Abstract
The mass defect of a substance can be used in mass spectral analysis to identify peaks as likely belonging to a compound class, such as peptides, if the mass defect is within the known range for that compound class. For peptides, a range of possible mass defects was calculated previously, using a set of theoretical peptides, where all possible amino acid combinations were considered (Mann, M. Abstract from the 43rd Annual Conference on Mass Spectrometry and Allied Topics; 1995, ASMS). We compare that range of theoretical peptide mass defects to new values obtained from in silico tryptic digests of proteins that are abundant in human serum and human seminal fluid. The range of mass defect values encompassing 95% of peptides for the human protein data sets was found to be up to 50% smaller than the previously reported mass defect range for the theoretical peptides. The smaller range established for human tryptic peptides can be used to improve peptide mass defect filters by excluding more species that are not likely to be peptides, thus improving filter selectivity for peptides during proteomic data analysis.
Keywords: mass spectrometry, mass defect, mass defect filter, tryptic peptide, human serum, human seminal fluid, proteomics, peptide mass fingerprinting
INTRODUCTION
Mass spectral data analysis can be a daunting task, but peak identification can be enhanced by incorporating mass defect (MD) analysis into the work flow. MD analysis is used to predict the elemental composition and the identification of organic compounds,1 metabolites,2–4 and petrochemicals,5,6 by using values that are explicit to each class of substances. The elemental composition of some peptides can be determined using MD calculations, but the utility of the method is generally limited to smaller peptides (<800 Da).7 MD analysis is also used to identify and classify multiple types of peptide modifications, such as phosphorylation8–10 or cross-linking.11 Peptide MD can also be used to deconvolute overlapping peaks,12 support charge state determination algorithms,13 and assist in high throughput protein identification, such as in peptide mass fingerprinting (PMF) techniques. PMF data analysis can be enhanced by excluding extrinsic peaks from analysis, whereas those peaks arise from substances other than the protein(s) of interest.14–15 Masses are excluded, or filtered, when the MD value is not within the window that is characteristic for that analyte. The expected window, or range, of mass defect values for peptides is established from theoretical peptide masses. This range is known to encompass only selected regions, or “clusters” on the mass scale.16–18 The values in between the clusters, referred to as the “forbidden zones”, are where unmodified peptides are not found. Any peaks located within these forbidden zones are indicative of peptide modifications or non-peptide interferents. Peptide modification can occur with functional groups that force the total mass into the forbidden zone, so that the MD is noticeably outside of the anticipated range for peptides, for example with iodine tags,19 or in cases of phosphorylation.8–10 PMF experiments benefit from mass defect data filtering, as evidenced by higher protein identification probability scores, fewer false positives, and increased number of identified peaks.15,20–22 Improvements to peptide MD filters can be made, using data sets with actual amino acid usage in place of theoretical peptides, because using these data will result in more accurate peptide MD values.18,23
Herein, we calculate mass defect values for two sets of human tryptic peptides and compare the data to theoretical peptide MD calculations. The human data sets were composed using results from in silico tryptic digestions, which were performed on human serum and seminal fluid proteins. Analysis of the human datasets defined the range of MD values that encompasses the middle 95% of unmodified peptides. The breadth of this range was compared to that of the range based on theoretical peptide calculations. Equations describing the refined MD range are presented for use in PMF studies.
EXPERIMENTAL
Peptide Generation
Proteins that are abundant in human serum24 and human seminal fluid 25 were chosen for tryptic peptide mass defect analysis. The sequences of the serum and seminal fluid protein sets were collected from the UniProt Knowledgebase, using search options or published accession numbers, respectively. Sequences containing signal peptides and propeptides were truncated so that proteins were analyzed in their relevant forms. The sequences were imported into ProteinProspector, version 5.1.8 Basic, and an in silico digestion was performed using the MS-Digest function. Trypsin was chosen for the protease, multiple charges and variable modifications to amino acids were omitted, and zero missed cleavages were allowed. Peptides with a minimum of five amino acids and within the mass range of 500–8000 Da were chosen for analysis.
Mass Defect Analysis
The mass defect was calculated for each tryptic peptide. The mass defect per unit mass (MaDPUM) was then calculated by dividing the mass defect by the monoisotopic peptide mass, and the values were sorted into 100 Da bins, based on the nominal masses of the peptides. The mean and the range of MaDPUM values that encompassed the middle 95% of peptides were established for each bin and plotted against the peptide mass. The MaDPUM calculations were performed separately for the human serum and seminal fluid data sets. A third set of MaDPUM ranges were established for theoretical peptides, based on previously published mass defect data for theoretical peptides.16,20
Filtering Proteomics Data with MaDPUM ranges
One thousand peaks from a depleted human serum LC/MS/MS experimental file26 were selected for analysis using the newly established MaDPUM ranges. The peaks were located between retention time 38.00 and 43.57 and had been selected for MS/MS analysis.26 The monoisotopic mass for each peak was collected and then classified as likely corresponding to a human tryptic peptide or not, using the MaDPUM ranges established using human serum peptides. The classification analysis was repeated using the MaDPUM ranges based on theoretical peptides.
RESULTS & DISCUSSION
Mass defect filters benefit proteomic data analysis by removing non-peptide peaks. The removal is performed prior to submitting MS data to an analysis program such as MASCOT, and is achieved by eliminating peaks whose numerical mass defect values are not within an expected window. Peaks located in the mass ranges outside of the expected window, often referred to as the "forbidden zone", are then excluded from further analysis.
MaDPUM Calculations
In these experiments, we defined the range of mass defect per unit mass (MaDPUM) values that incorporates the middle 95% of tryptic peptides from two human protein data sets. The sequences of the 50 most abundant seminal fluid proteins25 and nearly 300 human serum proteins24 were collected from the UniProt Knowledgebase. Each protein was subjected to an in silico tryptic digestion using the MS-Digest function within ProteinProspector. The outputs were used to calculate the mass defect (MD) for each peptide (equation I), which is defined as the difference between the nominal (NM) and monoisotopic masses (MI).1 Nominal mass is an integer sum consisting of the integer masses of the most abundant isotopes for each element, e.g. C = 12, H = 1, and O = 16.27 The monoisotopic mass is a sum of the exact masses of the most abundant isotope for each element in the sample of a substance, e.g. C = 12.0000, H = 1.0078, O = 15.9949.27 The MaDPUM was calculated by dividing the mass defect by the monoisotopic mass of the peptide, as in equation II.
| (I) |
| (II) |
Determination of Mean
The peptides within the human serum data set were grouped into 100 Da bins for analysis, according to the original nominal mass of the peptide. Within each 100 Da bin, the mean MaDPUM value was calculated. The global mean was then calculated and determined to be 0.00050 for the serum data set. In Figure 1a, the MaDPUM mean values are plotted for each nominal mass 100 Da bin for serum peptides. The mean MaDPUM value of 0.00050 is higher than the literature value of 0.00048,16,20 which incorporates theoretical peptide compositions. Since analysis of the human serum tryptic peptides produced results that differ from the literature values, the set of human seminal fluid peptides was analyzed as a second data set, to validate that the observations were not characteristic of human serum peptides alone. This data is shown in Figure 1b. The seminal fluid data set also showed that the global mean value was 0.00050, the same as the human serum data set. The agreement in these two data sets suggests that this mean mass defect value (0.00050) is appropriate to use for any large set of human proteins.
Figure 1.
MaDPUM mean values as calculated for each nominal peptide mass 100 Da bin. Global mean for both (a) human serum peptides and (b) human seminal fluid peptides was determined at 0.00050, indicated by the dark, horizontal line.
In Figure 1, the smallest peptides are noted to have a mean MaDPUM value greater than the average due to a mathematical bias that is based on protease specificity.28 All tryptic peptides, except possibly the C-terminal peptide, possess an R or K residue at the peptide’s C-terminus.29 The MaDPUM for both R and K is greater than the average MaDPUM for all common amino acids, thus creating a slight bias for an increased MaDPUM for the small tryptic peptides. Peptides with missed cleavages were purposely excluded from this study, as the missed cleavage peptides will be larger and thus possess a mass defect value that shows little effect from the mathematical R or K bias that is evident in small peptides.
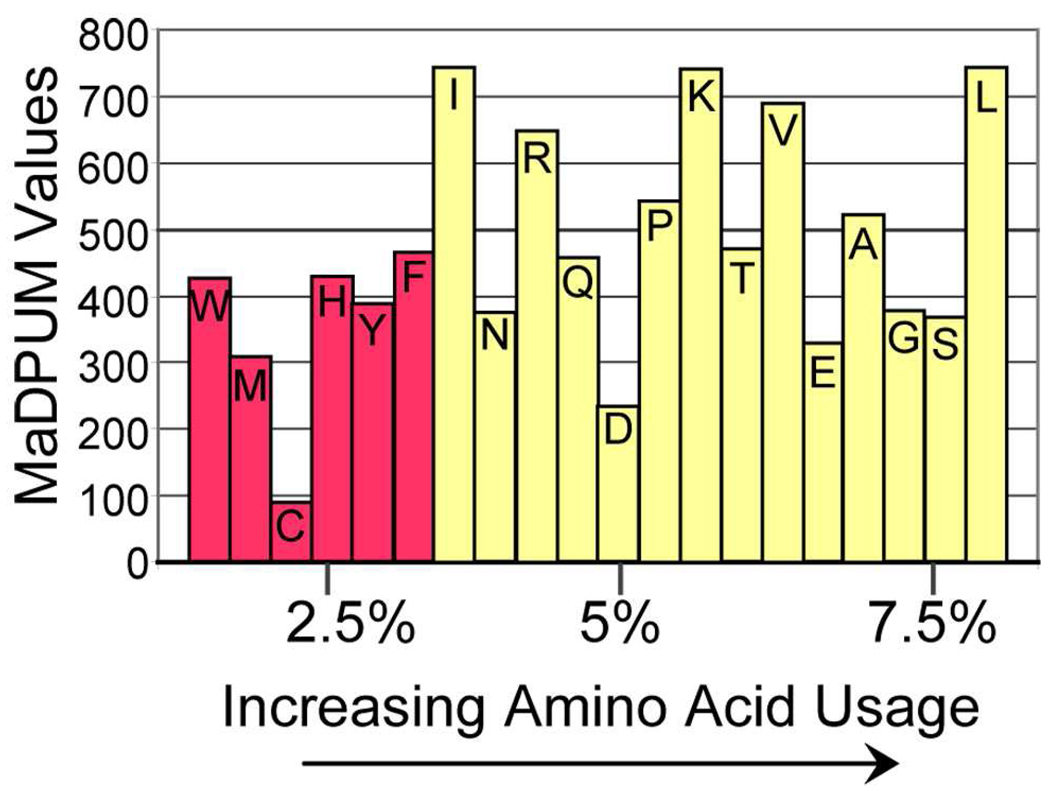
To understand why global mean values for both the serum and seminal fluid data was larger than anticipated (0.00050 instead of 0.00048), the peptide compositions, including amino acid usage, were analyzed in the seminal fluid data. The theoretical peptide data set is comprised of peptides where the 20 common amino acids are utilized equally, accounting for 5% of each peptide. In Figure 2, amino acid usage is shown, as calculated for the seminal fluid data set, ranked by increasing usage. The MaDPUM value for each amino acid is indicated by bar height. The ten amino acids to the right, in Figure 2, would be expected to comprise 50.0% of polypeptide compositions if each amino acid were used equally, but instead they account for 65.8% of amino acid usage in the seminal fluid data. The five least utilized amino acids, to the left in the figure, have MaDPUM values less than the average for amino acids. These observations demonstrate that amino acid usage will determine the MaDPUM value for each peptide, and the differential amino acid utilization also forces the global average MaDPUM value to 0.00050, higher than previously reported. These results are consistent with Wolski et al., who also demonstrated, using computational methods, that accounting for amino acid usage shifts the MaDPUM to >0.480.30 Amino acid usage clearly is an important consideration when refining mass defect filters for peptides, as usage varies across species.31
Figure 2.
Amino acid usage for the 20 common amino acids. The MaDPUM value for each amino acid is indicated by bar height in ppm. Usage is shown for the human seminal fluid data set. The 20 common amino acids are ranked by increasing usage from left to right. The 5 least utilized amino acids, shown in red, possess a MaDPUM value that is less than the average value for all amino acids. Amino acid usage in human seminal fluid proteins determines the mean MaDPUM value of 500 ppm (or 0.00050)
MaDPUM Width for Theoretical Peptides
The theoretical peptide masses and corresponding MaDPUM values were calculated using relationships based on nominal peptide mass, as described previously.16,20
| (III) |
| (IV) |
The MI (monoisotopic) masses for theoretical peptides were generated using equation III, and the width (W) of the range that encompassed 95% of peptides was determined using equation IV.16,20 We used these equations to calculate MaDPUM for the theoretical peptides, for each 100 Da bin. Results were plotted against nominal peptide mass (Figure 3a). These data are used to assess how similar the width is for the experimental MaDPUM data.
Figure 3.
Figure 3a. MaDPUM values in ppm vs. nominal peptide mass in Da. The red area represents the values that are excluded from the human serum tryptic peptide dataset, but included in theoretical peptide calculations. The area in yellow represents the human serum tryptic peptide MaDPUM values. The differences in MaDPUM widths for the nominal masses shown were found to be between 23–50% for the analyzed mass range.
Figure 3b. Mass defect vs. nominal peptide mass in Da. The area representing mass defect values that encompass 95% of human serum tryptic peptides is yellow, which is a significantly more narrow range of mass defect values than those corresponding to the theoretical peptide data set, shaded in red. The upper and lower boundaries incorporating 95% of the serum peptides was calculated for each 100 Da bin and data points are indicated by blue symbols, ▲ upper boundary and ■ lower boundary. Best fit lines through the points were added to the data, and the equations can be found in equations V and VI for the upper and lower boundaries of the serum peptides.
MaDPUM Width (range) for Human Biofluid Peptides
Extensive studies of the width, or the range, of mass defect values for unmodified peptides were conducted on both the serum and seminal fluid proteins. These results indicated that no differences existed between the two human data sets for the range of MaDPUM values encompassing the middle 95% of peptides. The width (range) determination studies below are presented exclusively using the serum peptides, as that set has more data points
After calculating the mass defect for each serum peptide in each 100 Da bin, we measured the mass defect width for the biofluid data, which can be defined as a range of MaDPUM values that incorporated 95% of all the human serum peptides, for each 100 Da bin. This width was calculated at each 100 Da bin by sorting the MaDPUM values in ascending order within the bin, and by manually observing the middle 95% of values. The yellow area in the center of Figure 3a represents the experimental width. The red areas in Figure 3a represent MaDPUM values that are excluded from the serum data’s width, but are included in the theoretical width, according to equation IV.
In Figure 3b, the data is plotted differently. Here, MD values are plotted vs. nominal peptide mass divided into 100 Da bins. The MD widths are shown for the theoretical peptides (in red) and the human serum peptides (in the center shown in yellow). The theoretical peptide width was established using equation IV. The width for the serum peptides was defined using the experimental data to determine data points for the upper and lower boundaries containing 95% of peptides within each 100 Da bin. In figure 3b, symbols indicate the individual data points for each bin, and linear trend lines to fit the data points are shown. Equations that define the upper and lower boundary best fit lines are found in equations V and VI below, respectively.
| (V) |
| (VI) |
The R2 values for the best fit lines are 0.99921 or greater.
Width Analysis
The differences in width between the human serum data set and the theoretical data were analyzed to determine how significantly this data will affect the size of MD mass analysis filters. The MaDPUM width established for theoretical peptide compositions are significantly wider than the width in this study on human serum and seminal fluid tryptic peptides. For peptides with a mass of 1000 Da, the width of MaDPUM values that would include the middle 95% of serum peptides is 42.3% smaller than the width established using theoretical peptides. See Table 1. At a mass of 3000 Da, the difference in width is smaller, 25.6%. These differences in the calculated size of the “forbidden zone” for mass defect analysis are largest where it makes the most impact, on the lower end of the mass scale; a majority of tryptic peptides are less than 3000 Da in mass.32 The differences in width indicate that a significant number of additional peaks may be excluded from analysis, without losing data from unmodified peptides.
Table 1.
Observed MaDPUM ranges for peptide data sets. Where ws = width of the observed MaDPUM range for human serum tryptic peptides, wt = width of the observed MaDPUM range for theoretical peptides. The MaDPUM values are presented in ppm.
| Peptide Mass (Da) | MaDPUMws | MaDPUMwt | Size Difference |
|---|---|---|---|
| 1000 | 162 | 281 | 42.3% |
| 2000 | 132 | 193 | 31.6% |
| 3000 | 114 | 162 | 25.6% |
Filtering Proteomics Data with MaDPUM Equations
One thousand peaks from a human serum LC/MS/MS file26 were collected and then filtered using the MaDPUM ranges established from human serum tryptic peptides. Of the 1,000 masses selected for filtering, 268, or 26.8%, were excluded (or filtered) because the mass defect values did not correspond to that of an unmodified human tryptic peptide. When the theoretical peptide range equations were used to filter the data, only 99, or 9.9%, of the peaks were excluded. In this experiment, using the MaDPUM ranges based on human serum peptides, described herein, produced a three-fold increase in the number of non-peptide peaks that can be excluded from further data analysis.
CONCLUSIONS
To refine peptide mass defect filters, the mass defect and mass defect per unit mass (MaDPUM) values were analyzed for two sets of human proteins and compared to data from the literature based on theoretical peptide compositions. The global human MaDPUM mean value was found to be 0.00050, larger than previously reported, and analysis of amino acid usage indicates that organism usage should be considered when refining mass defect filters for peptide analysis. The width of MaDPUM values encompassing 95% of peptides within the data sets analyzed were found to be up to 50% smaller than the width previously established using the theoretical peptide data. The selectivity of peptide mass defect filters can be improved by using the refined equations presented here, describing the upper and lower boundaries of the mass defect values for unmodified peptides. Use of these equations increase the number of excluded extraneous (non-peptide or modified peptide) peaks.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the National Institutes of Health, grant number R01RR026061 for financial support.
Footnotes
SUPPORTING INFORMATION AVAILABLE: MaDPUM range equations that describe the middle 90%, 97%, and 99% of human serum tryptic peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Kendrick E. Anal. Chem. 1963;35:2146–2154. [Google Scholar]
- 2.Mortishire-Smith RJ, Castro-Perez JM, Yu K, Shockcor JP, Goshawk J, Hartshorn MJ, Hill A. Rapid Commun. Mass Spectrom. 2009;23:939–948. doi: 10.1002/rcm.3951. [DOI] [PubMed] [Google Scholar]
- 3.Zhu M, Ma L, Zhang D, Ray K, Zhao W, Humphreys WG, Skiles G, Sanders M, Zhang H. Drug Metab. Dispos. 2006;34:1722–1733. doi: 10.1124/dmd.106.009241. [DOI] [PubMed] [Google Scholar]
- 4.Rousu T, Pelkonen O, Tolonen A. Rapid Commun. Mass Spectrom. 2009;23:843–855. doi: 10.1002/rcm.3953. [DOI] [PubMed] [Google Scholar]
- 5.Fu J, Kim S, Rodgers RP, Hendrickson CL, Marshall AG. Energy & Fuels. 2006;20:661–667. [Google Scholar]
- 6.Hughey CA, Hendrickson CL, Rodgers RP, Marshall AG. Anal. Chem. 2001;73:4676–4681. doi: 10.1021/ac010560w. [DOI] [PubMed] [Google Scholar]
- 7.Zubarev RA, Hakansson P, Sundqvist B. Anal. Chem. 1996;68:4060–4063. [Google Scholar]
- 8.Shi Y, Bajrami B, Morton M, Yao X. Anal. Chem. 2008;80:7614–7623. doi: 10.1021/ac801355u. [DOI] [PubMed] [Google Scholar]
- 9.Bruce C, Shifman MA, Miller P, Gulcicek EE. Anal. Chem. 2006;78:4374–4382. doi: 10.1021/ac060046w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Artemenko KA, Zubarev AR, Samgina TY, Lebedev AT, Savitski MM, Zubarev RA. Anal. Chem. 2009;81:3738–3745. doi: 10.1021/ac802532j. [DOI] [PubMed] [Google Scholar]
- 11.Pourshahian S, Limbach PA. J. Mass Spectrom. 2008;43:1081–1088. doi: 10.1002/jms.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renard BY, Kirchner M, Steen H, Steen JAJ, Hamprecht FA. BMC Bioinformatics. 2008;9:355. doi: 10.1186/1471-2105-9-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Marshall AG. J. Am. Soc. Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 14.Levander F, Rognvaldsson T, Samuelsson J, James P. Proteomics. 2004;4:2594–2601. doi: 10.1002/pmic.200300804. [DOI] [PubMed] [Google Scholar]
- 15.Ding Q, Xiao L, Xiong S, Jia Y, Que H, Guo Y, Liu S. Proteomics. 2003;3:1313–1317. doi: 10.1002/pmic.200300452. [DOI] [PubMed] [Google Scholar]
- 16.Mann M. Useful Tables of Possible and Probable Peptide Masses. Abstract from the 43rd Annual Conference on Mass Spectrometry and Allied Topics; Atlanta, GA. (American Society for Mass Spectrometry); 1995. [Google Scholar]
- 17.Gay S, Binz P-A, Hochstrasser DF, Appel RD. Electrophoresis. 1999;20:3527–3534. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3527::AID-ELPS3527>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Zubarev RA, Bonddarenko PV. Rapid Commun. Mass Spectrom. 1991;5:276–277. [Google Scholar]
- 19.Yao X, Diego P, Ramos AA, Shi Y. Anal. Chem. 2008;80:7383–7391. doi: 10.1021/ac801096e. [DOI] [PubMed] [Google Scholar]
- 20.Dodds ED, An HJ, Hagerman PJ, Lebrilla CB. J. Proteome Res. 2006;5:1195–1203. doi: 10.1021/pr050486o. [DOI] [PubMed] [Google Scholar]
- 21.Mann M, Kelleher NL. PNAS. 2008;105:18132–18138. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding J, Shi J, Poirier GG, Wu F-X. Proteome Science. 2009;7:9. doi: 10.1186/1477-5956-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senko MW, Beu SC, McLafferty FW. J. Am. Soc. Mass Spectrom. 1995;6:229–233. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 24.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. Mol. Cell. Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Fung KYC, Glode LM, Green S, Duncan MW. Prostate. 2004;61:171–181. doi: 10.1002/pros.20089. [DOI] [PubMed] [Google Scholar]
- 26.Ma Z-Q, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, Tabb DL. J. Proteome Res. 2009;8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siuzdak G. The Expanding Role of Mass Spectrometry in Biotechnology. 2nd Ed. San Diego, CA: MCC Press; 2006. p. 240. [Google Scholar]
- 28.Barsnes H, Eidhammer I, Cruciani V, Mikalsen S-O. Eur. J. Mass Spectrom. 2008;14:311–317. doi: 10.1255/ejms.934. [DOI] [PubMed] [Google Scholar]
- 29.Olsen JV, Ong S-E, Mann M. Mol. Cell. Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Wolski WE, Farrow M, Emde A-K, Lehrach H, Lalowski M, Reinert K. Proteome Science. 2006;4 doi: 10.1186/1477-5956-4-18. [from: http://www.proteomesci.com/content/4/1/18.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlin S, Blaisdell BE, Bucher P. Protein Engineering. 1992;5:729–738. doi: 10.1093/protein/5.8.729. [DOI] [PubMed] [Google Scholar]
- 32.Huang P, Wall DB, Parus S, Lubman DM. J Am. Soc. Mass Spectrom. 2000;11:127–135. doi: 10.1016/S1044-0305(99)00128-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.