Abstract
Diverse mitochondrial (mt) genetic systems have evolved independently of the more uniform nuclear system and often employ modified genetic codes. The organization and genetic system of dinoflagellate mt genomes are particularly unusual and remain an evolutionary enigma. We determined the sequence of full-length cytochrome c oxidase subunit 1 (cox1) mRNA of the earliest diverging dinoflagellate Perkinsus and show that this gene resides in the mt genome. Apparently, this mRNA is not translated in a single reading frame with standard codon usage. Our examination of the nucleotide sequence and three-frame translation of the mRNA suggest that the reading frame must be shifted 10 times, at every AGG and CCC codon, to yield a consensus COX1 protein. We suggest two possible mechanisms for these translational frameshifts: a ribosomal frameshift in which stalled ribosomes skip the first bases of these codons or specialized tRNAs recognizing non-triplet codons, AGGY and CCCCU. Regardless of the mechanism, active and efficient machinery would be required to tolerate the frameshifts predicted in Perkinsus mitochondria. To our knowledge, this is the first evidence of translational frameshifts in protist mitochondria and, by far, is the most extensive case in mitochondria.
INTRODUCTION
Mitochondria, the energy-producing organelles in eukaryotic cells, possess their own genomes. Mitochondrial (mt) genomes have been reduced relative to those of their bacterial ancestors by a series of evolutionary events, including massive gene transfers to the nuclear genome and gene loss (1). Most of the mt genomes sequenced to date are single, circular, double-stranded DNA molecules that typically encode dozens of genes for respiratory electron-transport chain proteins, ATP synthase proteins, ribosomal RNA (rRNA) and transfer RNA (tRNA). However, due to independent evolutionary events across eukaryotic taxa, mt genomes are very diverse with regard to physical structure, genome size and gene content. For example, mt genomes of land plants are highly expanded (up to 2.4 Mbp in muskmelon) (2), and the smallest mt genome reported is a 6-kb long linear molecule in apicomplexan parasites (3). An mt genome with unusual organization—several hundred linear DNA molecules coding one or a few genes—is found in the icthyosporean Amoebidium (4). In Euglenozoan flagellate Diplonema, one mt gene is separated into multiple fragments, each encoded on a different mini circlular molecule (5,6).
mt gene expression is distinct from that in the nucleus, and mitochondria are notable for having alternative genetic codes. One well-known code alteration is codon reassignment in which codons are not decoded as designated in the standard codon table. For example, the UGA codon in mitochondria of many eukaryotes (other than land plants) codes for tryptophan rather than a stop (7); AGR codons (R = A or G) in inchordata mitochondria code for glycine rather than arginine (8); and CUN codons (N = A, U, G or C) in yeast mitochondria code for threonine instead of leucine (9). Some codon reassignments, even those that result in the same coding change, are suggested to have evolved independently in separate taxa; one example is the reassignment of UAG codon to leucine in chlorophycean and in fungal mitochondria (7,10–12).
Dinoflagellate mt genomes are known for their remarkable organization and genetic systems. Although the overall mt genome structure is not yet determined, these are suggested to be composed of a number of heterogeneous DNA molecules that resulted from rampant homologous recombination (13,14). The entire mt genome size is estimated to be at least 30 kb but is probably much larger (15). The genome encodes a strictly limited set of genes: three protein-coding genes, cox1, cox3 and cob, and several fragmented rRNA genes. Curiously, the three protein-coding genes lack canonical start (AUG) and stop (UAA, UAG and UGA) codons in the 5′ and 3′ terminal regions, respectively (13–17). Transfer RNA genes have not been detected in any of the dinoflagellate mt genomes, and most of these dinoflagellate mt genomes comprise non-coding and pseudogene sequences (13,17,18). Recent studies on two basally-branching dinoflagellates have further highlighted the complexity of these mt genomes; the mt genomes of both Oxyrrhis marina and Amphidinium carterae are comprised of a number of DNA molecules bearing multiple copies of the three protein-coding genes with different intergenic contexts to one another (15,19). Particularly in the latter species, long intergenic sequences containing extensive inverted repeats are predicted to form many stem-loop structures (15).
Some of the unusual characteristics of dinoflagellate mt genomes are shared with those of parasitic apicomplexans, albeit with significant differences in mt genome organization (14,20). Apicomplexa is the sister lineage to dinoflagellates and is composed of a variety of protozoan parasites, including the malaria parasites Plasmodium spp. Generally, the mt genomes of apicomplexans are linear and ∼6 kb long, the smallest of the known mt genomes (3). The genomes are tightly packed and have the same three protein-coding genes as dinoflagellates, as well as fragmented rRNA genes; the protein-coding genes also lack canonical start and stop codons (21,22). Although the mt genomes of these two sister lineages, which share unusual features, have not been fully characterized for the mechanisms of gene expression, the shared gene content suggests that the drastic gene reduction in genome content occurred before the divergence of these lineages. In contrast, the significant difference in dinoflagellate and apicomplexa mt genome structures indicates that drastic mt genome reorganization events occurred after the two lineages split and independently diverged from their common ancestor (14).
To further understand the uniformity and diversity of mt genomes of dinoflagellates and apicomplexans, we are characterizing the mt genome of Perkinsus spp., which are well-known, aquatic unicellular parasites of various commercially important bivalve mollusks. In particular, the most studied species, P. marinus, parasitizes the eastern oyster, Crassostrea virginica, causing mass mortality in the host species (23). Genus Perkinsus is assumed to be the most basal of the dinoflagellate lineages discovered to date, branching just after the split between dinoflagellates and apicomplexans (24). Molecular studies on this organism are currently limited, but due to its industrial and phylogenetical significance, the genome project for P. marinus is being undertaken by scientists at the J. Craig Venter Institute (JCVI; formerly, The Institute for Genomic Research, TIGR) and scientists at the Department of Microbiology and Immunology, University of Maryland School of Medicine/Institute of Marine and Environmental Technology (IMET; formerly, at the Center of Marine Biotechnology, UMBI) (25). Although we have observed DNA in the mitochondria of P. marinus using DNA- and mitochondria-specific dyes (26), critical molecular data and annotated mt gene sequences are not been available from either the National Center for Biotechnology Information (NCBI) database or the previously available TIGR draft genome database (note that the P. marinus genomic data set is currently being curated at JCVI).
In this article, we report the first cloning and characterization of a Perkinsus mt gene. We used PCR with degenerate primers, ultracentrifugal isolation both of mitochondria and mt genome, and pulsed-field gel electrophoresis. Although these initial attempts detected neither the partial nor whole mt genome, we identified short fragments of mt gene remnants inserted into the nuclear genome of P. marinus in the previous TIGR database. We obtained the full-length mRNA sequence for cox1, which codes for mt cytochrome c oxidase subunit 1, by PCR and RACE. The primary sequence of this mRNA shared several features with orthologs from related species, and together with Southern hybridization data, the codon usage suggested that this gene resides in the P. marinus mt genome. Unexpectedly, multiple sequence alignments and a three-frame translation indicated that the translation of this mRNA employs a modified decoding system. We discussed the primary sequence features of this mRNA and further described the possibility of a unique, modified translational decoding system in Perkinsus mitochondria.
MATERIALS AND METHODS
Strains and culture conditions
The P. marinus strain CRTW-3HE was purchased from the American Type Culture Collection (ATCC, no. 50439) and maintained at 26°C in ATCC medium 1886. Discontinued products were substituted as follows: Lipid Mixture (1000×; L5146; Sigma) replaced Lipid Concentrate (100×; 21900-014; Gibco) and Instant Ocean Sea Salt (Aquarium Systems) replaced artificial seawater (S1649; Sigma). Strains of P. honshuensis and P. olseni were provided by Dr Tomoyoshi Yoshinaga (The University of Tokyo) and maintained in the same manner.
Nucleic acid preparation
Perkinsus cells were collected by centrifugation at 800g for 5 min and re-suspended in extraction buffer [100 mM Tris, 100 mM boric acid and 50 mM ethylenediamine tetraacetic acid (EDTA), pH 8.0]. Cell suspensions were treated with sodium dodecyl sulfate at 60°C for 30 min. Total DNA was purified using standard phenol–chloroform extraction and ethanol precipitation methods. Total RNA was prepared using TRIzol Reagent (Invitrogen) according to the manufacturer’s protocols, followed by the poly(A)+-RNA enrichment with PolyATract mRNA Isolation System III (Promega). Complementary DNA (cDNA) was synthesized with SMART RACE cDNA amplification kit (Clontech) following manufacturer’s instruction.
PCR, RACE, cloning and sequencing
PCR was performed using Takara Ex Taq (Takara Bio) or PfuUltra II HS DNA polymerase (Stratagene). We prepared reaction mixtures according to the manufacturers’ instructions. Amplification was performed as follows: denaturation at 94°C for 4 min followed by 35 cycles of 94°C for 30 s, a primer annealing gradient from 40 to 50°C for 30 s, and extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min. Primer set Pmcox1F1 and Pmcox1R3, based on the cox1-like sequences of nuclear DNA of mt origin (Numt) found in database ('Results' section), was used to amplify the partial sequence of the Perkinsus mt cox1 (Supplementary Figure S1). Pmcox1R3 was then used in combination with a degerenate primer cox1-3f, which was designed based on cox1 orthologs from related species (Supplementary Figure S1), to additionally sequence the upstream region of Perkinsus mt cox1. After determining the full P. marinus cox1 (Pmcox1) mRNA sequence, we designed the primer set Pmcox1fullF and Pmcox1fullR for use in PCR of the nearly full-length Pmcox1 both from genomic DNA (gDNA) and cDNA. Primer sequences are listed in Supplementary Table S1.
We performed RACE experiments using Takara Ex Taq with P. marinus cDNA as the template. Reaction mixtures were prepared according to the instructions of the cDNA synthesis kit manufacturer. Reaction conditions were 35 cycles of 94°C for 30 s, 48°C for 30 s, and 72°C for 3 min, and a final extension at 72°C for 7 min. Primers for 5′ and 3′ RACE were Pmcox1-5RACE and Pmcox1-3RACE, respectively (Supplementary Table S1).
PCR and RACE products were separated by electrophoresis on 1.2% agarose gel containing 1× Tris–Borate EDTA (TBE) buffer and target products were extracted with the MagExtractor PCR & Gel Clean up kit (Toyobo). The gel-purified products were then cloned using the TOPO TA cloning kit for Sequencing (Invitrogen). The recombinant plasmids containing PCR or RACE products were extracted from transformed E. coli (strain DH5α) using MagExtractor Plasmid (Toyobo). Both strands of cloned products were sequenced with the DYEnamic ET Terminator Cycle Sequencing kit (GE Healthcare) on an ABI310 automatic sequencer (Applied Bioscience). Sequences were determined from more than three clones, unless otherwise stated. For nearly full-length gene fragments, direct sequencing was performed on four independently obtained PCR products. Consensus sequences were determined from the alignments of multiple sequences. The assembled full-length mRNA sequence was deposited to the DNA Data Bank of Japan (accession no. AB513789).
Sequence analysis
Sequences were aligned with Clustal X 1.83 (27) and amino acid sequences were predicted using the ExPASy translate tool (http://www.expasy.org/tools/dna.html). Codon usage in several P. marinus genes was calculated using the Countcodon program (Kazusa DNA Res. Inst., http://www.kazusa.or.jp/codon/countcodon.html). Accession numbers for P. marinus nuclear genes are as follows: ispC (AB284362), sod1 (AY095212), sod2 (AY095213) and act1 (AY436364).
Southern hybridization
DNA fragments for use as probes were amplified by PCR using the following primer sets (for primer sequences, see Supplementary Table S1): Pmcox1pF and Pmcox1pR for Pmcox1, nucLSU-7f and nucLSU-7r for large subunit ribosomal DNA (LSU rDNA), PmNumt1F and PmNumt1R for cox1-like Numt1 and its flanking regions, and PmNumt2F and PmNumt2R for Numt2 and its flanking regions. The amplified fragments were cloned as described earlier. The extracted plasmids were digested with EcoRI overnight except for two Numt-plasmids, which were digested with both NotI and PstI, and the fragments were purified, labeled and hybridized to P. marinus genomic DNA with or without restriction enzyme digestion. The probes were used for detection with the AlkPhos Direct Labelling and Detection System with CDP-Star (GE Healthcare) as follows. First, 1 μg of P. marinus genomic DNA was digested with each restriction enzyme overnight at 37°C. Digested and uncut DNA was subjected to electrophoresis on a 0.3% agarose gel and transferred onto Hybond N+ nylon membrane (GE Healthcare) overnight. Purified probe (100 ng) was labeled with alkaline phosphatase and hybridized to the membrane-linked genomic DNA overnight at 42°C. The membrane was washed and incubated with the substrate CDP-Star, and the chemiluminescence signal was detected using LAS-4000 (Fujifilm).
RESULTS
Primary sequence of P. marinus cox1
Preliminary searches for mt genome fragments of P. marinus in the NCBI databases of May 2008 and the P. marinus draft genome database at TIGR using mt gene sequences of dinoflagellates and apicomplexans as queries did not produce any sequences that were supported with statistical significance (E < 0.01). The identified sequences were checked carefully by eye while referring to the amino acid alignment of COX1 from related species to identify highly conserved amino acid residues in the partial sequences, and two contigs were found to harbor cox1-like fragments, albeit these were only partial and tiny fragments (Supplementary Figure S1). Contig no. 22713 (available as part of AAXJ01000589 in Genbank/EMBL/DDBJ) contained a fragment with 75.0% AT, that showed 68% predicted-amino acid identity (17/25 residues) with O. marina COX1 (ABK57983) and was found to include functionally essential amino acid residues His276 and Glu278 [amino acid numbers according to Iwata et al. (28)]. Another fragment in contig no. 22822 (available as part of AAXJ01000147) had 70.7% AT and showed 52% predicted-amino acid identity (20/38 residues) with O. marina COX1 and conserved His325 and His326 (Supplementary Figure S1A). We realized that the base composition of these cox1-like fragments differed from those of the flanking regions (<55% AT). The flanking regions did not show sequence similarity to cox1 and were discovered to harbor nuclear genes like RNA helicase gene and clathrin-associated protein gene, the former of which contained the cox1-like fragment in one of its intronic regions (Supplementary Figure S1B). These observations imply that these cox1-like, AT-rich fragments are nuclear DNA of mt origin (Numts), which are DNA fragments that had been transferred from mt genomes into the nucleus and, in many cases, have become transcriptionally inactive.
Because the cox1-like Numts and the true mt cox1 are likely to have similar sequences, we used two primer sets for PCR: (i) Pmcox1F1 and Pmcox1R3, both of which were derived from the Numt sequences and (ii) Pmcox1R3 and a degenerate primer cox1-3f, which was designed based on cox1 sequences of closely related species. In each case, there was a distinct single DNA amplification from total P. marinus DNA template. Sequencing of these PCR products confirmed the lengths at 167 and 434 bp, respectively, with the former being completely included in the latter. To obtain the full-length sequence of this gene, we performed 5′ and 3′ RACE using internal primers Pmcox1-5RACE and Pmcox1-3RACE, respectively, with P. marinus cDNA as the template. After cloning and sequencing five clones for each of the RACE products (∼700 bp each), we amplified the nearly full-length sequence (∼1400 bp) of both gDNA and cDNA using specific primer sets (Pmcox1fullF and Pmcox1fullR) followed by direct sequencing of multiple independent PCR products. The sequences of the RACE products and the nearly full-length sequence were manually assembled to determine the full-length mRNA sequence (1434 bp) of this gene, which was confirmed to contain sequences identical to the PCR and RACE fragments obtained above. Conversely, this mRNA contained regions which are similar to, but not identical to Numt sequences, and their flanking regions were completely different from each other (Supplementary Figure S1C). There were no substitutions, insertions and deletions between sequences from gDNA and cDNA, suggesting that RNA editing does not occur in this gene. The overall AT content of this gene was 80.9%. As a whole, this gene was similar to cox1 of dinoflagellates and apicomplexans with an E < 10−70; hereafter, we refer to this sequence as Pmcox1 mRNA.
Genomic localization of Pmcox1
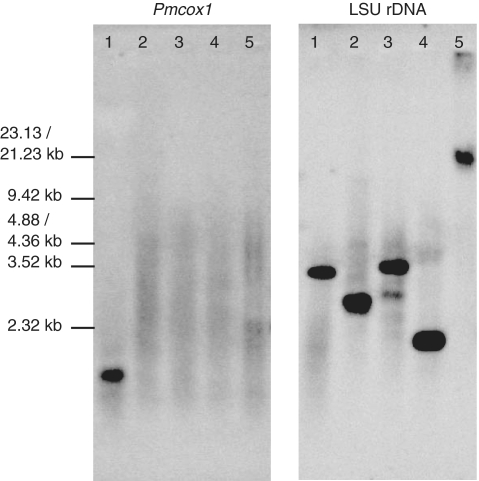
To determine the localization of Pmcox1 in P. marinus genomes (nucleus or organelles), we conducted Southern hybridization using total DNA because it was difficult to isolate pure mt DNA or intact mitochondria from P. marinus. Pmcox1 signals constituted a smear in the low molecular-weight region (<10 kb) of uncut genomic DNA, which is far lower than the expected position for chromosomal DNA (Figure 1). Similarly, Pmcox1 signals formed a smear for the digestion of total DNA with BamHI, EcoRI or HindIII. A distinct signal was only observed (1–2 kb region) for the digestion of total DNA with AccI. Given the high AT content of Pmcox1, it is natural that AccI was the only restriction enzyme tried here which cut P. marinus mt genome sequences around Pmcox1.
Figure 1.
Southern hybridization with Pmcox1 and a nuclear control probe. Southern hybridization using Pmcox1 (left) and nuclear LSU rDNA (right) probes. Lanes 1–4, P. marinus genomic DNA digested with AccI (1), BamHI (2), EcoRI (3) and HindIII (4); lane 5, uncut genomic DNA.
In sharp contrast to the Pmcox1 probe, the probe for the nuclear LSU rDNA hybridized to the stacked, high molecular-weight, chromosomal DNA in the uncut DNA sample (Figure 1). Moreover, one or two distinct LSU rDNA band(s) were detected in genomic DNA digested with AccI, BamHI, EcoRI or HindIII. The LSU rDNA signals indicate the high quality of genomic DNA and that the restriction digests were complete. The smear signals from the Pmcox1 probe suggest that Pmcox1 resides on small (<10 kb) heterogeneous non-chromosomal DNA. Like the LSU rDNA probe, probes for Numts and its flanking regions hybridized to the undigested chromosomal DNA without a smear signal, indicating that they reside on chromosomal DNA (Supplementary Figure S2).
Prediction of amino acid sequence
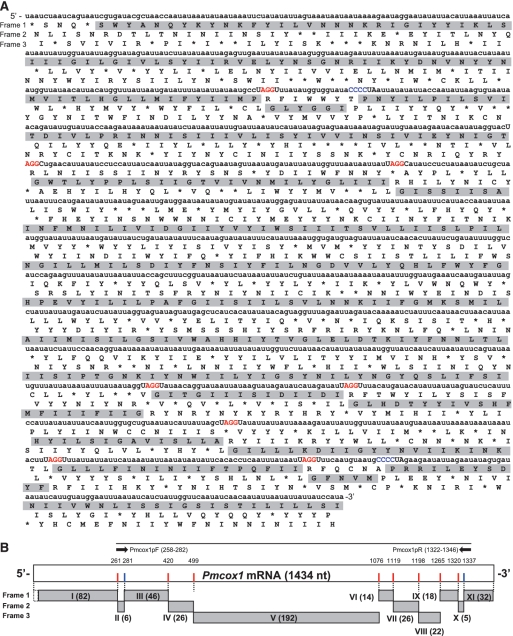
The amino acid sequence predicted to be encoded by the primary Pmcox1 mRNA sequence unexpectedly could not be translated in its entirety using the standard codon table in a single reading frame; several stop codons appeared in all three frames (Figure 2A). Performing Blastx-based search using the entire Pmcox1 mRNA sequence as a query identified several partial COX1-like amino acid sequences that appeared separately in all three reading frames (gray boxes in Figure 2A). In total, we found eleven COX1-like ‘coding-blocks’ (gray boxes numbered I–XI in Figure 2B) that cover almost the entire sequence of Pmcox1, though discontinuously.
Figure 2.
(A) Nucleotide sequence of Pmcox1 and its three frame translation. Gray boxes denote COX1-like amino acid sequences. Nucleotides in uppercase represent the sequence motifs found in transition areas of the reading frame where COX1-like amino acid sequences are deduced. AGG and CCC codons in the motifs are highlighted in red and blue, respectively. (B) Schematic model of Pmcox1 mRNA and the COX1-like blocks. COX1-like amino acid sequences distributed across three reading frames are represented by gray boxes labeled with Roman numerals. Numbers in parenthesis adjacent to the Roman numerals indicate the number of the amino acid residues. Red bars at the end of COX1-like blocks indicate UAGGY motifs containing in-frame AGG, and blue bars indicate CCCCUA motifs including in-frame CCC. Numbers above the open box indicate the positions of the first nucleotide in each motif. The uppermost bar indicates the probe region used for Southern hybridization with arrows indicating the primer locations.
To understand the discontinuity in the COX1-like amino acid sequences, we aligned the Pmcox1 mRNA sequence with cox1 sequences of related species (Supplementary Figure S3). Among the four cox1 sequences, Pmcox1 was the most divergent and contained the largest number of insertions and deletions (indels). Curiously, there were 10 one- or two-base indels specifically in the Pmcox1 mRNA that occurred in the context of UAGGY (8 of 10) or CCCCUA (2 of 10) motifs (shown on a black background in Supplementary Figure S3). Most intriguingly, these 10 regions appear to coincide with the transitions between coding-blocks, and AGG and CCC appear in-frame preceded by the predicted COX1-coding blocks (Figure 2).
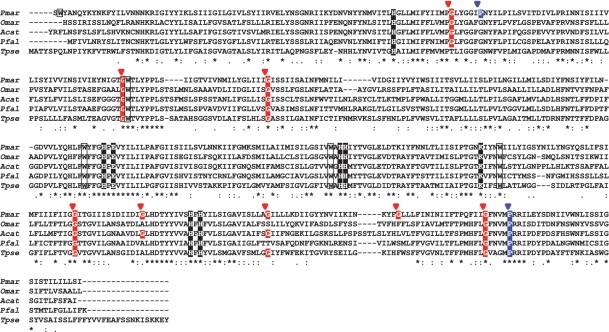
Based on these observations, we postulated that modified decoding, which could shift the reading frame, occurs in the translation of Pmcox1 mRNA. In our hypothesis, specifically, when an in-frame AGG or CCC appears, the reading frame should be shifted forward by one or two bases, respectively. Accordingly, we prepared a putative PmCOX1 amino acid sequence in the following manner. We eliminated the A residues of the UAGGY motifs and made a +1 frameshift, making GGY instead of AGG in-frame. We also deleted the first two C residues of CCCCUA motifs and made a +2 frameshift, making CCU instead of CCC in-frame. This model accounts for all the Perkinsus-specific one- and two-base indels and connects the 11 ‘blocks’ into one consecutive coding sequence. The alignment of our putative PmCOX1 sequence with counterparts from related organisms shows the conservation of functionally important amino acid residues (Figure 3, black boxes). This sequence also conserves the glycine and proline residues, which are most common in the proximity of the UAGGY and CCCCUA motifs. The potential mechanisms for these frameshifts will be further discussed later.
Figure 3.
Alignment of multiple COX1 amino acid sequences of P. marinus and related protists. The sequence for Perkinsus was predicted based on the frameshift model. Asterisks, colons and dots indicate identical residues, conserved and semi-conserved substitutions, respectively. Residues highlighted in black are conserved amino acid essential for cytochrome c oxidase function: His94 (ligand for heme a), His276 (ligand for CuB), Glu278 (D-channel), His 325, His326 (ligand for CuB), Lys354 (for K-channel), His411 (ligand for heme a3) and His413 [ligand for heme a; numbers are according to Paracoccus denitrificans homolog, (28)]. The red and blue arrowheads indicate the motifs (UAGGY and CCCCUA, respectively) where the reading frame is hypothetically shifted. Note that the glycine and proline residues at the frameshift sites are often highly conserved (highlighted in red and blue, respectively). The open boxes indicate tryptophans coded by UGA codons in Pmcox1 and those conserved at the homologous positions in related species. cox1 sequences were obtained from the NCBI database for Oxyrrhis marina (Omar, EF680822), Alexandrium catenella (Acat, AB374235), Plasmodium falciparum (Pfal, AY282930) and Thalassiosira pseudonana (Tpse, DQ186202). Note that the ciliate genes are not included because they are highly divergent and the gene length also differs greatly from those of related alveolates.
Based on this amino acid sequence, we identified the following characteristics of codon usage in Pmcox1. Around the 5′ terminal regions, no AUG codon that is likely to act as start codon was identified. Canonical stop (UAA, UAG and UGA) codons were not observed in 3′ terminal regions, as is often the case with mt genes of dinoflagellates and apicomplexans. Comparison of the COX1 amino acid alignment and nucleotide sequence also showed well-conserved tryptophan residues among related species that appeared to be coded by UGA codons in Pmcox1 (Figure 2A and open boxes in Figure 3). On the whole, Pmcox1 utilizes only 35 different codons whereas nuclear genes use 53–60 (Supplementary Table S2).
DISCUSSION
Pmcox1 is located in the mt genome
Using the newly determined sequence of Pmcox1 mRNA and nearly the full-length of its genomic counterpart, we find evidence to suggest that this gene is located in the mt genome. First, Southern hybridization of total DNA from P. marinus shows the localization of the Pmcox1 gene that is distinct from that of the nuclear LSU rDNA. Signal from a Pmcox1 probe formed a smear in the relatively low molecular-weight regions of uncut total DNA, while LSU rDNA probe hybridized to stacked, uncut DNA with high molecular weight, i.e. chromosomal DNA (Figure 1). These results indicate that Pmcox1 resides on the relatively small DNA molecules distinct from chromosomal, nuclear DNA. The present hybridization data (Figure 1) is congruent with previously reported results on other dinoflagellates (16,29,30), suggesting that Pmcox1 is encoded on multiple heterogeneous DNA molecules, which is similar to the structure found for other dinoflagellate mt genomes.
Second, canonical start and stop codons are not found in the terminal regions of Pmcox1 (Figure 2A). As the mt genes of dinoflagellates and apicomplexans do not possess AUG start codon and stop codons, these are assumed to utilize alternative start and stop mechanisms (16,17,22,29,30). All of the Perkinsus nuclear genes examined here had AUG start and stop codons in the expected positions based on comparisons to orthologs from related species. These observations support that Pmcox1 resides in the mt genome.
Lastly, overall codon usage showed significant differences between Pmcox1 and Perkinsus nuclear genes (Supplementary Table S2). Moreover, several UGA codons, which typically function as stop codons in nuclear genes but often code for tryptophan in mt genes, were present in the Pmcox1 mRNA and appeared to code for tryptophan (Figures 2A and 3). While we have no direct evidence that Pmcox1 is located in the mt genome, these multiple lines of evidence strongly support the conclusion that this gene is not located in the nuclear but in the mt genome.
mt gene translation of Perkinsus involves multiple frameshifts
Surprisingly, the Pmcox1 mRNA is apparently not translated in a single reading frame. Because we detected the cyanide-sensitive enzyme activity of cytochrome c oxidase according to the method described previously (31), functional COX1 protein most likely exists in Perkinsus mitochondria. Furthermore, we obtained partial sequences of Pmcox1 orthologs from two other Perkinsus species, P. olseni and P. honshuensis (Supplementary Figure S4). Their nucleotide sequence identity to Pmcox1 was >96%, and the UAGGY motif was conserved. There were no gaps in the alignment and all substitutions were synonymous, indicating the selective pressure to conserve the amino acid sequence in Pmcox1 and these orthologs. Taken together with there being no cox1-like sequence other than Pmcox1, these results further emphasize that Pmcox1 is functional and is translated with the aid of an unusual mechanism that requires multiple frameshifts (Figures 2 and 3).
At present, we are unable to show direct evidence that translation of Pmcox1 mRNA requires frameshifts because we have not directly sequenced the PmCOX1 protein. However, the predicted PmCOX1 amino acid sequence reinforces the validity of our frameshift model. As a major functional component of mt cytochrome c oxidase, COX1 reduces molecular oxygen to water using electrons from cytochrome c and transports protons from the mt matrix to the intermembrane space. The amino acid sequence of PmCOX1 predicted by the frameshift model retains the conserved residues that are essential for these reactions (see Figure 3 and its legend) (28). The reading frame is possibly shifted back by one base (−1 frameshift) at the CCCCUA motif, but this is less likely because it would require the insertion of one extra amino acid residue into the alignment.
Moreover, this frameshift motif may be conserved in another mt gene. We identified a cob-like fragment from P. marinus whole-genome shotgun assemblies (AAXJ01022806) in a Blast-based search using dinoflagellate mt gene sequences. This fragment included four conserved UAGGY motifs and one GAGGY motif where the reading frame appeared to be shifted forward by one base to connect discontinuous COB-like amino acid sequences to form a plausible COB protein (Supplementary Figure S5). In contrast, the deduced amino acid sequences for Perkinsus nuclear genes shown in Supplementary Table S1 did not include such translational frameshifts. These observations strongly indicate that an unconventional event occurred during translation, specifically in mitochondria of P. marinus, and also of other Perkinsus species. Our data are the first evidence of a frameshift-dependent translation system in protist mitochondria.
Possible mechanisms suggested for frameshift in Perkinsus mt genes
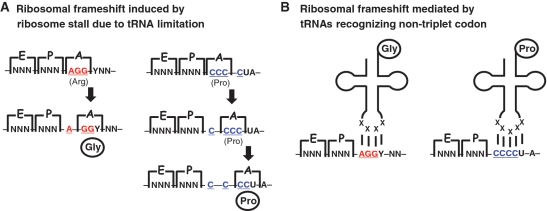
If Pmcox1 mRNA is read in all three frames to generate PmCOX1, an unconventional mechanism must exist in the Perkinsus mt translation system to shift the reading frame systematically. One possible mechanism is a ribosomal frameshift, a phenomenon observed in a wide range of organisms which results in a shift forward or backward in the reading frame during translation (32). In the case of +1 ribosomal frameshift, a rarely used codon or a stop codon in the ribosome A site is suggested to induce the ribosome to stall and allow the reading frame to be subsequently shifted forward by skipping one base (33,34). Ribosomal frameshifts have also been found in mt genes from various animals, and a +1 frameshift is suggested at specific codons (35–40). Based on previous studies, we hypothesized that ribosomes in Perkinsus mitochondria skip the A residue in the first position of the in-frame AGG in the shared UAGGY motif and the first two C residues in the CCCCUA motif by shifting forward by one base at in-frame CCC (Figure 4A). These two types of frameshifts at the rarely used AGG and CCC codons change the reading frame and allow the discontinuous COX1-like amino acid sequences to be joined, which produces the preferred amino acid residues at the frameshift sites (Figure 3).
Figure 4.
Possible mechanisms of frameshift-dependent translation at AGG and CCC codons of Perkinsus mt genes. (A) A ribosome stalled by tRNA limitation induces a ribosomal frameshift. When an in-frame AGG or CCC moves into the ribosome A site during translation, the ribosome stalls due to the limitation of the corresponding tRNA molecules, and the reading frame is subsequently shifted to the +1 frame. Translation then restarts in the new frame. In the case of CCCCUA, two consecutive frameshifts occur. (B) Specialized tRNAs recognize non-triplet codons, AGGY and CCCCU, and the reading frame shifts forward by one (AGGY) or two (CCCCU) bases.
Alternatively, specialized tRNAs that recognize non-triplet codons may be utilized at frameshift sites during translation. Naturally occurring deviant tRNAs recognize four-base codons and act as suppressors of non-sense mutations, and artificial tRNAs bearing modified loops can recognize quadruplet and even quintuplet codons (41–44). In the case of Pmcox1, specialized tRNAs may recognize AGGY (for glycine) and CCCCU (for proline) to enable the proposed frameshifts (Figure 4B). With these tRNAs, the reading frame would be shifted by one and two base(s), respectively, and one contiguous COX1 protein would be translated. Specialized tRNAs with altered decoding capacity may be used in Perkinsus mitochondria, although such mt tRNAs have not yet been identified from any organism.
Regardless of the mechanism, it should be noted that the efficiency of translational frameshift depends on the nucleotide sequence and the abundance of tRNAs, but 100% efficiency has never been observed (45). Lower frameshift efficiencies are not lethal to organisms known to have frameshift-dependent genes because there is only one (most cases) or at most two [for nuclear genes of some ciliates like Euplotes (46)] ribosomal frameshifts per gene. In contrast, frameshift must occur at as many as 10 sites to produce a complete COX1 protein in Perkinsus, which is a surprisingly high number. If one frameshift failure occurs at any of the 10 sites due to low efficiency, only a truncated COX1 protein, and not the full-length protein, will be synthesized to deleterious effect on respiratory function of Perkinsus. It is known that ‘stimulatory’ elements such as upstream Shine-Dalgarno-like sequences or downstream pseudoknot structures promote efficient frameshifts (47,48). There are, however, no such sequences associated with the frameshift in Pmcox1.
Based on these observations, we suggest that the complete translation of Pmcox1, a Perkinsus mt gene, requires a mechanism that is quite accurate for high frequency and high efficiency frameshifts. ‘Ten times per gene’ is by far the highest frequency among the reported ribosomal frameshifts. We suggest that the function of the frameshift mechanism in Perkinsus mitochondria is far more efficient and active than that of the frameshifts in other organisms. Elucidation of the amino acid sequence of Pmcox1 is still ongoing and is required to confirm the frameshift model and also to identify the start and stop codons within Pmcox1 mRNA. We will also investigate the translational machinery in Perkinsus mitochondria to understand the mechanisms that promote these ‘extensive’ frameshifts.
ACCESSION NUMBER
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Creative Scientific Research (18GS0314 to K.K.) and for JSPS Fellows (2105920 to I.M.) from the Japan Society for the Promotion of Science (JSPS). I. M. is a JSPS research fellow. Funding for open access charge: Grants-in-Aid for JSPS Fellows (2105920 to I.M.) from the Japan Society for the Promotion of Science (JSPS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Y. Watanabe (The University of Tokyo) for helpful comments and discussions about codon recognition, Dr T. Mogi (The University of Tokyo) for valuable information on COX1, and Dr R. Kamikawa (University of Tsukuba) for providing critical comments on mt genomes of dinoflagellates. We are also grateful to Dr T. Yoshinaga (The University of Tokyo) for providing the isolates of P. honshuensis and P. olseni.
REFERENCES
- 1.Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Ward BL, Anderson RS, Bendich AJ. The mitochondrial genome is large and variable in a family of plants (cucurbitaceae) Cell. 1981;25:793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]
- 3.Feagin JE. The 6-kb element of Plasmodium falciparum encodes mitochondrial cytochrome genes. Mol. Biochem. Parasitol. 1992;52:145–148. doi: 10.1016/0166-6851(92)90046-m. [DOI] [PubMed] [Google Scholar]
- 4.Burger G, Forget L, Zhu Y, Gray MW, Lang BF. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc. Natl Acad. Sci. USA. 2003;100:892–897. doi: 10.1073/pnas.0336115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marande W, Lukes J, Burger G. Unique mitochondrial genome structure in diplonemids, the sister group of kinetoplastids. Eukaryot. Cell. 2005;4:1137–1146. doi: 10.1128/EC.4.6.1137-1146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marande W, Burger G. Mitochondrial DNA as a genomic jigsaw puzzle. Science. 2007;318:415. doi: 10.1126/science.1148033. [DOI] [PubMed] [Google Scholar]
- 7.Sengupta S, Yang X, Higgs PG. The mechanisms of codon reassignments in mt genetic codes. J. Mol. Evol. 2007;64:662–688. doi: 10.1007/s00239-006-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondow A, Suzuki T, Yokobori S, Ueda T, Watanabe K. An extra tRNAGly(U*CU) found in ascidian mitochondria responsible for decoding non-universal codons AGA/AGG as glycine. Nucleic Acids Res. 1999;27:2554–2559. doi: 10.1093/nar/27.12.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osawa S, Collins D, Ohama T, Jukes TH, Watanabe K. Evolution of the mitochondrial genetic code. III. Reassignment of CUN codons from leucine to threonine during evolution of yeast mitochondria. J. Mol. Evol. 1990;30:322–328. doi: 10.1007/BF02101886. [DOI] [PubMed] [Google Scholar]
- 10.Ohama T, Inagaki Y, Bessho Y, Osawa S. Evolving genetic code. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2008;84:58–74. doi: 10.2183/pjab.84.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kück U, Jekosch K, Holzamer P. DNA sequence analysis of the complete mitochondrial genome of the green alga Scenedesmus obliquus: evidence for UAG being a leucine and UCA being a non-sense codon. Gene. 2000;253:13–18. doi: 10.1016/s0378-1119(00)00228-6. [DOI] [PubMed] [Google Scholar]
- 12.Laforest MJ, Roewer I, Lang BF. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘stop’ codons recognized as leucine. Nucleic Acids Res. 1997;25:626–632. doi: 10.1093/nar/25.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nash EA, Nisbet RE, Barbrook AC, Howe CJ. Dinoflagellates: a mitochondrial genome all at sea. Trends Genet. 2008;24:328–335. doi: 10.1016/j.tig.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Waller RF, Jackson CJ. Dinoflagellate mt genomes: stretching the rules of molecular biology. Bioessays. 2009;31:237–245. doi: 10.1002/bies.200800164. [DOI] [PubMed] [Google Scholar]
- 15.Nash EA, Barbrook AC, Edwards-Stuart RK, Bernhardt K, Howe CJ, Nisbet RE. Organization of the mitochondrial genome in the dinoflagellate Amphidinium carterae. Mol. Biol. Evol. 2007;24:1528–1536. doi: 10.1093/molbev/msm074. [DOI] [PubMed] [Google Scholar]
- 16.Jackson CJ, Norman JE, Schnare MN, Gray MW, Keeling PJ, Waller RF. Broad genomic and transcriptional analysis reveals a highly derived genome in dinoflagellate mitochondria. BMC Biol. 2007;5:41. doi: 10.1186/1741-7007-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamikawa R, Nishimura H, Sako Y. Analysis of the mitochondrial genome, transcripts, and electron transport activity in the dinoflagellate Alexandrium catenella (Gonyaulacales, Dinophyceae) Phycol. Res. 2009;57:1–11. [Google Scholar]
- 18.Norman JE, Gray MW. A complex organization of the gene encoding cytochrome oxidase subunit 1 in the mitochondrial genome of the dinoflagellate, Crypthecodinium cohnii: homologous recombination generates two different cox1 open reading frames. J. Mol. Evol. 2001;53:351–363. doi: 10.1007/s002390010225. [DOI] [PubMed] [Google Scholar]
- 19.Slamovits CH, Saldarriaga JF, Larocque A, Keeling PJ. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mt genomes. J. Mol. Biol. 2007;372:356–368. doi: 10.1016/j.jmb.2007.06.085. [DOI] [PubMed] [Google Scholar]
- 20.Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu. Rev. Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- 21.Conway DJ, Fanello C, Lloyd JM, Al-Joubori BM, Baloch AH, Somanath SD, Roper C, Oduola AM, Mulder B, Povoa MM, et al. Origin of Plasmodium falciparum malaria is traced by mitochondrial DNA. Mol. Biochem. Parasitol. 2000;111:163–171. doi: 10.1016/s0166-6851(00)00313-3. [DOI] [PubMed] [Google Scholar]
- 22.Rehkopf DH, Gillespie DE, Harrell MI, Feagin JE. Transcriptional mapping and RNA processing of the Plasmodium falciparum mitochondrial mRNAs. Mol. Biochem. Parasitol. 2000;105:91–103. doi: 10.1016/s0166-6851(99)00170-x. [DOI] [PubMed] [Google Scholar]
- 23.Villalba A, Reece KS, Ordás MC, Casas SM, Figueras A. Perkinsosis in molluscs: a review. Aquat. Living. Resour. 2004;17:411–432. [Google Scholar]
- 24.Saldarriaga JF, McEwan ML, Fast NM, Taylor FJ, Keeling PJ. Multiple protein phylogenies show that Oxyrrhis marina and Perkinsus marinus are early branches of the dinoflagellate lineage. Int. J. Syst. Evol. Microbiol. 2003;53:355–365. doi: 10.1099/ijs.0.02328-0. [DOI] [PubMed] [Google Scholar]
- 25.Joseph SJ, Fernández-Robledo JA, Gardner MJ, El-Sayed NM, Kuo C-H, Schott EJ, Wang H, Kissinger JG, Vasta GR. The alveolate Perkinsus marinus: biological insights from EST gene discovery. BMC Genom. 2010;11:228. doi: 10.1186/1471-2164-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuzaki M, Kuroiwa H, Kuroiwa T, Kita K, Nozaki H. A cryptic algal group unveiled: a plastid biosynthesis pathway in the oyster parasite Perkinsus marinus. Mol. Biol. Evol. 2008;25:1167–1179. doi: 10.1093/molbev/msn064. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 29.Chaput H, Wang Y, Morse D. Polyadenylated transcripts containing random gene fragments are expressed in dinoflagellate mitochondria. Protist. 2002;153:111–122. doi: 10.1078/1434-4610-00090. [DOI] [PubMed] [Google Scholar]
- 30.Norman JE, Gray MW. The cytochrome oxidase subunit 1 gene (cox1) from the dinoflagellate, Crypthecodinium cohnii. FEBS Lett. 1997;413:333–338. doi: 10.1016/s0014-5793(97)00938-1. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto J, Sakamoto K, Shinjyo N, Kido Y, Yamamoto N, Yagi K, Miyoshi H, Nonaka N, Katakura K, Kita K, et al. Anaerobic NADH-fumarate reductase system is predominant in the respiratory chain of Echinococcus multilocularis, providing a novel target for the chemotherapy of alveolar echinococcosis. Antimicrob. Agents Chemother. 2008;52:164–170. doi: 10.1128/AAC.00378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farabaugh PJ. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:131–170. doi: 10.1016/s0079-6603(00)64004-7. [DOI] [PubMed] [Google Scholar]
- 33.Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vimaladithan A, Farabaugh PJ. Special peptidyl-tRNA molecules can promote translational frameshifting without slippage. Mol. Cell. Biol. 1994;14:8107–8116. doi: 10.1128/mcb.14.12.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Härlid A, Janke A, Arnason U. The mtDNA sequence of the ostrich and the divergence between paleognathous and neognathous birds. Mol. Biol. Evol. 1997;14:754–761. doi: 10.1093/oxfordjournals.molbev.a025815. [DOI] [PubMed] [Google Scholar]
- 36.Mindell DP, Sorenson MD, Dimcheff DE. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol. Biol. Evol. 1998;15:1568–1571. doi: 10.1093/oxfordjournals.molbev.a025884. [DOI] [PubMed] [Google Scholar]
- 37.Beckenbach AT, Robson SK, Crozier RH. Single nucleotide +1 frameshifts in an apparently functional mitochondrial cytochrome b gene in ants of the genus Polyrhachis. J. Mol. Evol. 2005;60:141–152. doi: 10.1007/s00239-004-0178-5. [DOI] [PubMed] [Google Scholar]
- 38.Russell RD, Beckenbach AT. Recoding of translation in turtle mitochondrial genomes: programmed frameshift mutations and evidence of a modified genetic code. J. Mol. Evol. 2008;67:682–695. doi: 10.1007/s00239-008-9179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milbury CA, Gaffney PM. Complete mitochondrial sequence of the eastern oyster Crassostrea virginica. Mar. Biotechnol. 2008;7:697–712. doi: 10.1007/s10126-005-0004-0. [DOI] [PubMed] [Google Scholar]
- 40.Rosengarten RD, Sperling EA, Moreno MA, Leys SP, Dellaporta SL. The mitochondrial genome of the hexactinellid sponge Aphrocallistes vastus: evidence for programmed translational frameshifting. BMC Genomics. 2008;9:33. doi: 10.1186/1471-2164-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magliery TJ, Anderson JC, Schultz PG. Expanding the genetic code: selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli. J. Mol. Biol. 2001;307:755–769. doi: 10.1006/jmbi.2001.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atkins JF, Björk GR. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. An expanded genetic code with a functional quadruplet codon. Proc. Natl Acad. Sci. USA. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 45.Sundararajan A, Michaud WA, Qian Q, Stahl G, Farabaugh PJ. Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshift in yeast. Mol. Cell. 1999;4:1005–1015. doi: 10.1016/s1097-2765(00)80229-4. [DOI] [PubMed] [Google Scholar]
- 46.Klobutcher LA. Sequencing of random Euplotes crassus macronuclear genes supports a high frequency of +1 translational frameshifting. Eukaryot. Cell. 2005;4:2098–2105. doi: 10.1128/EC.4.12.2098-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gesteland RF, Atkins JF. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 48.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.