groups of target genes.
Inducible nuclear uptake of IKKε
In unstimulated cells, most of the endogenous IKKε protein is found in the cytosol, while a minor fraction localizes to nuclear speckles which most probably correspond to PML-NBs. This constitutively nuclear fraction of IKKε can be found at very low levels in the promoter regions of specific genes such as Saa3 or Vcam1. The relevance of the kinase activity for basal nuclear IKKε localization was seen by the analysis of the kinase inactive IKKε variant which was completely excluded from the nucleus. But the kinase function of IKKε is also important for the TNF-induced nuclear import of IKKε. A sequence inspection of IKKε did not reveal any obvious sequences with the potential to mediate nuclear import or export, suggesting that IKKε reaches the nucleus upon binding to a binding partner by a piggyback mechanism. Accordingly, TNF stimulation allowed the phosphorylation-dependent association of IKKε to p65 (see Figure 6B). The stimulus-induced interaction between IKKε and p65, and an absent interaction in unstimulated cells is also compatible with the leptomycin B experiments. These revealed nuclear accumulation of p65 but cytosolic localization of IKKε after blockage of nuclear export and in the absence of a stimulatory signal. The role of p65 for nuclear import of IKKε was suggested by a lacking nuclear translocation of the kinase in p65−/− cells, but also by the parallel kinetics of IKKε and p65 during TNF-induced nuclear import and export. Accordingly, a recent study showed the necessity of p65 for the recruitment of IKKε to the Inos promoter (31). In addition to p65, IKKε also phosphorylates further transcription factors including c-Rel, c-Jun, IRF3 and IRF7 (52–56). It will therefore be interesting to reveal a possible contribution of these proteins for the regulated nuclear uptake of IKKε. Within the nucleus, IKKε can be found in the nucleoplasm and also partially in PML-NBs. The integrity of these subnuclear structures depends on a PML domain which allows non-covalent binding to SUMO (57). Intriguingly, the vast majority of constitutive or inducible PML-NB resident proteins can be modified by SUMOylation, thus allowing the formation of protein meshworks that are glued together upon interaction of SUMO and SUMO-binding domains (58). The reversible retention of IKKε in PML-NBs may thus be explained by the recently discovered SUMOylation of this kinase (59).
A nuclear function for IKKε
This study shows that Saa3 activation is dependent on p65 phosphorylation and also sensitive to IKKε expression, thus fitting a model where IKKε-mediated p65 phosphorylation is relevant for gene activation. On the contrary, while Vcam1 similarly requires IKKε for optimal activation, loss of phosphorylation sites actually results in greater induction. This suggests that there are yet further targets of IKKε phosphorylation that are relevant at least for Vcam1 expression or more generally, for the effect of IKKε mediated activation, as schematically summarized in Figure 8D. Accordingly, recent data show that inducible chromatin recruitment of IKKε allows the c-Jun-dependent clearance of the corepressor NCoR from target promoters (31). This would also explain the occurrence of IKKε at loci which control the expression of IKKε-independent genes. The recent years have mounted evidence that some cytosolic components of the NF-κB signaling cascade (including IKKα, p38, GSK3β, COMMD1, etc.) can also be found at selected NF-κB target genes where they participate in modification of the DNA-binding subunits or chromatin (7,60). The IKKε-related IKKα protein, for example, regulates gene expression upon phosphorylation of histone H3 at serine 10 (61,62), the corepressor SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) (63) and the acetyl transferase CBP (64). Also IKKα was found to associate with chromatin at control regions for the cIAP-2, Il-8 or Maspin genes (63,65), but the general mechanisms employed by IKKs to allow chromatin association remain to be elaborated. Our Re-ChIP experiments suggest that chromatin association via binding to p65 might only be relevant for a subset of binding sites, as they occur at the Saa3 and Vcam1 promoters. Consistently and as exemplified by the Saa3 promoter, phosphorylation-dependent binding of IKKε to p65 is required for chromatin recruitment of IKKε (see also Figure 6C). A detailed understanding of the mechanisms underlying chromatin recruitment of IKKs will therefore require unbiased and systematic genome-wide approaches. This study also revealed the relevance of PML for appropriate targeting of IKKε to the nucleus. Accordingly, cells expressing the oncogenic and dominant negative PML-RARα fusion protein failed to display inducible nuclear localization of IKKε. This defect is rather attributable to delayed nuclear import and not to enhanced nuclear export, as PML-RARα-dependent blockage of TNF-triggered IKKε translocation occurred even in the presence of leptomycin B (Supplementary Figure S4). This finding might explain the recently revealed dysregulation of the IKKε target gene Vcam1 that occurs in acute promyelocytic leukemia cells (66). Aberrant gene expression of further IKKε-controlled genes would not be unexpected in acute promyelocytic leukemia cells, thus adding another example for the interplay between two oncogenes. Given the role of IKKε intracellular localization revealed here, it will also be relevant to investigate whether cancer cells overexpressing IKKε also show changes in the intracellular localisation of this breast cancer oncogene.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutscher akademischer Austauschdienst (DAAD), Deutsche Forschungsgemeinschaft projects SCHM 1417/4-2; SCHM 1417/7-1; SCHM 1417/8-1; GRK 1566/1 and the Excellence Cluster Cardio-Pulmonary System (ECCPS). C.S. and J.-M.S. were supported by a grant of the Deutsche Forschungsgemeinschaft (Schu 1412/2-1). Funding for open access charge: Justus-Liebig University Giessen.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Dr S. Herold (Giessen) for the preparation of mouse alveolar macrophages, Dr S. Akira (Osaka, Japan) for IKKε−/− MEFs and Dr T. Stamminger (Erlangen, Germany) for a vector encoding a PML specific shRNA.
REFERENCES
- 1.Gaestel M, Kotlyarov A, Kracht M. Targeting innate immunity protein kinase signalling in inflammation. Nat. Rev. Drug Discov. 2009;8:480–499. doi: 10.1038/nrd2829. [DOI] [PubMed] [Google Scholar]
- 2.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 3.Natoli G, De Santa F. Shaping alternative NF-kappaB-dependent gene expression programs: new clues to specificity. Cell Death Differ. 2006;13:693–696. doi: 10.1038/sj.cdd.4401880. [DOI] [PubMed] [Google Scholar]
- 4.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci. STKE. 2006;2006:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat. Rev. Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim CA, Yao F, Wong JJ, George J, Xu H, Chiu KP, Sung WK, Lipovich L, Vega VB, Chen J, et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-kappaB upon TLR4 activation. Mol. Cell. 2007;27:622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 10.Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc. Natl Acad. Sci. USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J. Exp. Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews JR, Botting CH, Panico M, Morris HR, Hay RT. Inhibition of NF-kappaB DNA binding by nitric oxide. Nucleic Acids Res. 1996;24:2236–2242. doi: 10.1093/nar/24.12.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 14.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 15.Ea CK, Baltimore D. Regulation of NF-kappa B activity through lysine monomethylation of p65. Proc. Natl Acad. Sci. USA. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naumann M, Scheidereit C. Activation of NF-kappa B in vivo is regulated by multiple phosphorylations. EMBO J. 1994;13:4597–4607. doi: 10.1002/j.1460-2075.1994.tb06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 18.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz ML, Mattioli I, Buss H, Kracht M. NF-kappaB: a multifaceted transcription factor regulated at several levels. Chembiochem. 2004;5:1348–1358. doi: 10.1002/cbic.200400144. [DOI] [PubMed] [Google Scholar]
- 21.Neumann M, Naumann M. Beyond IkappaBs: alternative regulation of NF-kappaB activity. FASEB J. 2007;21:2642–2654. doi: 10.1096/fj.06-7615rev. [DOI] [PubMed] [Google Scholar]
- 22.Dong J, Jimi E, Zhong H, Hayden MS, Ghosh S. Repression of gene expression by unphosphorylated NF-kappaB p65 through epigenetic mechanisms. Genes Dev. 2008;22:1159–1173. doi: 10.1101/gad.1657408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buss H, Dorrie A, Schmitz ML, Hoffmann E, Resch K, Kracht M. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-{kappa}B at serine 536 is mediated by multiple protein kinases including I{kappa}B kinase (IKK)-{alpha}, IKK{beta}, IKK{epsilon}, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 2004;279:55633–55643. doi: 10.1074/jbc.M409825200. [DOI] [PubMed] [Google Scholar]
- 24.Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, Repaka A, Mayo MW, Burstein E. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA. Genes Dev. 2009;23:849–861. doi: 10.1101/gad.1748409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. Phosphorylation of NF-kappa B p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 2009;10:381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat. Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 27.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 28.Adli M, Baldwin AS. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. J. Biol. Chem. 2006;281:26976–26984. doi: 10.1074/jbc.M603133200. [DOI] [PubMed] [Google Scholar]
- 29.Mattioli I, Sebald A, Bucher C, Charles RP, Nakano H, Doi T, Kracht M, Schmitz ML. Transient and selective NF-kappa B p65 serine 536 phosphorylation induced by T cell costimulation is mediated by I kappa B kinase beta and controls the kinetics of p65 nuclear import. J. Immunol. 2004;172:6336–6344. doi: 10.4049/jimmunol.172.10.6336. [DOI] [PubMed] [Google Scholar]
- 30.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W, Ghisletti S, Perissi V, Rosenfeld MG, Glass CK. Transcriptional integration of TLR2 and TLR4 signaling at the NCoR derepression checkpoint. Mol. Cell. 2009;35:48–57. doi: 10.1016/j.molcel.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang SH, Bazuine M, Lumeng CN, Geletka LM, Mowers J, White NM, Ma JT, Zhou J, Qi N, Westcott D, et al. The protein kinase IKKepsilon regulates energy balance in obese mice. Cell. 2009;138:961–975. doi: 10.1016/j.cell.2009.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 34.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, Mack M, Kuziel WA, Corazza N, Brunner T, et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J. Exp. Med. 2008;205:3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattioli I, Geng H, Sebald A, Hodel M, Bucher C, Kracht M, Schmitz ML. Inducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J. Biol. Chem. 2006;281:6175–6183. doi: 10.1074/jbc.M508045200. [DOI] [PubMed] [Google Scholar]
- 36.Gresko E, Ritterhoff S, Sevilla-Perez J, Roscic A, Frobius K, Kotevic I, Vichalkovski A, Hess D, Hemmings BA, Schmitz ML. PML tumor suppressor is regulated by HIPK2-mediated phosphorylation in response to DNA damage. Oncogene. 2009;28:698–708. doi: 10.1038/onc.2008.420. [DOI] [PubMed] [Google Scholar]
- 37.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 2006;80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 2009;10:381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 2009;284:14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gloire G, Dejardin E, Piette J. Extending the nuclear roles of IkappaB kinase subunits. Biochem. Pharmacol. 2006;72:1081–1089. doi: 10.1016/j.bcp.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 41.Vanden Berghe W, Ndlovu MN, Hoya-Arias R, Dijsselbloem N, Gerlo S, Haegeman G. Keeping up NF-kappaB appearances: epigenetic control of immunity or inflammation-triggered epigenetics. Biochem. Pharmacol. 2006;72:1114–1131. doi: 10.1016/j.bcp.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Harhaj EW, Sun SC. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Mol. Cell Biol. 1999;19:7088–7095. doi: 10.1128/mcb.19.10.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becskei A, Mattaj IW. Quantitative models of nuclear transport. Curr. Opin. Cell Biol. 2005;17:27–34. doi: 10.1016/j.ceb.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Wu WS, Xu ZX, Chang KS. The promyelocytic leukemia protein represses A20-mediated transcription. J. Biol. Chem. 2002;277:31734–31739. doi: 10.1074/jbc.M201648200. [DOI] [PubMed] [Google Scholar]
- 45.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature. 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 46.Seldon MP, Silva G, Pejanovic N, Larsen R, Gregoire IP, Filipe J, Anrather J, Soares MP. Heme oxygenase-1 inhibits the expression of adhesion molecules associated with endothelial cell activation via inhibition of NF-kappaB RelA phosphorylation at serine 276. J. Immunol. 2007;179:7840–7851. doi: 10.4049/jimmunol.179.11.7840. [DOI] [PubMed] [Google Scholar]
- 47.Nowak DE, Tian B, Jamaluddin M, Boldogh I, Vergara LA, Choudhary S, Brasier AR. RelA Ser276 phosphorylation is required for activation of a subset of NF-kappaB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes. Mol. Cell Biol. 2008;28:3623–3638. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prasad RC, Wang XL, Law BK, Davis B, Green G, Boone B, Sims L, Law M. Identification of genes, including the gene encoding p27Kip1, regulated by serine 276 phosphorylation of the p65 subunit of NF-kappaB. Cancer Lett. 2009;275:139–149. doi: 10.1016/j.canlet.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anrather J, Racchumi G, Iadecola C. cis-acting, element-specific transcriptional activity of differentially phosphorylated nuclear factor-kappa B. J. Biol. Chem. 2005;280:244–252. doi: 10.1074/jbc.M409344200. [DOI] [PubMed] [Google Scholar]
- 50.O'Shea JM, Perkins ND. Thr435 phosphorylation regulates RelA (p65) NF-kappaB subunit transactivation. Biochem. J. 2010;426:345–354. doi: 10.1042/BJ20091630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buerki C, Rothgiesser KM, Valovka T, Owen HR, Rehrauer H, Fey M, Lane WS, Hottiger MO. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris J, Oliere S, Sharma S, Sun Q, Lin R, Hiscott J, Grandvaux N. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J. Immunol. 2006;177:2527–2535. doi: 10.4049/jimmunol.177.4.2527. [DOI] [PubMed] [Google Scholar]
- 53.Sweeney SE, Hammaker D, Boyle DL, Firestein GS. Regulation of c-Jun phosphorylation by the I kappa B kinase-epsilon complex in fibroblast-like synoviocytes. J. Immunol. 2005;174:6424–6430. doi: 10.4049/jimmunol.174.10.6424. [DOI] [PubMed] [Google Scholar]
- 54.Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 55.Chau TL, Gioia R, Gatot JS, Patrascu F, Carpentier I, Chapelle JP, O'Neill L, Beyaert R, Piette J, Chariot A. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 2008;33:171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 57.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renner F, Moreno R, Schmitz ML. SUMOylation-dependent localization of IKKε in PML nuclear bodies is essential for protection against DNA damage-triggered cell death. Mol. Cell. 2010;37:503–515. doi: 10.1016/j.molcel.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 60.Chariot A. The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–413. doi: 10.1016/j.tcb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 61.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 63.Hoberg JE, Popko AE, Ramsey CS, Mayo MW. IkappaB kinase alpha-mediated derepression of SMRT potentiates acetylation of RelA/p65 by p300. Mol. Cell Biol. 2006;26:457–471. doi: 10.1128/MCB.26.2.457-471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang WC, Ju TK, Hung MC, Chen CC. Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NF-kappaB. Mol. Cell. 2007;26:75–87. doi: 10.1016/j.molcel.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, Gonias SL, Cheresh DA, Karin M. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–694. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- 66.Yuan W, Payton JE, Holt MS, Link DC, Watson MA, DiPersio JF, Ley TJ. Commonly dysregulated genes in murine APL cells. Blood. 2007;109:961–970. doi: 10.1182/blood-2006-07-036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.