Abstract
Within healthy human somatic cells, retrotransposition by long interspersed nuclear element-1 (also known as LINE-1 or L1) is thought to be held in check by a variety of mechanisms, including DNA methylation and RNAi. The expression of L1-ORF1 protein, which is rarely found in normal tissue, was assayed using antibodies with a variety of clinical cancer specimens and cancer cell lines. L1-ORF1p expression was detected in nearly all breast tumors that the authors examined, and the protein was also present in a high percentage of ileal carcinoids, bladder, and pancreatic neuroendocrine tumors, as well as in a smaller percentage of prostate and colorectal tumors. Tumors generally demonstrated cytoplasmic L1-ORF1p; however, in several breast cancers, L1-ORF1p was nuclear. Patients with breast tumors displaying nuclear L1-ORF1p had a greater incidence of both local recurrence and distal metastases and also showed poorer overall survival when compared with patients with tumors displaying cytoplasmic L1-ORF1p. These data suggest that expression of L1-ORF1p is widespread in many cancers and that redistribution from cytoplasm to nucleus could be a poor prognostic indicator during breast cancer. High expression and nuclear localization of L1-ORF1p may result in a higher rate of L1 retrotransposition, which could increase genomic instability.
Keywords: line expression, line movement, breast cancers
Introduction
Many organisms harbor retrotransposons, which are genetic elements capable of reverse transcribing their own mRNAs and inserting DNA copies of themselves into new places within the genome. In humans, the predominant retrotransposon is known as L1 or LINE-1. Numerous retrotransposition events by LINEs have turned them into highly repetitive elements. LINEs now constitute 21% of the human genome.1 An active L1 element is composed of about 6,000 base pairs of DNA with an internal promoter element and 1 open reading frames termed ORF-1 and ORF-2.2 A bicistronic RNA transcript is made from an inserted copy of LINE-1, which is then translated into ORF-1 and ORF-2 proteins. L1-ORF1p forms a homotrimer3 that binds to the L1 RNA to create a ribonucleoprotein complex. L1-ORF1p has nucleic acid chaperone activity and is required for the retrotransposition reaction.4,5 The ORF-2 protein, which has reverse transcriptase and endonuclease activity, cuts human genomic DNA to provide a 3′ end primer from which the L1 mRNA is copied into DNA, and an insertion then is made at this site.2 In many cases, an attenuated L1 element is produced due to incomplete reverse transcription or to inversions that often occur during reverse transcription.6 Over time, viable L1 copies also accumulate mutations and become defective. In the human genome, there are more than 500,000 copies of defective or inactive elements.1,7 Approximately 100 copies of L1 remain functional and have retained the ability to move about the genome.8
As part of the retrotransposition reaction, the L1 ribonucleoprotein complex needs access to genomic DNA, but precisely how this occurs is not clear. L1-ORF2p contains a nuclear localization sequence, but expression of this protein is very low, and the presence of L1-ORF2p has not been detected within the L1 ribonucleoprotein complex. L1-ORF1p, which packages the L1 mRNA, does not contain an obvious nuclear localization sequence. In most cases, researchers have detected only cytoplasmic L1-ORF1p.9-13 Indeed, it has been suggested that the ability to prevent proteins such as L1-ORF1p from entering the nucleus is an important cellular defense against genomic changes caused by retrotransposons14 and that the L1-ORF1p ribonucleoprotein complex may gain access only upon nuclear envelope disintegration during mitosis, at which time the chromatin is protected from retrotransposition by its highly condensed state. Arguing against this, retrotransposition of L1s has been shown to occur in nonmitotic cells.15 A small percentage of epitope-tagged L1-ORF1p has been shown to localize to the nucleolus of a cell line upon overexpression from a highly active viral promoter,16 but the physiological significance of that finding has not been clear given that the use of both epitope tags and strong, heterologous promoters has been known to alter targeting of proteins.
In humans, retrotransposition of L1s is thought to occur primarily in germline tissues. Of the previously characterized mutations caused by L1 retrotransposition, nearly all occurred in the germline.17 Although expression of L1-ORF1p has been detected within somatic cells of the reproductive tract of both mouse18 and human,11 for the most part L1 genes appear to be transcriptionally silent in somatic tissue. Somatic inactivation of L1 transcription is thought to be largely due to the hypermethylation of cytosine residues in the DNA of these elements in somatic tissues.19-22 RNAi also plays a role in downregulating L1 expression.23
During tumorigenesis, cells can undergo both hyper- and hypomethylation of cytosine residues in different regions of genomic DNA, which can result in epigenetic changes that result in an altered gene expression pattern at the level of transcription.24 Thus, L1 expression may be derepressed by changes in DNA methylation during tumor formation. Indeed, the L1-ORF1 protein has previously been detected in 12 out of 12 breast tumors as well as in a smaller percentage of tumors derived from the germline.10,12 Given the potential consequences of genomic instability that accompany active retrotransposition, we set out to characterize L1-ORF1p expression in a more diverse set of human tumors and to determine whether that expression might be of clinical importance. Here we show that expression is quite common in many tumor types, including breast cancers, ileal carcinoids, pancreatic neuroendocrine tumors, and prostate, bladder, and colorectal cancers. Interestingly, L1-ORF1p can be found in the nucleus of certain tumors, particularly in breast cancers, and nuclear localization of this protein is associated with a poor prognostic outcome in patients with breast cancers.
Results
L1-ORF1p Western blot analysis on human cell lines
Polyclonal antisera were prepared against bacterially expressed, full-length human L1-ORF1 protein as described in the Materials and Methods section. Antisera were affinity purified and then tested to determine if they interact specifically with the L1-encoded protein. The representative Western blots are shown in Figure 1.
Figure 1.
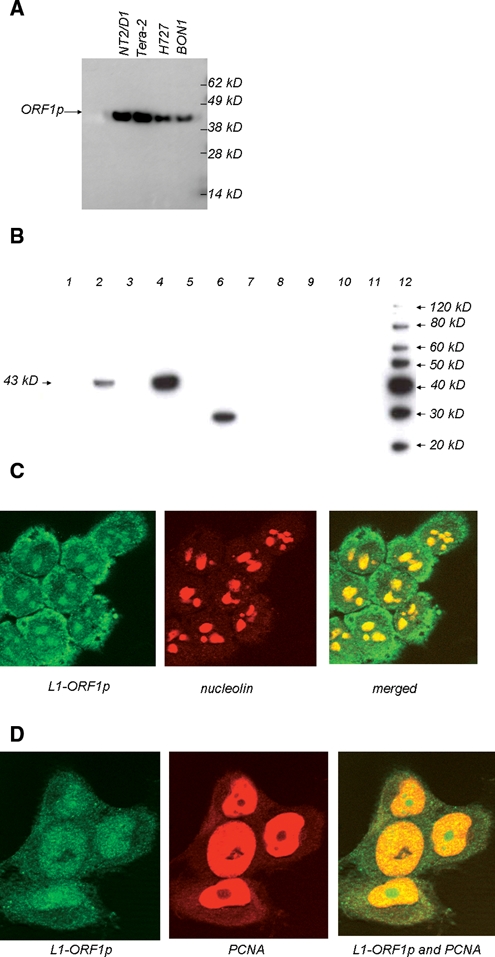
Western blot and immunofluorescence experiments using anti-L1-ORF1p sera. (A) and (B) Protein extracts were prepared from human cell lines or from normal or cancerous human tissue prior to separation by SDS-PAGE and Western blotting. In (A), extracts were from cell lines Tera-2, NTera2/D1,NCI-H727, and BON1. In (B), lanes 1 and 2 are extracts from normal adjacent tissue and breast tumor from patient “A”; lanes 3 and 4 are from normal adjacent tissue and breast tumor from patient “B”; lanes 5-10 are from normal uterine, spleen, ileal, lung, colon, and pancreatic tissue, respectively; lane 11 contains no protein extract; and lane 12 contains molecular weight markers. (C) and (D) Immunofluorescence experiments using lung cell line H460 (C) or colon cell line H1299 (D). In (C), nucleolin is used to image nucleoli, and L1-ORF1p appears to locate in the cytoplasm and nucleoli of the H460 cell line. In (D), PCNA antisera were used as these can stain nuclei but not nucleoli in HCT116 (data not shown), and the L1-ORF1 protein appears to localize in both nuclei and nucleoli but not in the cytoplasm of this cell line.
A total protein extract from the cell line N-Tera2D1, which derives from a tumor of germ cells, was analyzed because this cell line has previously been reported to be a strong expresser of L1-ORF1p.25 As shown in Figure 1A, N-Tera2D1 expresses a 43-kD protein that reacts with the L1-ORF1p antisera. This size is within the range of 38 to 46 kD that has been reported for L1-ORF1p by other researchers using a variety of gel systems.10,13,25 Long-term exposure of this Western blot revealed no other cross-reacting protein bands (data not shown). We also observed a protein of the same size within a related cell line, Tera-2, as well as the pancreatic neuroendocrine cell lines QGP-1 and BON-1 (Fig. 1A). Indeed, we detected L1-ORF1p in a large percentage of the cell lines that we tested, with exceptions being the leukemic cell line MOLT-4 and the pituitary cell line HP75 (data not shown). Figure 1B demonstrates that these antisera also recognize a single, 43-kD species within protein extracts from certain breast tumors. Previous studies have shown that breast tumors express L1-ORF1p.10,26 As negative controls, L1-ORF1p expression was tested using normal somatic tissue. We could find no evidence of L1-ORF1p in extracts of normal breast, colon, liver, lung, ileum, spleen, uterus, pituitary, or pancreatic islets (Fig. 1B). Together, these data indicate that the antisera can recognize L1-ORF1p with high specificity in cell lines and tumors.
Expression and localization of L1-ORF1p within human cell lines
Although L1-ORF1p has previously been reported to be a cytoplasmic protein, we were particularly interested in whether there might be expression within the nucleus of certain cell lines. A nuclear localization of the L1-ORF1p/L1-mRNA ribonucleoprotein particle should be important for the transposition of L1s into the human genome. We examined localization of L1-ORF1p using immunofluorescence. Depending on the cell line, L1-ORF1p gave two distinct patterns of localization: cytoplasmic/nucleolar and nuclear/nucleolar. An example of a cell line with cytoplasmic/nucleolar L1-ORF1p is shown in Figure 1C. In this experiment, the large-cell lung cancer cell line H460 was treated with the anti–L1-ORF1p as well as with antisera raised against human nucleolin. Although there is clearly L1-ORF1p labeling within the cytoplasm of H460, there is also labeling within regions of the nucleus. Within these nuclear regions, L1-ORF1p and nucleolin signals overlap, indicating that L1-ORF1p is a nucleolar protein in this cell line. These data are consistent with the previous finding that virally encoded, epitope-tagged L1-ORF1p can be found in the cytoplasm and nucleolus18 and demonstrate that nucleolar localization can occur when the protein is produced from its endogenous promoter. L1-ORF1p shows a very different localization in the colon cell line HCT116 (Fig. 1D). In this experiment, antisera raised against both L1-ORF1p and also proliferating cell nuclear antigen (PCNA) were added. Under the conditions employed, the PCNA antisera labeled nuclei but not nucleoli of these cell lines (data not shown). As shown in the figures, PCNA (orange) and L1-ORF1p (green) labeling overlap, appearing yellow in much of the nucleus, but in some nuclear regions, only an L1-ORF1p signal was observed. These data demonstrate that L1-ORF1p can be found in nuclei as well as in nucleoli in this cell line.
Localization of L1-ORF1p within human tissue sections and cancers
The L1-ORF1p antisera were also used for immunohistochemistry on paraffin-embedded human tissue. As a first test, we stained several normal (noncancerous) human tissue sections, such as the prostate, colon, kidney, and spleen sections shown in Figure 2A. None of the normal sections stained with L1-ORF1p antisera except for the occasional background label whose pattern differs considerably from a positive result (see below). These antisera therefore appeared suitable for immunohistochemistry in a number of tissues.
Figure 2.
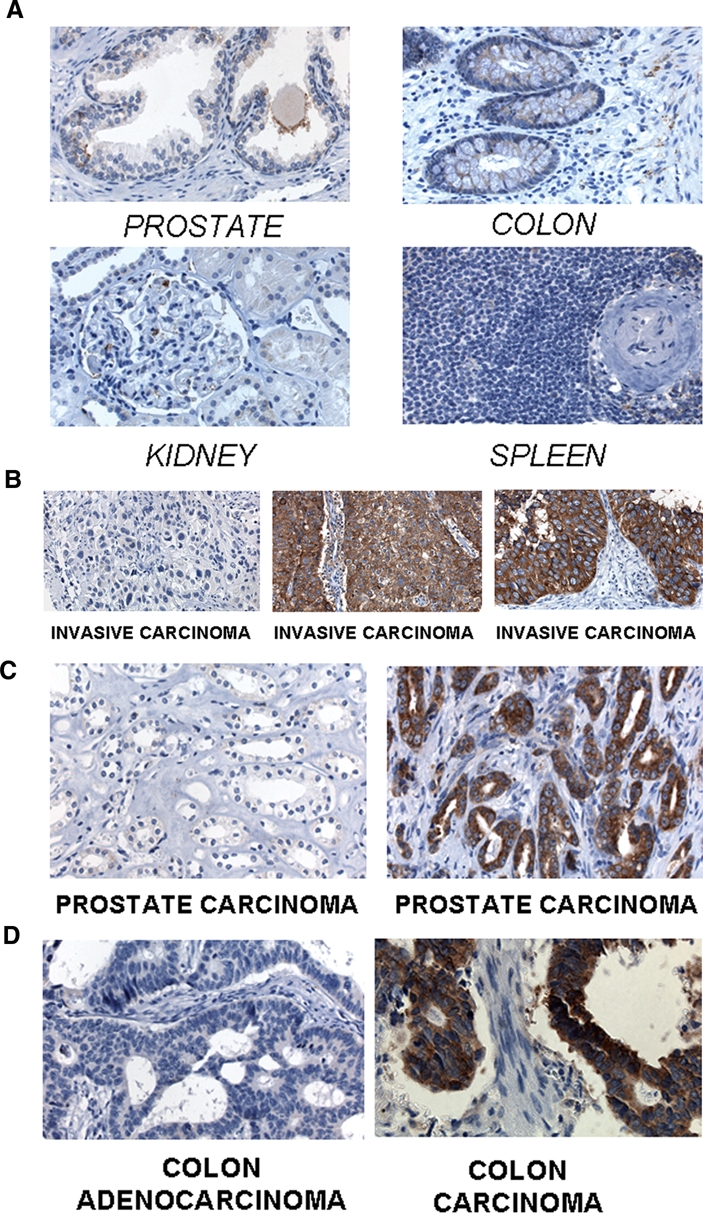
Cytoplasmic localization of L1-ORF1p in human tumors. The following tissues were stained with L1-ORF1p antisera: (A) Healthy prostate, colon, kidney, and spleen samples, none of which stain significantly for L1-ORF1p, thus revealing the specificity of the L1-ORF1p antisera; (B) tumors originating from human bladder tissue, with the first panel displaying a tumor that does not stain for L1-ORF1p and the other two panels staining strongly for cytoplasmic L1-ORF1p; (C) two prostate tumors, in which the left panel does not stain but the right panel stains for cytoplasmic L1-ORF1p; (D) two colorectal tumors, the first of which does not stain for L1-ORF1p whereas the other stains for cytoplasmic L1-ORF1p.
We then set out to determine whether L1 expression might play a role in a variety of human cancer types. Figure 2 presents sections of bladder, prostate, and colorectal tumors stained for the L1-ORF1 antigen. Many of these sections stain positively for the protein, indicating that L1-ORF1p is expressed in each of these tumor types. Within the tumor sections examined, the L1-ORF1p sera stained only the cancer cells and not the stromal cells. Expression of the L1-ORF1p protein was quite variable and depended on the tumor tissue type. A large number of bladder, prostate, and colorectal tumors were analyzed for their expression of the L1-ORF1p protein. A summary of these results is shown in Table 1. Bladder tumors were highly likely to express L1-ORF1p, with 73% of the cases studied staining for this protein. Conversely, only about one third of colorectal cancers stained for L1-ORF1p. Within the set of prostate tumors, about half of the samples stained for L1-ORF1p. L1-ORF1p was cytoplasmic in each of the bladder, colorectal, and prostate tumors for which staining was detected. In other sets of tumors, we were able to detect nuclear localization of L1-ORF1p. These are shown in Figure 3. In Figure 3A, 2 pancreatic neuroendocrine tumors are shown, one of which has nuclear L1-ORF1p and the other of which has a cytoplasmic protein. In Figure 3B, 2 ileal carcinoids are shown, including one with nuclear L1-ORF1p. In both pancreatic neuroendocrine tumors and midgut carcinoids, the overall incidence of L1-ORF1p was very common (Table 1). However, nuclear L1-ORF1p was decidedly rare in these two classes of tumors. Of 65 cases of pancreatic neuroendocrine tumors that stained for L1-ORF1p, only the 1 case shown in Figure 3A showed nuclear protein. In the ileal carcinoids, 39 cases stained for the protein, but only 2 had nuclear L1-ORF1p.
Table 1.
Summary of L1-ORF1p Staining in Human Tumors
| Tumor type | Negative | Positive (%) | Location (%) |
|---|---|---|---|
| Breast | 4 | 437 (99) | Cytoplasm (82) or nucleus (18) |
| Pancreatic neuroendocrine | 41 | 65 (61) | Cytoplasm or nucleus (1 case) |
| Ileal carcinoid | 9 | 39 (81) | Cytoplasm or nucleus (2 cases) |
| Bladder | 22 | 61 (73) | Cytoplasm |
| Colorectal | 25 | 11 (31) | Cytoplasm |
| Prostatic | 29 | 28 (49) | Cytoplasm |
Figure 3.
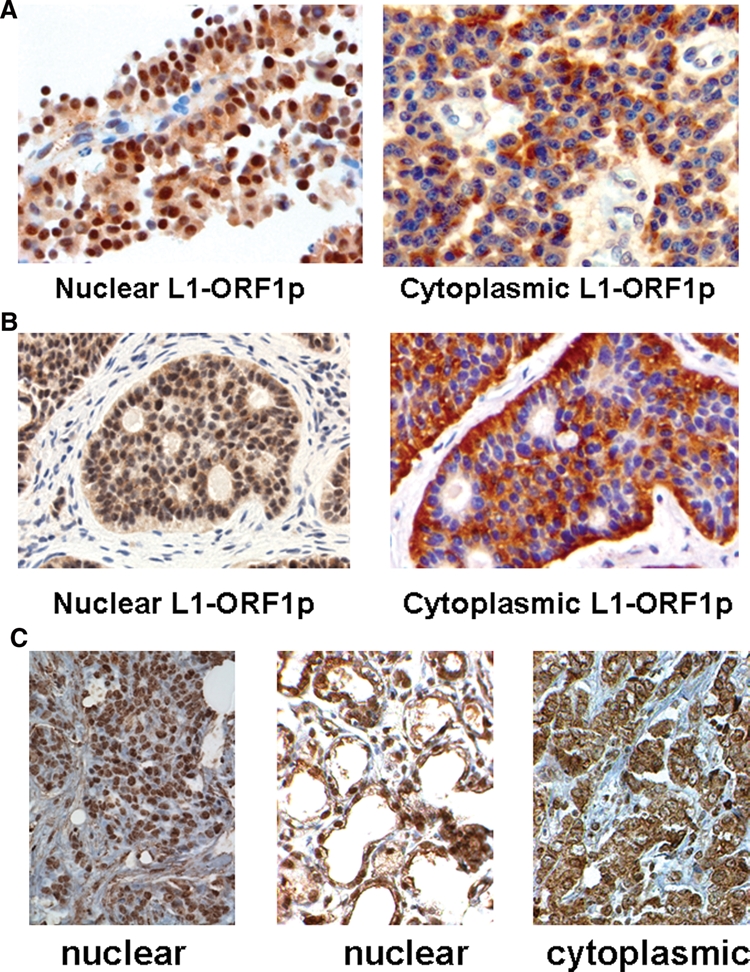
Nuclear localization of L1-ORF1p in selected human tumors. (A) two different pancreatic neuroendocrine tumors stained with L1-ORF1p antisera, showing nuclear (left panel) and cytoplasmic localization of the protein; (B) two different ileal carcinoid tumors, showing nuclear (left panel) and cytoplasmic localization of L1-ORF1p; (C) three different breast tumors, showing nuclear (left and middle panels) and cytoplasmic localization of L1-ORF1p.
Many more examples of tumors with nuclear L1-ORF1p were detected by staining for this protein in breast tumors. As was previously seen in a smaller study of 12 tumors,10 expression of L1-ORF1p in breast cancer is extremely common (Table 1). However, our study employed a much larger set of 441 tumors, and within this very large set, we were able to find 81 tumors that expressed this protein in the nucleus. Two examples of tumors with nuclear L1-ORF1p are shown in Figure 3C.
Potential clinical importance of nuclear L1-ORF1p
These breast cancer samples are part of a study in which extensive clinical information is available. The median age of patients in this study was 50 y. Of the tumors, 87.05% were ductal, 7.27% were lobular, and 5.68% were other types. Of the patients, 82.95% were Caucasian, 13.86% were African American, and 3.18% were other races. Unsurprisingly, given that nearly all of the breast tumors stained positive with anti–L1-ORF1p, no correlation between expression of the protein and patient outcome was observed. However, when the localization of the L1-ORF1p protein in a cell and tissue section was examined, a clinical correlation was uncovered. This is shown in Figure 4. Patients with nuclear L1-ORF1p survived a shorter amount of time after diagnosis than did patients with cytoplasmic protein (Fig. 4A). Patients with nuclear L1-ORF1p also showed a higher incidence of both local recurrence (Fig. 4B) and distal metastases (Fig. 4C) than did patients with cytoplasmic protein. These clinical outcomes were statistically significant.
Figure 4.
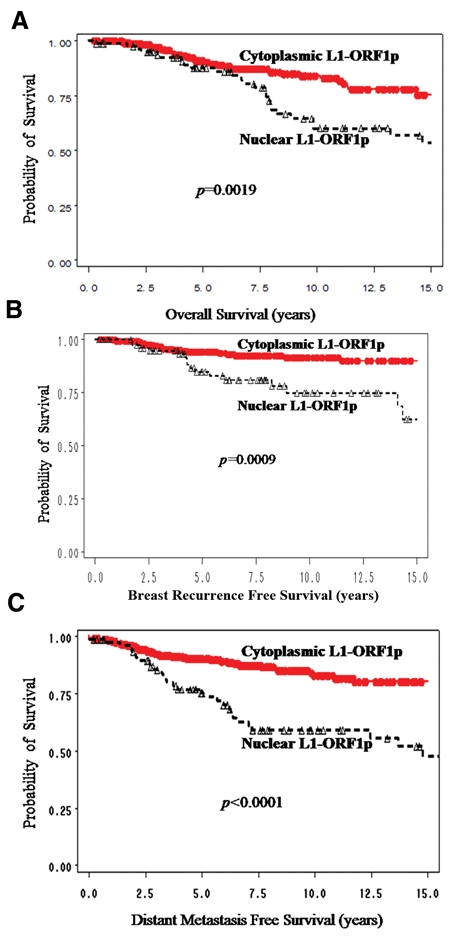
Potential clinical impact of nuclear L1-ORF1p on breast cancer. (A) overall patient survival according to presence of nuclear or cytoplasmic L1-ORF1p; (B) incidence of local recurrence of primary breast tumor according to presence of nuclear or cytoplasmic L1-ORF1p; (C) incidence of formation of distal metastases according to presence of nuclear or cytoplasmic L1-ORF1p. Each curve shows statistical significance as determined by p values, which were generated by log-rank test.
As shown in Table 2, nuclear localization L1-ORF1p also correlates with other breast cancer prognostic factors. Tumors deriving from premenopausal women were more likely to display nuclear L1-ORF1p, as were tumors expressing estrogen receptor or progesterone receptor. Ductal tumors were not more likely to show nuclear L1-ORF1p; rather, nuclear localization was very common in lobular tumors. All of these correlations (premenopausal patients and lobular, estrogen-and progesterone-positive tumors) were statistically significant. Tumors not amplified for the Her2 locus showed a clear trend toward having nuclear L1-ORF1p, but within this data set, Her2-negativity was not quite statistically significant. Nuclear localization of L1-ORF1p was not linked to race, tumor size, nodal status, or familial history of breast cancer.
Table 2.
Correlation between Localization of L1-ORF1p and Clinical Predictors of Breast Cancer
| Prognostic factor | Cytoplasmic L1-ORF1p | Nuclear L1-ORF1p | P value |
|---|---|---|---|
| Age, y | |||
| <50 | 166 | 49 | |
| ≥50 | 194 | 32 | 0.02* |
| Race/ethnicity | |||
| White | 292 | 73 | |
| Black | 53 | 8 | 0.29 |
| Histology | |||
| Ductal | 319 | 64 | |
| Lobular | 17 | 15 | |
| Other | 24 | 1 | <0.001* |
| Nodal status | |||
| Negative | 171 | 39 | |
| Positive | 66 | 13 | 0.73 |
| Tumor size | |||
| T1 | 250 | 57 | |
| ≥T2 | 80 | 15 | 0.65 |
| Family history | |||
| Unknown/none | 280 | 60 | |
| Moderate-strong | 48 | 13 | 0.48 |
| ER status | |||
| Negative | 169 | 22 | |
| Positive | 178 | 52 | 0.003* |
| PR status | |||
| Negative | 188 | 23 | |
| Positive | 161 | 52 | <0.001* |
| Her2 status | |||
| Negative | 285 | 68 | |
| Positive | 65 | 7 | 0.06 |
Statistically significant.
Among the 106 patients with pancreatic neuroendocrine tumors, L1-ORF1p expression did not appear to correlate with the patient’s age, site of the tumor, or the clinical stage of the disease (data not shown). Survival of these patients will continue to be followed, as this data set is a relatively new one (mean follow-up time of 44 mo), and pancreatic neuroendocrine tumors progress slowly. As mentioned above, one patient within this group showed nuclear L1-ORF1p, and this patient was found to have bone metastases, which is an unusually aggressive form of this disease. But as a single result, this is not statistically meaningful.
Analysis of whether L1-ORF1p expression is of clinical importance for ileal carcinoids, prostate, bladder, or colon cancer will have to await another study. We did not have access to patient data for any of the bladder, prostate, and colon cancer samples, whereas the ileal carcinoid database was not suitable for clinical analysis because most of these patients presented initially with metastasis, as is common for this type of tumor,27,28 and most of the patients within the study remain alive with this slowly progressing disease. Disease progression within the 2 ileal carcinoid patients with nuclear L1-ORF1p has been unremarkable to date.
Discussion
The experiments presented here demonstrate that expression of the L1 retrotransposon protein, ORF1p, can be detected in a variety of cancers and that its nuclear localization correlates with poor outcomes in patients with breast cancers. Understanding whether nuclear localization of L1-ORF1p is causative or coincidental will require additional study. It is clear, however, that L1s display many properties that can alter the evolution of a cancer and affect its outcome.29 For instance, L1s can insert into genes and inactivate them,30 they can increase the transcription rate of neighboring genes,31 and they can promote genomic rearrangements.32,33
Previously, L1s have been linked to cancer through studies on the hypomethylation of L1 promoters in a number of tumors including breast,34 bladder,35 liver,36 prostate,37 chronic lymphocytic leukemia,38 ileal carcinoids,39 and chronic myeloid leukemia.40 In some of these studies, a clinical consequence of L1 hypomethylation has been noted, including a link to genomic instability.37,40 Certainly hypomethylation of L1 promoters, which has been shown to correlate with L1-ORF1p expression in cell lines,22 would be consistent with the increases in L1-ORF1p expression that we see in a variety of tumors (Table 1). But hypomethylation of L1s could affect the genome in ways other than increasing transcription of the retrotransposon. For example, as highly repetitive elements, L1s could serve as substrates for ectopic recombination events between heterologous portions of the human genome, which would lead to genetic events seen in many cancers including translocations and chromosome loss. Ectopic recombination is presumably inhibited by DNA methylation, which condenses DNA. But it remains unclear whether the extent of L1 hypomethylation observed in tumors is sufficient to enable ectopic recombination. Hypomethylation of L1 elements may also simply be a marker for hypomethylation of other genes, such as oncogenes.
These data presented here, in which breast cancer outcomes are linked to the nuclear localization of L1-ORF1p (Fig. 4), suggest that the L1 retrotransposition reaction itself, rather than global changes in DNA methylation, may play a role in the progression of certain breast tumors. Nuclear localization of L1-ORF1p would presumably bring the L1 mRNA into position for its reverse transcription by L1-ORF2p, as required for L1 retrotransposition. There have been other studies that have suggested that the L1 retrotransposition reaction can affect tumor formation. For instance, it has been reported that tumorigenesis of cell lines in mouse xenograft models can be attenuated by reducing L1-ORF2p activity.41 In one breast tumor in particular, an L1 insertion was discovered that may have upregulated the c-myc oncogene.42 However, it should also be pointed out that, despite intensive sequencing efforts aimed at finding mutations within oncogenes and tumor suppressor genes in many different tumors, only two L1 insertions have ever been found, with the other being an L1 that inactivated the critical tumor suppressor, APC, in a patient with colon cancer.43 This failure to detect a higher rate of L1 retrotransposition in tumors may be due to the fact that sequencing efforts traditionally focus on exons, whereas L1 insertions may be capable of exerting effects if inserted within introns30 by creating new promoters31 altering the rate of transcription or bringing in new polyadenylation sites.44 In addition, it has become clear that not all tumors express L1-ORF1p, and only a subclass of these express the protein in the nucleus. Thus, we have begun efforts to look for L1 insertions within breast tumors that express nuclear L1-ORF1p.
If nuclear localization of L1-ORF1p affects patient outcome through catalysis of L1 retrotransposition, then patients expressing nuclear L1-ORF1p might benefit from treatments that decrease the rate of the retrotransposition reaction. Nevirapine is a small molecule that has been used to treat other diseases and that has been shown to inhibit the L1 reverse transcriptase in whole cell assays.41 Perhaps nevirapine or other small molecules would prove beneficial to patients with nuclear L1-ORF1p. It is also notable that radiation has been shown to accelerate L1 retrotransposition45 and that all of the patients in the breast tumor study were treated with radiation following breast surgery. Within the set of patients expressing nuclear L1-ORF1p, less aggressive radiation therapies, or alternative postoperative treatments, might be considered.
Although nuclear L1-ORF1p appears to have a more severe clinical impact than the cytoplasmic protein in breast cancers, it is not clear that cytoplasmic L1-ORF1p is truly benign. We could not test the clinical effects of cytoplasmic L1-ORF1p in breast tumors, where it is ubiquitous, and we saw no effects of cytoplasmic protein on pancreatic neuroendocrine tumors, but that particular data set may be too recent to yield meaningful information. For this reason, the clinical importance of the expression of this protein in tumors such as bladder or colorectal tumors remains of interest. We also do not know if nucleolar L1-ORF1p, which appears in many cell lines, has any clinical importance. Because of technical limitations, it has not been possible to demonstrate that nucleolar localization of the L1-ORF1p protein occurs in tumors.
Changes in cellular localization of certain proteins, as we show for L1-ORF1p (Fig. 3), are a common theme in tumorigenesis. One well-studied example is β-catenin, which, like L1-ORF1p, is associated with tumor progression upon nuclear localization.46,47 In the case of β-catenin, a variety of mutations and/or extracellular signals can result in nuclear localization.48,49 At this time, we do not know the mechanism by which L1-ORF1p becomes nuclear. Because it does not have a nuclear localization sequence of its own, we assume that it interacts with another protein to enter the nucleus, and expression of this protein may vary among tumors. The L1-ORF2p protein would seem to be the best candidate, and it would be interesting to determine whether the breast tumors that demonstrate nuclear L1-ORF1p also express higher levels of L1-ORF2p. The identification of cell line models in which L1-ORF1p is nuclear (Fig. 1D), as well as other cell lines in which it is cytoplasmic (Fig. 1C), will help us to address the mechanism by which nuclear localization of the protein occurs and to better understand the consequences of that event.
Materials and Methods
L1-ORF1p isolation, antisera production, and affinity purification
The plasmid 99PUR RPS eGFP (Ostertag et al.50) was a generous gift from the laboratory of H. H. Kazazian Jr. The sequence encoding the L1-ORF1 protein was isolated from 99PUR RPS eGFP and subcloned into pGEX-6P2 (GE Healthcare, Piscataway NJ) to create a gene encoding a GST/ORF1 fusion protein. This plasmid was transformed into the Escherichia coli strain BL21-codonplus-RP (Agilent Technologies, La Jolla, CA). Transformed E. coli were grown at 37°C, harvested, and lysed. A GST/ORF1 protein band was excised following SDS-polyacrylamide gel electrophoresis and directly injected into rabbits to prepare antisera against L1-ORF1p. The GST/ORF1 protein was also isolated on glutathione columns (GE Healthcare), and the GST moiety was removed by proteolysis using PreScission Protease (GE Healthcare). The column-purified L1-ORF1p was covalently linked to CnBr-sepharose 4B (GE Healthcare) and then used for affinity purification of the rabbit antisera as described by the manufacturer.
Cell culture
Unless otherwise noted, cell lines were obtained from ATCC (Manassas, VA) and were grown as recommended by ATCC. The BON-1 cell line51 was a gift from C. Townsend (University of Texas Medical Branch, Galveston) and was grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. QGP-1 was obtained from the Japan Health Sciences Foundation and was grown in RPMI-1640 supplemented with 10% fetal bovine serum. Cell lines were maintained at 37°C with 5% CO2. Cell culture reagents were purchased from Invitrogen (Carlsbad, CA).
Protein extraction and Western blots
Cell lines were grown to 50% confluence, at which time the cells were washed 3 times with phosphate-buffered saline (PBS). Cells were scraped from plate directly into freeze-thaw buffer and subjected to three rounds of freezing in liquid nitrogen followed by thawing on ice. Snap-frozen normal and tumor tissues were supplied by the Cooperative Human Tissue Network (Philadelphia, PA). Whole cell extracts were prepared by subjecting the frozen tissues to multiple rounds of freezing and thawing, followed by Dounce homogenization using a Type A pestle. Protein samples were normalized for total protein amounts and separated using a 4% to 12% Bis-Tris polyacrylamide gel (Invitrogen), then transferred onto Immobilon-P membranes (Millipore, Billerica, MA). Rabbit polyclonal anti–L1-ORF1p sera were added at 1:5,000 dilution, followed by addition of goat anti-rabbit sera linked to horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA). Supersignal West Pico Chemiluminescent reagent (Thermo Fisher Scientific, Rockford, IL) was used to visualize the protein bands. To confirm protein normalization, membranes were stripped and reprobed with monoclonal sera raised against human β-actin (data not shown).
Immunofluorescence
Monoclonal PCNA antisera, FITC-tagged goat anti-mouse IgG, and Alexa 488-tagged goat anti-rabbit IgG were purchased from Santa Cruz Biotechnology. Human nucleolus antisera were purchased from Meridian Life Science (Saco, ME). Cells grown on coverslips were fixed with 2% formaldehyde in PBS for 15 min at room temperature, then washed for 30 min in 3 changes of PBS. Methanol was then added for 10 min at −20°C. Blocking was performed for 30 min in 3 changes of 2% milk/0.05% Tween 20/PBS. Primary antibodies were added in blocking buffer for 1 h at room temperature, followed by 3 washes in blocking buffer. Secondary antisera were added in blocking buffer for 1 h at room temperature. Cells were again washed in 2 changes of blocking buffer. Antifade was applied to all slides prior to microscopy.
Immunohistochemistry of human tumor sections
Affinity purified L1-ORF1p polyclonal antisera were used on formalin-fixed and paraffin-embedded human tissue sections. For breast tumor microarrays, staining was performed as previously described.52 For all other tumor sets, standard avidin-biotin immunoperoxidase reaction and heat-induced antigen retrieval were applied with 0.01M citric acid at pH 6.0 and microwave oven. Optimal dilution for the antibody was determined after titration experiment, and slides were incubated with the primary antibody at 1:500 dilution overnight. Anti-rabbit IgG made in goat was used as a secondary reagent at 1:1,000 dilution, followed by avidin-biotin complex at 1:25 dilution. DAB was used as a chromogen and hematoxylin as a counterstain. Two breast tumor microarrays were studied, one of which included 158 premenopausal women52 and the second of which included 283 women with a wider range of ages, which will be described in another article (Haffty, Yang, et al., submitted). Breast tumor microarray data were analyzed as previously described.52 Pancreatic neuroendocrine tumors and ileal carcinoids were from the lab of L. Tang. Prostatic, bladder, and colorectal tumor microarrays were from the lab of C. Cordon-Cardo. Germ cell tumors were also from the laboratory of C. Cordon-Cardo. Tissue staining was analyzed by trained pathologists. All experiments using human tissues were performed with patients’ consent and with the approval of Institutional Review Boards from Memorial Sloan-Kettering Hospital or from the University of Medicine and Dentistry of New Jersey–Robert Wood Johnson Medical School/Cancer Institute of Jersey.
Acknowledgments
The authors wish to thank Lei Cong and Julia Friedman from the Cancer Institute of New Jersey Immunohistochemistry Shared Resource Laboratory for their technical expertise. We also thank Drs Evan Vosburgh and Abram Gabriel for valuable discussions on the article.
Footnotes
The authors declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
This study was funded by the Verto Institute.
References
- 1. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921 [DOI] [PubMed] [Google Scholar]
- 2. Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat 2007;28:527-39 [DOI] [PubMed] [Google Scholar]
- 3. Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci U S A 2003;100:13815-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol 2001;21:467-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet 2005;14:3237-48 [DOI] [PubMed] [Google Scholar]
- 6. Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res 2001;11:2059-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ovchinnikov I, Rubin A, Swergold GD. Tracing the LINEs of human evolution. Proc Natl Acad Sci U S A 2002;99:10522-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A 2003;100:5280-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martin SL, Branciforte D. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol Cell Biol 1993;13:5383-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asch HL, Eliacin E, Fanning TG, Connolly JL, Bratthauer G, Asch BB. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol Res 1996;8:239-47 [PubMed] [Google Scholar]
- 11. Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem 2004;279:27753-63 [DOI] [PubMed] [Google Scholar]
- 12. Bratthauer GL, Fanning TG. Active LINE-1 retrotransposons in human testicular cancer. Oncogene 1992;7:507-10 [PubMed] [Google Scholar]
- 13. Bratthauer GL, Fanning TG. LINE-1 retrotransposon expression in pediatric germ cell tumors. Cancer 1993;71:2383-6 [DOI] [PubMed] [Google Scholar]
- 14. Rashkova S, Karam SE, Pardue ML. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc Natl Acad Sci U S A 2002;99:3621-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kubo S, Seleme MC, Soifer HS, Perez JL, Moran JV, Kazazian HH, Jr, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci U S A 2006;103:8036-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr A potential role for the nucleolus in L1 retrotransposition. Hum Mol Genet 2004;13:1041-8 [DOI] [PubMed] [Google Scholar]
- 17. Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet 2001;35:501-38 [DOI] [PubMed] [Google Scholar]
- 18. Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol 1994;14:2584-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Woodcock DM, Williamson MR, Doherty JP. A sensitive RNase protection assay to detect transcripts from potentially functional human endogenous L1 retrotransposons. Biochem Biophys Res Commun 1996;222:460-5 [DOI] [PubMed] [Google Scholar]
- 20. Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 1997;13:335-40 [DOI] [PubMed] [Google Scholar]
- 21. Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 1997;189:227-34 [DOI] [PubMed] [Google Scholar]
- 22. Thayer RE, Singer MF, Fanning TG. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene 1993;133:273-7 [DOI] [PubMed] [Google Scholar]
- 23. Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol 2006;13:763-71 [DOI] [PubMed] [Google Scholar]
- 24. Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet 1999;21:163-7 [DOI] [PubMed] [Google Scholar]
- 25. Leibold DM, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG. Translation of LINE-1 DNA elements in vitro and in human cells. Proc Natl Acad Sci U S A 1990;87:6990-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bratthauer GL, Cardiff RD, Fanning TG. Expression of LINE-1 retrotransposons in human breast cancer. Cancer 1994;73:2333-6 [DOI] [PubMed] [Google Scholar]
- 27. Oberg K. Carcinoid tumors: molecular genetics, tumor biology, and update of diagnosis and treatment. Curr Opin Oncol 2002;14:38-45 [DOI] [PubMed] [Google Scholar]
- 28. Vosburgh E. Neoplasms of the diffuse endocrine system. In: Kufe D, editor. Cancer medicine 7. Ontario (Canada): B.C. Decker; 2006. p. 1109-30 [Google Scholar]
- 29. Schulz WA. L1 retrotransposons in human cancers. J Biomed Biotechnol 2006;2006:83672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays 2005;27:775-84 [DOI] [PubMed] [Google Scholar]
- 31. Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 2001;21:1973-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 2002;110:327-38 [DOI] [PubMed] [Google Scholar]
- 33. Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell 2002;110:315-25 [DOI] [PubMed] [Google Scholar]
- 34. Alves G, Tatro A, Fanning T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene 1996;176:39-44 [DOI] [PubMed] [Google Scholar]
- 35. Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer 1999;80:1312-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin CH, Hsieh SY, Sheen IS, Lee WC, Chen TC, Shyu WC, et al. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. Cancer Res 2001;61:4238-43 [PubMed] [Google Scholar]
- 37. Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate 1999;39:166-74 [DOI] [PubMed] [Google Scholar]
- 38. Dante R, Dante-Paire J, Rigal D, Roizes G. Methylation patterns of long interspersed repeated DNA and alphoid repetitive DNA from human cell lines and tumors. Anticancer Res 1992;12:559-63 [PubMed] [Google Scholar]
- 39. Choi IS, Estecio MR, Nagano Y, Kim do H, White JA, Yao JC, et al. Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors). Mod Pathol 2007;20:802-10 [DOI] [PubMed] [Google Scholar]
- 40. Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervante F, Sanchez J, Garate L, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene 2005;24:7213-23 [DOI] [PubMed] [Google Scholar]
- 41. Sciamanna I, Landriscina M, Pittoggi C, Quirino M, Mearelli C, Beraldi R, et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene 2005;24:3923-31 [DOI] [PubMed] [Google Scholar]
- 42. Morse B, Rotherg PG, South VJ, Spandorfer JM, Astrin SM. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature 1988;333:87-90 [DOI] [PubMed] [Google Scholar]
- 43. Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 1992;52:643-5 [PubMed] [Google Scholar]
- 44. Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 2004;429:268-74 [DOI] [PubMed] [Google Scholar]
- 45. Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res 2006;34:1196-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Park WS, Oh RR, Park JY, Kim PJ, Shin MS, Lee JH, et al. Nuclear localization of beta-catenin is an important prognostic factor in hepatoblastoma. J Pathol 2001;193:483-90 [DOI] [PubMed] [Google Scholar]
- 47. Garcia-Rostan G, Tallini G, Herrero A, D’Aquila TG, Carcangiu ML, Rimm DL. Frequent mutation and nuclear localization of beta-catenin in anaplastic thyroid carcinoma. Cancer Res 1999;59:1811-5 [PubMed] [Google Scholar]
- 48. Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 1997;275:1787-90 [DOI] [PubMed] [Google Scholar]
- 49. Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res 1998;58:1130-4 [PubMed] [Google Scholar]
- 50. Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res 2000;28:1418-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Evers BM, Townsend CM, Jr, Upp JR, Allen E, Hurlbut SC, Kim SW, et al. Establishment and characterization of a human carcinoid in nude mice and effect of various agents on tumor growth. Gastroenterology 1991;101:303-11 [DOI] [PubMed] [Google Scholar]
- 52. Parikh RR, Yang Q, Higgins SA, Haffty BG. Outcomes in young women with breast cancer of triple-negative phenotype: the prognostic significance of CK19 Expression. Int J Radiat Oncol Biol Phys 2008;70:35-42 [DOI] [PubMed] [Google Scholar]


