Abstract
IRE1α is an endoplasmic reticulum (ER)-located transmembrane RNase that plays a central role in the ER stress response. Upon ER stress, IRE1α is activated and cleaves specific exon–intron sites in the mRNA encoding the transcription factor X-box-binding protein 1 (XBP1). In addition, previous studies allow us to predict that IRE1α targets several RNAs other than the XBP1. In fact, we have identified CD59 mRNA as a cleavage target of IRE1α. However, it is not yet clear how IRE1α recognizes and cleaves target RNAs. To address this question, we devised a unique method that combines an in vitro cleavage assay with an exon microarray analysis, and performed genome-wide screening for IRE1α cleavage targets. We identified 13 novel mRNAs as candidate IRE1α cleavage targets. Moreover, an analysis of the novel cleavage sites revealed a consensus sequence (CUGCAG) which, when accompanied by a stem-loop structure, is essential for IRE1α-mediated cleavage. The sequence and structure were also conserved in the known IRE1α cleavage targets, CD59 and XBP1. These findings provide the important clue to understanding the molecular mechanisms by which IRE1α recognizes and cleaves target RNAs.
INTRODUCTION
IRE1 is a transmembrane-RNase with an endoplasmic reticulum (ER) luminal sensor domain and cytosolic kinase and ribonuclease domains (1,2). IRE1 plays a central role in the ER stress response (3). Upon ER stress, IRE1 is activated and cleaves specific mRNAs. In yeast, the activated Ire1 cleaves the Hac1 mRNA at two sites, which are then rejoined by the tRNA ligase Rlg1 (4). These reactions convert the Hac1 mRNA into its mature form and allow the efficient translation of the Hac1 protein, which functions as a transcription factor that recognizes the UPR element, located in the upstream regions of target genes (5,6). In mammals, there are two IRE1 paralogues, IRE1α and IRE1β (7–9). IRE1α is expressed ubiquitously throughout the body, while the expression of IRE1β is restricted to the epithelium of the gastrointestinal tract (10). Similar to the yeast Ire1, mammalian IRE1α is also activated upon ER stress (11,12), and cleaves specific exon–intron sites in the mRNA that encodes the transcription factor X-box-binding protein 1 (XBP1) (13,14). This cleavage initiates an unconventional splicing reaction, which leads to production of an active transcription factor and induction of various adaptive genes.
IRE1α-mediated RNA processing has been studied most extensively using XBP1. The unconventional splicing of XBP1 mRNA occurs in the cytoplasm, while most mRNA splicing occurs in the nucleus (15,16). It was recently reported that the XBP1 mRNA is tethered to the cytosolic side of the ER membrane via nascent peptides, and that this tethering is important for effective processing (17). Moreover, we found that the region corresponding to nucleotides (nts) 506–579 of the XBP1 mRNA is necessary and sufficient for XBP1-splicing, and that cleavage and ligation occur as a coupled reaction (18).
The sole output of activated yeast Ire1 appears to be the splicing of the Hac1 mRNA (19). However, several studies have suggested that IRE1 in metazoans has additional functions not mediated by XBP1 (20,21). Indeed, it was reported that IRE1α targets a specific subset of mRNAs in flies (22), and in mammals, IRE1α cleaves its own mRNA in addition to XBP1 (23). We found that the human CD59 mRNA is also cleaved by IRE1α, and attenuated in a stress-dependent manner (24). Moreover, recent studies revealed the stress-dependent decay of a subset of mRNAs in mammals (25,26), including the insulin mRNA (27). Meanwhile, IRE1β can cleave the 28S ribosomal RNA to induce translational repression (9), or the MTP mRNA to regulate chylomicron production in the intestines (28). Although post-transcriptional gene regulation mediated by the IRE1 proteins has received a lot of attention in recent years, it has not been fully elucidated how these proteins recognize and cleave their targets.
In this study, we devised a novel genome-wide screening method by combining an in vitro cleavage assay (18,24) with micro-array analysis. By this approach, we identified 13 novel targets that were cleaved in vitro by IRE1α. An analysis of the cleavage sites revealed a consensus sequence (CUGCAG) accompanied by a stem-loop structure. The sequence and structure were also conserved in the known cleavage targets, CD59 and XBP1. Our results provide the important clue toward an understanding of the molecular mechanisms by which IRE1α recognizes and cleaves its target RNAs.
MATERIALS AND METHODS
Cell culture, transfection and treatment
HeLa cells and HEK293T cells were cultured at 37°C in DMEM supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin and 10% fetal bovine serum, in an atmosphere containing 5% CO2. The calcium phosphate–DNA precipitation method was used to introduce plasmid DNA into the cells. To induce ER stress, cells were treated with tunicamycin (2.5 µg/ml) for the indicated times (Supplementary Figure S7).
Northern blot analysis
ISOGEN (Nippon Gene) was used to prepare total RNA and IRE1α-cleaved RNA fragments. RNAs were loaded on 1% denaturing agarose gels and transferred onto Hybond-N membranes (Amersham-Pharmacia). Hybridization was performed at 65°C in Church buffer. After three washes in Church buffer, signals were detected using the BAS 2500 system (Fuji Film). The following probes were used to detect XBP1, BiP and CD59: XBP1, +4 to +493; BiP, +1 to +1959; and CD59, −97 to +486 relative to the ATG. Information about the other probes used to detect cleavage candidates (Figure 2A; Supplementary Figures S2, S4 and S7) is given in Supplementary Table S2. These probes were radio labeled using the Random Primer DNA Labeling Kit version 2.0 (Takara).
Figure 2.
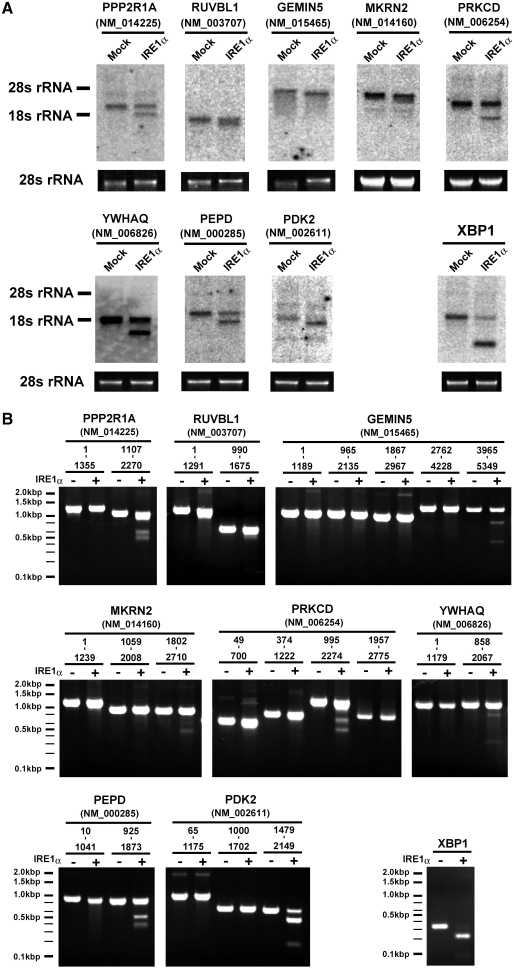
IRE1α-dependent cleavage of novel candidates. (A) Production of faster migrating species by treatment with IRE1α. Total RNAs (5 µg) were treated with IRE1α or GFP and analyzed by northern blotting with probes specific for each cleavage candidate (Supplementary Table S2). Below each northern blot is a photograph of the stained gel containing the 28S rRNA. The northern blot with the XBP1 probe (5′ exon) is included as a control. (B) In vitro cleavage of the newly identified IRE1α targets. RNA fragments derived from each candidate mRNA were incubated with or without IRE1α, resolved on a 2% denaturing agarose gel, and stained with ethidium bromide. A partial fragment of XBP1 was used as a control. The mRNA region corresponding to each fragment is indicated above the panels.
RNA cleavage assay
In vitro cleavage reactions were performed as described previously (18,24). Briefly, the cytoplasmic domain of human IRE1α was expressed in HEK293T cells and immunoprecipitated with an anti-Flag antibody. For the preparation of full-length IRE1α (Supplementary Figure S6), IRE1α-HA (12) was expressed and immunoprecipitated with an anti-HA antibody. The substrate RNA was incubated with the IRE1α preparation at 37°C, then the resulting fragments were purified using ISOGEN (Nippon Gene). The substrates were total RNA from HEK293T cells (Figures 1B and 2A; Supplementary Figures S2 and S4), or in vitro transcribed RNA fragments (Figure 2B; Supplementary Figures S3 and S5). To prepare the RNA fragments, cDNA fragments were subcloned into pBlueScript II SK (−), and transcribed using the Riboprobe in vitro Transcription System (Promega), with the T7 primer. Information about the cloned cDNAs is given in Supplementary Table S4. In the cleavage reaction with total RNAs, mock-treated reference sample was prepared using identical reaction conditions, except that IRE1α was substituted with GFP. The resulting RNA samples were resolved on 2% denaturing agarose gels for Figure 2B and Supplementary Figure S3, or 6% TBE-Urea gels (Novex TBE-Urea Gels; Invitrogen) for Supplementary Figure S5, and stained with ethidium bromide. The XBP1 RNA fragment (266–602) was used as a control (18). An RNA ladder (100–2000 bp; Invitrogen) was used as size marker.
Figure 1.
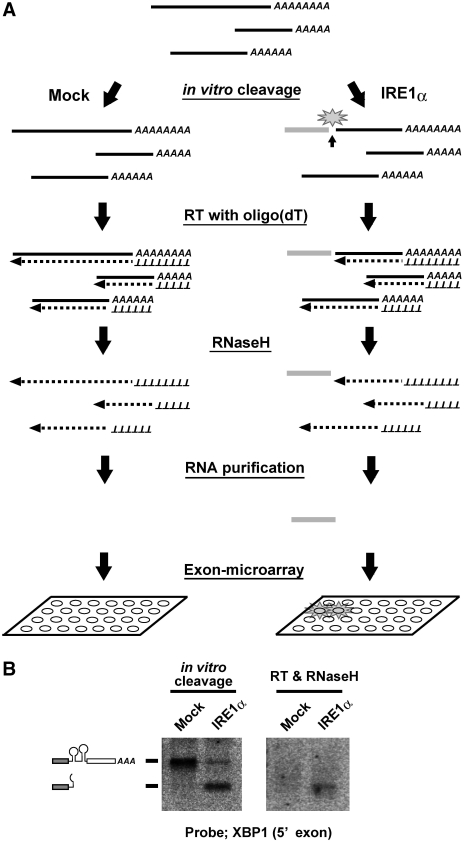
Scheme of the screening approach used to identify cleavage targets of IRE1α. (A) Schematic representation of the screening procedure. Cellular RNAs were subjected to in vitro cleavage by the cytoplasmic domain of human IRE1α, or by GFP as a mock treatment. Then the RNAs were reverse transcribed with oligo-dT primers and treated with RNase H. The resulting RNA fragments, corresponding to the 5′ sides of the cleaved RNAs (represented by the gray bar in the figure), were purified and used as probes in the exon microarray analysis. (B) Efficient cleavage and production of a probe fragment in the case of the XBP1 mRNA. Left panel: total RNA (5 µg) was incubated with IRE1α or GFP and analyzed by northern blotting with a probe specific for the 5′ fragment of the XBP1 mRNA. Right panel: the RNA samples, as described for the left panel, were reverse transcribed and treated with RNaseH before blotting.
Screening procedure
The screen illustrated in Figure 1A was performed, with details as follows. Total RNAs (5 µg) from HEK293T cells were subjected to the in vitro cleavage assay using either the cytoplasmic domain of human IRE1α or GFP. Then the RNAs were purified using ISOGEN, reverse transcribed using an oligo-dT primer and the Superscript First-Strand Synthesis System (Invitrogen), and treated with RNase H. The resulting RNA fragments were purified using ISOGEN and subjected to the Exon microarray analysis. Samples were prepared for the Exon microarrays as follows. The RNA samples were treated with RiboMinus (Invitrogen) to remove rRNA, and then reverse-transcribed with random primers to produce cDNAs. Then the cDNAs were in vitro transcribed to synthesize cRNAs. Finally, ssDNAs were synthesized from the cRNAs, fragmented and endo-labeled. The sample preparation, hybridizations to the microarrays, detection and data analysis were performed by KURABO using a GeneChip Exon Array (Human Exon 1.0 ST Array).
Primer extension analysis
Reverse transcription (RT) was performed using SuperScript II reverse Transcriptase (Invitrogen). For each reaction, cleaved or uncleaved RNAs were used as templates. The primers were [32P]-labeled with MEGALABEL (Takara). The resulting products were resolved on a sequencing gel [7 M urea, 6% acrylamide with Long Ranger Gel Solution (Takara)]. To map the products onto each mRNA (Figure 3A), the same radiolabeled primers were used to sequence the corresponding cDNAs. For CD59 and XBP1, the previously reported RNA fragments and cDNA templates were used (18,24). Sequencing was carried out using the T7 Sequencing Kit (USB) or the SequiTherm EXCEL II DNA Sequencing Kit (EPICENTRE Biotechnologies). The sequencing reactions were run in lanes adjacent to the primer extension reactions, and the resolved gels were dried for autoradiography. The following primers were used: CD59 (NM_203331), 5′-GTGACGTCGTTGAAATTGCAATGC-3′; PPP2R1A (NM_014225), 5′-GACGTCCACATCCTGGTCC-3′; RUVBL1 (NM_003707), 5′-CACACCAGGTAAGGAAGG-3′; GEMIN5 (NM_015465), 5′-ACACTTCTGATCAGATGAAG-3′; YWHAQ (NM_006826), 5′-GTAAGGGAGGGAATGTAG-3′; PEPD (NM_000285), 5′-CAATCTCTTCCACAGTGCG-3′; PDK2 (NM_002611), 5′-GATGGACTAGGCTTGTGG-3′; MKRN2 (NM_014160), 5′-GAGCTCACAACTTGAATGC-3′; PRKCD (NM_006254), 5′-GAATATGTTCTCTTTGCACATCC-3′; XBP1 (NM_005080), 5′-GAATCTGAAGAGTCAATACC-3′.
Figure 3.
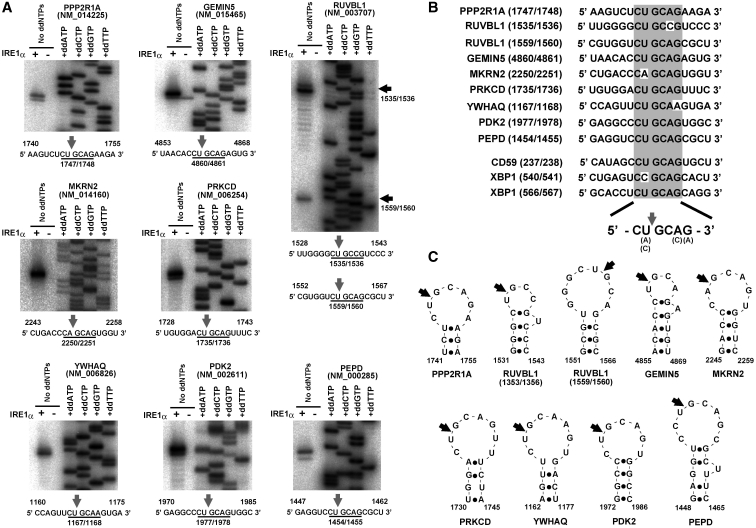
Mapping of the cleavage sites, and their secondary structure prediction. (A) Mapping of the IRE1α cleavage sites on each mRNA by primer extension analysis. RNA fragments identified as cleavage targets in the in vitro cleavage assay (Supplementary Figure S5) were used as templates for the primer extension analysis. The direct sequencing analysis of each mRNA is shown on the right side of each panel. Below the panels are partial sequences containing the cleavage sites (indicated by arrows). (B) Conservation of the consensus sequence in the cleavage targets of IRE1α. The sequences around the cleavage sites in ten cleavage targets of IRE1α are shown. The consensus sequence is indicated by the gray box. (C) Schematic representation of the IRE1α cleavage sites with secondary structures. The secondary structures around the cleavage sites were predicted using M-FOLD or CentroidFOLD. The cleavage sites identified in (A) are indicated by arrows.
Prediction of cellular localization and secondary structure of mRNAs
For Figure 3B, secondary structure was predicted using M-FOLD (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=mfold) or CentroidFOLD (http://www.ncrna.org/centroidfold/).
RESULTS
Development of the novel screening procedure
To identify mRNAs cleaved by IRE1α, we devised the molecular screen shown in Figure 1A. In brief, total RNA was isolated from cells and subjected to in vitro cleavage by IRE1α (18,24). Then the RNAs were reverse transcribed with an oligo-dT primer and treated with RNase H, a ribonuclease which specifically degrades the RNA strands of RNA–DNA hybrids. This treatment removed the uncleaved RNAs and the 3′ fragments from the cleaved RNAs, because these RNAs would form hybrids with their own cDNAs. The remaining 5′ fragments of the cleaved RNAs were purified to remove the cDNA molecules. The sample containing these 5′ RNA fragments was used as a probe in the microarray analysis. A mock-treated reference sample was prepared in the same way but with GFP instead of IRE1α.
First, we checked the quality of IRE1α used for the in vitro cleavage (Supplementary Figure S1). As the XBP1, mRNA fragment was cleaved by wild-type IRE1α, not by the mutant IRE1α (K599A) or mock-treated, it was confirmed that the cleavage proceeds from IRE1α and not from a contaminating RNase (Supplementary Figure S1).
Next, we checked whether the designed reaction could be used to detect cleaved XBP1 mRNA. Total RNA was incubated with IRE1α or GFP, and treated with or without reverse transcriptase and RNaseH, and then analyzed by northern blotting with a probe specific to the 5′ exon of XBP1 (Figure 1B). After treatment with IRE1α, the mRNA was efficiently converted to a faster migrating species, which corresponded to the cleaved 5′ fragment (Figure 1B, left panel). The subsequent RT reaction and RNase H treatment eliminated the signals from the uncleaved mRNA in both the IRE1α-treated and mock-treated samples. Only the 5′ fragment of the cleaved mRNA could be detected in the IRE1α-treated sample (Figure 1B, right panel). These results indicated that the designed reaction can effectively produce cleavage-specific fragments, and enables us to detect the cleavage of XBP1 mRNA.
We also paid attention to choosing the appropriate microarray (Figure 1A). Currently, there are several popular types of expression microarrays, including 3′, whole Gene and Exon microarrays (Affymetrix). The 3′IVT array contains probes derived from the 3′-terminus of each mRNA, and is the most widely used expression array. However, this type of array is not suitable for our study, because we need to detect the 5′ fragments of cleaved RNAs. The Gene array contains probes derived from the entire length of each mRNA, and is designed to integrate all of the data for each gene. Therefore, this array would not allow us to analyze individual signals derived from probes for a single gene. The Exon microarray is designed to analyze each exon separately, and therefore we adopted the Exon microarray for our screen.
Screening for cleavage targets of IRE1α
The samples prepared from both the IRE1α-treated and the mock-treated reactions described above were hybridized to the Exon microarrays. More than 37 000 exons gave signals that were at least 2-fold stronger in the IRE1α-treated sample compared with the mock-treated sample (data not shown). The well-known target XBP1 was found among these exons and ranked 269th based on signal intensity. Thus, this screen appeared to capture the cleavage targets of IRE1α, including the previously identified target.
To select robust candidates from the enormous number of the exons, exons fulfilling the following requirements were identified: ranking within the top 300 based on signal intensity in the IRE1α-treated sample; showing a >2.5-fold higher signal in the IRE1α-treated sample compared with the mock-treated sample; and being registered at NCBI. The resulting 56 genes were selected as IRE1α cleavage candidates (Supplementary Table S1) and were subjected to further analysis.
First, each cleavage candidate (Supplementary Table S1) was evaluated by northern-blot analysis. Total RNA samples were incubated with IRE1α or GFP and analyzed using northern blots with a specific probe for each candidate gene. We were able to detect 46 of the 56 mRNAs, and among them, 19 showed faster migrating species after treatment with IRE1α (Figure 2A; Supplementary Figure S2). Next, the cleavage of each mRNA was further analyzed with purified RNA fragments, not with total RNA, to confirm that these candidates are themselves cleaved by IRE1α. The cDNA of each candidate gene was amplified by PCR as a set of smaller, partially overlapping fragments, which were subcloned and then transcribed in vitro to produce isolated mRNA fragments. These were treated with and without IRE1α (Supplementary Figure S3). This assay revealed that 13 of the mRNAs were cleaved upon treatment with IRE1α (Figure 2B; Supplementary Figure S3). Thus, the screening method led us to identify 13 novel targets that are cleaved by IRE1α in vitro (Table 1).
Table 1.
The newly identified cleavage targets of IRE1α
| Accession number | Size (bp) | Gene | ER-targeting signal (in protein)a | Cellular localization (of protein)b | mRNA region containing the cleavage sitec | Position of cleavage sited |
|---|---|---|---|---|---|---|
| NM_014225 | 2357 | PPP2R1A | – | Cytoplasm | 1336–1919 | 1747/1748 |
| NM_003707 | 1750 | RUVBL1 | – | Cytoplasm | 1290–1675 | 1535/1536, 1559/1560 |
| NM_001008883 | 2093 | CCT3 | – | Cytoplasm | 1249–1685 | – |
| NM_015465 | 5399 | GEMIN5 | – | Nuclear or cytoplasm | 4660–5349 | 4860/4861 |
| NM_001025242 | 3499 | IRAK1 | – | Nuclear | 1617–2364 | – |
| NM_014160 | 2778 | MKRN2 | – | Nuclear | 2088–2710 | 2250/2251 |
| NM_006254 | 2850 | PRKCD | – | Cytoplasm | 1337–1989 | 1735/1736 |
| NM_016098 | 988 | BRP44L | – | Cytoplasm | 471–905 | – |
| NM_004462 | 2155 | FDFT1 | – | Cytoplasm | 632–1265 | – |
| NM_007221 | 1082 | PMF1 | – | Nuclear or cytoplasm | 405–1082 | – |
| NM_006826 | 2166 | YWHAQ | – | Nuclear | 858–1392 | 1167/1168 |
| NM_000285 | 2018 | PEPD | – | Cytoplasm | 1242–1596 | 1454/1455 |
| NM_002611 | 2319 | PDK2 | – | Cytoplasm | 1802–2149 | 1977/1978 |
aThe presence of an ER-targeting signal in the translated protein was predicted using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/).
bThe cellular localization of the translated protein was predicted using PSORT II (http://psort.ims.u-tokyo.ac.jp/form2.html).
cThe mRNA region containing the cleavage site is based on the data in Supplementry Figure S5.
dThe position of the cleavage site was evaluated based on the data in Figure 3A.
In order to verify that this screen was specific in identifying IRE1α cleavage targets, we further selected 20 genes whose exons exhibited ratios near 1.0 in the microarray analysis (Supplementary Table S3). These mRNAs were tested for cleavage by IRE1α. Northern blotting with specific probes allowed us to detect 14 of the 20 mRNAs tested. Importantly, none of the 14 mRNAs showed faster migrating species after treatment with the IRE1α (Supplementary Figure S4). This indicated that the mRNAs identified as non-candidates in the screening are actually not cleaved by IRE1α. Thus, the screening method was effective in specifically identifying cleavage targets of IRE1α.
Identification of a consensus sequence in the cleavage targets of IRE1α
The identification of novel cleavage targets and the in vitro technique allowed us to look for common characteristics in the sequences cleaved by IRE1α. Therefore, we began by mapping the novel cleavage sites as follows.
First, the newly identified RNAs (Table 1) were further fragmented as described above and subjected to in vitro cleavage (Supplementary Figure S5). This confirmed that all of the 13 mRNAs were cleaved, and allowed us to identify smaller regions of 400–700 nt that contained the cleavage sites (indicated by red characters in Supplementary Figure S5). Next, eight candidates (PPP2R1A, GEMIN5, MKRN2, PRKCD, YWHAQ, PEPD, PDK2 and RUVBL1) were selected based on the cleavage efficiency in Supplementary Figure S5, and subjected to RT-driven primer extension to map the cleavage sites (Figure 3A). While most of the mRNAs (PPP2R1A, GEMIN5, MKRN2, PRKCD, YWHAQ, PEPD and PDK2) contained one cleavage site, the RUVBL1 mRNA contained two cleavage sites located close to each other (Figure 3A). In the primer extensions, some of the mRNAs showed a 1-bp shifted band in addition to the major primer extension signal (see the IRE1α-treated lanes in Figure 3A). This was likely caused by an accidental shortening or partial degradation after the IRE1α cleavage.
Surprisingly, this mapping identified a consensus sequence that was common to all the cleavage sites. Of the nine newly identified sites, six [in mRNAs PPPR1A, RUVBL1 (1559/1560), GEMIN5, PRKCD, PEPD and PDK2] were located in the consensus sequence CUGCAG, and the remaining three sites [in RUVBL1 (1535/1536), MKRN2 and YWHAQ] were in sequences that differed by only one base from the consensus (Figure 3B). Also, all of the cleavage sites were positioned between the second and third base in the consensus (Figure 3B). Moreover, stem-loop structures were predicted to accompany the sites, and the cleavage sites were located in the loop portion (Figure 3C), which is similar to a stem-loop structure in XBP1 (13,14).
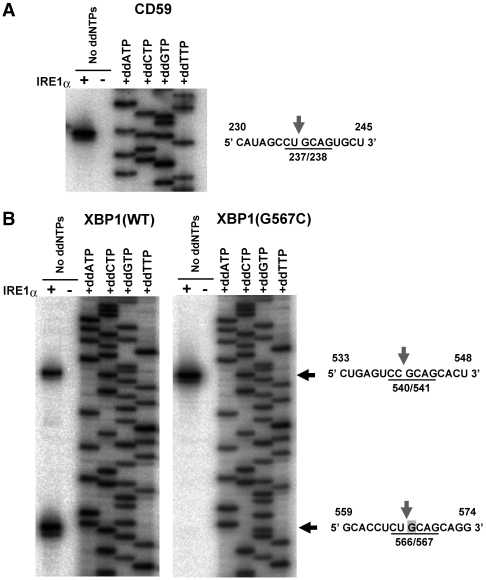
The consensus sequence was also found in the known targets of IRE1α. As shown in Figure 4A, the cleavage site on CD59 occurs in the CUGCAG consensus. Two cleavage sites were mapped in XBP1 by primer extension, one between the second and third bases in the consensus 565CUGCAG570, and one between the second and third bases in 539CCGCAG544 (Figure 4B). These mapped positions in XBP1 were shifted by 1 bp from those suggested in the previous reports (13,14). However, the sequence of the spliced XBP1 mRNA is thought to be identical in either case because both of the sites were shifted equally. The 1-bp shift of the cleavage sites would not be an artifact caused by the use of cytosolic domain of IRE1α, because the full-length IRE1α also cleaved XBP1 mRNA at the identical positions (Supplementary Figure S6). The substitution of the G at the third position of the consensus with a C (G567C) in XBP1 (13) specifically eliminated cleavage at the 566/567 point (Figure 4B). The G in the third position is strictly conserved among all the targets (Figure 3B), therefore, this residue may be one of the determinants of specific cleavage by IRE1α.
Figure 4.
Conservation of the consensus sequence in the cleavage targets of IRE1α. (A) Mapping of the IRE1α cleavage site in the CD59 mRNA. A CD54 mRNA fragment was prepared and treated with or without IRE1α as described in a previous report (24), and used as a template for primer extension analysis. The direct sequencing analysis of the mRNA is shown on the right side of the gel. A partial sequence of the CD59 mRNA, including the cleavage site (indicated by an arrow) is shown on the right. (B) Mapping of the IRE1α cleavage sites in the XBP1 mRNA. XBP1 mRNA fragments were treated with or without IRE1α as described in a previous report (18), and used as templates for primer extension analysis. The direct sequencing analysis of the XBP1 mRNA is shown on the right side of each gel. Partial sequences of the XBP1 mRNA, including the cleavage sites (indicated by arrows), are shown to the right. The substituted residue (G567C) is indicated by a gray box.
DISCUSSION
In this study, we devised a genome-wide screening method to identify cleavage targets of IRE1α (Figure 1), and identified 13 novel targets (Table 1) that are cleaved in vitro by IRE1α (Figure 2; Supplementary Figures S2, S3 and S5). Moreover, an analysis of the novel cleavage sites resulted in the identification of a consensus sequence (CUGCAG), which is accompanied by a stem-loop structure (Figure 3). The consensus sequence and structure are also conserved in the previously identified IRE1α targets, CD59 and XBP1 (Figure 4). This is the first report identifying a consensus element that is recognized and cleaved by IRE1α, based on a comparison analysis with multiple targets.
The constructed screening method was useful in the case of IRE1α. However, there is room for some improvements. One improvement would be to eliminate the possible 3′ bias in the identified cleavage sites. In this screening, most of the newly identified cleavage sites were located within 1000 bp of the mRNA 3′ termini (Figure 3; Supplementary Figure S5). It is unlikely that such a bias would be seen in cells. For example, the cleavage sites in XBP1 and CD59 are located in the middle or the 5′ ends of their mRNAs, respectively. One likely reason for this artificial bias is the limitation in the length of cDNA that can be transcribed in vitro by reverse transcriptase under normal circumstances. Any regions of the mRNA that are not reverse transcribed, such as regions far from the poly-A tail, would remain undigested by the RNase H in both the IRE1α-treated and mock-treated samples. These would not be identified as possible cleavage targets in the Exon microarray analysis. To identify a wider range of targets, this problem should be resolved in some way. Nevertheless, the screening method has important advantages. One advantage is its versatility, as the method could be adopted for use with any other site-specific RNase that has an in vitro reaction. Moreover, this method enables us to search the RNase-targets more directly than the other approaches, as previous methods including the profiling of cellular transcripts detect many byproducts from various side effects as well as the inherent targets. With these advantages, the screening method has the potential to be a useful tool in the RNA research field.
The most important point in this study is the identification of the consensus element recognized and cleaved by IRE1α. The sequential and structural characters (Figure 3), which is also found in the XBP1 and the CD59 (Figure 4), are further conserved in other IRE1α-targets, including mammalian IRE1α mRNA itself (186/187 position) (23) or insulin mRNA (mIns2 site1) (25). Thus, the CUGCAG sequence and the stem-loop structure seem to be highly conserved among mammalian IRE1α-targets.
Recently, it was suggested that IRE1α-mediated RNA processing is divided into two pathways, one is the classical pathway represented by the XBP1, and the other is RIDD (regulated IRE1-dependent decay) (26). The former pathway requires only the activation of IRE1α’s RNase activity (26), and could be reconstituted in vitro only with the activated RNase and target RNA. Because the XBP1 mRNA (18), CD59 mRNA (24), insulin mRNA (25) and the newly identified targets in this study (Figure 3) were cleaved only with IRE1α in vitro, these mRNAs would be processed by the classical pathway. Similarly, the consensus element (CUGCAG sequence accompanied with the stem-loop structure) would be applicable to this pathway. Meanwhile, in the RIDD pathway that targets BLOS1 or SCARA3 mRNA in mammal (26), any consensus sequence or structural characters have not been identified. One reason behind this is that RIDD pathway would need some other factors than IRE1 RNase to process target RNA (26), and detailed analysis including the identification of the cleavage sites have not been performed. Therefore, it is unclear that the consensus element in the classical pathway is common in the RIDD pathway.
As mentioned in the ‘Introduction’ section, post-transcriptional gene regulation mediated by the IRE1 proteins has received a lot of attention, and various targets have been identified. Most of the cleavage targets of the mammalian IRE1α, other than XBP1, are rapidly degraded upon ER stress (24–27). Thus, we wondered whether the newly identified mRNAs (Table 1) also show stress-dependent decay. In particular, since the RUVBL1 mRNA contains two cleavage sites (Figure 3A), this transcript might be processed similarly to XBP1 with a size shift. To investigate the former possibility, we performed a northern-blot analysis of total RNA from stressed cells. However, no stress-dependent decay was detected in any of the 13 newly identified mRNAs (Supplementary Figure S7). Furthermore, no size sifting was detected in RT-PCR analyses of the 13 mRNAs, even after stress conditions (data not shown). Therefore, it is highly possible that these mRNAs are not cleaved in cells, even though they are cleaved in vitro. The putative lack of cleavage in cells may be explained by cellular localization. Indeed, a previous report indicated that the localization of the target mRNA to the ER membrane is an important factor in stress-dependent decay by IRE1α (22). Moreover, it was recently reported that the mammalian XBP1 mRNA is tethered to the ER-membrane via nascent peptides, and this tethering is important for effective processing (17). None of the newly identified IRE1α targets has an ER-targeting signal or a signal peptide for secretion or membrane localization (Table 1), and none of them seem to be tethered to the ER-membrane. Thus, the different localization between IRE1α and the candidate mRNAs may mean that these mRNAs are never cleaved in cells.
However, it is still unclear whether these candidates do not have any function under stressed conditions, as any linkage of those contained genes with ER stress response have not been studied. Moreover, there are still many cellular IRE1α-targets as implied by a number of recent studies (25,26). To identify these mRNAs, we will need to refine this screening method, or use mRNAs isolated from various cells or tissues such as the placenta, where the IRE1α pathway is significantly activated (29). As an alternative, an in silico approach based on the consensus sequence (CUGCAG) might be useful. Further research would be needed to address these problems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Initiative Research Program of RIKEN; Japan Science and Technology Agency (JST); Ministry of Education, Culture, Sports, Science and Technology (MEXT; grant number 21790218); Mochida Memorial Foundation; Suzuken Memorial Foundation; Takeda Science Foundation; Naito Memorial Foundation; Japan Society for the Promotion of Science (JSPS). Funding for open access charge: JST.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Akai for technical assistance. We are grateful for the support of BSI’s Research Resources Center for DNA sequencing analysis.
REFERENCES
- 1.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 2.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 3.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 4.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 5.Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook J. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, Tsuru A, Kohno K. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 2001;3:158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 10.Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, West AB, Ron D. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J. Clin. Invest. 2001;107:585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 12.Oikawa D, Kimata Y, Kohno K, Iwawaki T. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp. Cell Res. 2009;315:2496–2504. doi: 10.1016/j.yexcr.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 14.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 15.Back SH, Lee K, Vink E, Kaufman RJ. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J. Biol. Chem. 2006;281:18691–18706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- 16.Uemura A, Oku M, Mori K, Yoshida H. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J. Cell Sci. 2009;122:2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]
- 17.Yanagitani K, Imagawa Y, Iwawaki T, Hosoda A, Saito M, Kimata Y, Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 18.Iwawaki T, Akai R. Analysis of the XBP1 splicing mechanism using endoplasmic reticulum stress-indicators. Biochem. Biophys. Res. Commun. 2006;350:709–715. doi: 10.1016/j.bbrc.2006.09.100. [DOI] [PubMed] [Google Scholar]
- 19.Niwa M, Patil CK, DeRisi J, Walter P. Genome-scale approaches for discovering novel nonconventional splicing substrates of the Ire1 nuclease. Genome Biol. 2005;6:R3. doi: 10.1186/gb-2004-6-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J. Clin. Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 23.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oikawa D, Tokuda M, Iwawaki T. Site-specific cleavage of CD59 mRNA by endoplasmic reticulum-localized ribonuclease, IRE1. Biochem. Biophys. Res. Commun. 2007;360:122–127. doi: 10.1016/j.bbrc.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipson KL, Ghosh R, Urano F. The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PLoS One. 2008;3:e1648. doi: 10.1371/journal.pone.0001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, Ron D, Tabas I, Hussain MM. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7:445–455. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc. Natl Acad. Sci. USA. 2009;106:16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.