Summary
The ordered packaging of DNA within the nucleus of somatic cells reflects a dynamic supportive structure that facilitates stable transcription interrupted by intermittent cycles of extreme condensation. This dynamic mode of packing and unpacking chromatin is intimately linked to the ability of the genome to specifically complex with both histones and non-histone proteins. Understanding the underlying mechanism that governs the formation of higher order chromatin structures is a key to understanding how local architecture modulates transcription. In part, the formation of these structures appears to be regulated through genomic looping that is dynamically mediated by attachment to the nuclear scaffold/matrix at S/MARs, i.e., Scaffold/Matrix Attachment Regions. Although the mechanism guiding the formation and use of these higher-ordered structures remains unknown, S/MARs continue to reveal a multitude of roles in development and the pathogenesis of disease.
Keywords: S/MAR attachment, Nuclear Matrix, disease, scaffold, model, gene regulation, LIS, NaCl
I. Introduction
While the sequencing of the human genome has provided an invaluable resource for high throughput genomic studies, sequence alone has not been able to explain how the genome encodes the cellular program. This ability appears to be linked to the push–pull of compaction and potentiation, i.e., the formation of an open chromatin domain from a closed state. These changes in state are intimately linked to the ability of the DNA to form regulatory protein complexes. The ordered packaging of DNA within the nucleus must reflect a dynamic mechanism that supports stable transcription interrupted by intermittent cycles of condensation. On one hand, the four core histones and linker act to physically organize the eukaryotic DNA. On the other hand, they provide a medium to epigenetically reflecting the interphase condensation states that are recapitulated through many cycles of replication. The addition of modifiers at dozens of sites [reviewed in (Bernstein et al. 2007)] provides long-term epigenetic memory. These modifications contribute to cellular differentiation by marking loci for multi-generational activation or silencing (Jenuwein and Allis 2001). Working in concert as part of a combinatorial mechanism they only influence ~25% of gene expression (Wang et al. 2008). The question remains: what other controls participate to determine cell- specificity through transcription? The mechanics of transitioning between different states of condensation contributing to transcriptional control must be carefully orchestrated to maintain order and inhibit the intertwining of DNA fibers. Functional condensation is aided considerably because chromatin can be periodically constrained by a dynamic mesh of non- histone nuclear scaffold or matrix proteins. The nuclear scaffold/matrix was described well over 50 years ago (Zbarskii and Debov 1951), however its characterization has remained elusive. As a result, the regions of the genome that bind to the nuclear scaffold/matrix, also known as S/MARs, are poorly defined at a genomic level. It has been suggested that S/MARs function to help maintain chromatin states while demarcating points of transition between states (Berezney and Coffey 1974; Mirkovitch et al. 1984). Interactions of the genome within this nuclear framework may coordinately act with histone modifications to generate the stable chromatin states that maintain expression or silencing. In concert they could ensure that this differentiated state is replicated.
II. Genome organization from nucleosome to chromosome territory
In somatic cells, the first order of DNA packaging is achieved by nucleosomes that wrap 146 bp in ~1.7 helical turns around an octamer of H2A2, H2B2, H32 and H42 proteins (Kornberg 1974; Luger et al. 1997). The nucleosomes are separated by 10 to 50 bp stretches of histone H1 bound linker DNA. Nucleosome positioning is influenced by several factors. These include the ability of the DNA to stretch and/or bend sharply around the histone core (Richmond and Davey 2003; Ong et al. 2007) and the direction of the bend of the major groove (Drew and Travers 1985). The energy of packaging is reduced within flexible AT rich regions and these sites of maximum curvature provide a means to precisely position nucleosomes. Local sequence composition can also influence nucleosome positioning despite the absence of a consensus binding sequence. Even with this level of understanding, only ~50% of the nucleosomal interactions can be predicted by sequence characteristics alone (Segal et al. 2006; Valouev et al. 2008).
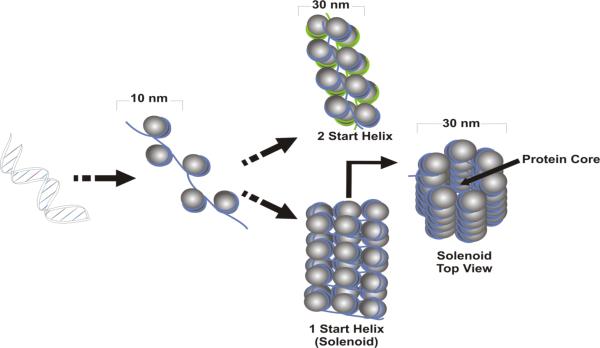
In solutions of low ionic strength, nucleosomes for a 10 nm fiber that is visualized as ‘beads on a string’. The higher order folding of the 10 nm fiber becomes evident in the presence of the HI linker (Yaneva et al. 1995) appearing as a 30 nm fiber (Shen et al. 1995). Several models of genome packaging are presented in Figure 1, although the one start and two start helical structures are still debated (Wu et al. 2007). The one-start helix, or solenoid, models are characterized by interactions between neighboring nucleosomes. This determines the dimensions of the coiled fiber as the DNA is bent into a solenoid (Finch and Klug 1976). The two start helix models are based on a nucleosomal arrangement that appears as a zigzag at low ionic strength (Worcel et al. 1981; Woodcock et al. 1984) with the linker maintaining a straight extended conformation (Williams et al. 1986; Smith et al. 1990). Recent evidence supporting the solenoid model suggests that nucleosome interdigitation among neighboring helical gyres creates a high-density nucleosome fiber (Robinson et al. 2006). However, these models fail to consider the effect of additional changes that are imposed upon condensation. For example, the effect of N-terminal acetylation of histone H4 that is necessary for the formation of the 30 nm fiber [reviewed in (Tremethick 2007)]. Irrespective, the 30 nm fiber is further condensed into a series of chromatin loops that are attached at their bases to non-histone nuclear scaffold or matrix proteins.
Figure 1. Folding of the 10 nm Fiber.
In the presence of linker histone H1, the 10 nm fiber folds into the 30 nm a fiber. Several models of folding have been proposed. The one start helix model (bottom) requires the coiling of the 10 nm fiber around a central protein core into a solenoid. In comparison, the two-start helix model (top) requires two 10 nm fibers to associate, and form a secondary 30 nm fiber.
The first evidence for constrained looping of the 30 nm fiber in mammalian cells was based on sedimentation rates of nucleoids, i.e., nuclei depleted of proteins, through sucrose gradients (Cook and Brazell 1976). A similar study in Drosophila melanogaster suggested that there were approximately 400 nucleosomes or ~80 kb of DNA per loop (Benyajati and Worcel 1976). Looping was visualized by electron microscopy of histone depleted metaphase nuclei (Paulson and Laemmli 1977; McCready et al. 1979).
Several models have been proposed for the native organization of the chromosomal loops into higher order metaphase structures including the radial loop model of metaphase chromosome organization (Marsden and Laemmli 1979). Electron micrographs clearly showed the attachment of radially arranged loops to a central scaffold. Building upon this model, others proposed minibands formed as the 30 nm solenoid filaments were organized into 60 kb loops grouped 18 per turn (Pienta and Coffey 1984). Each mitotic chromatid was then derived from ~106 minibands arranged along a central axis.
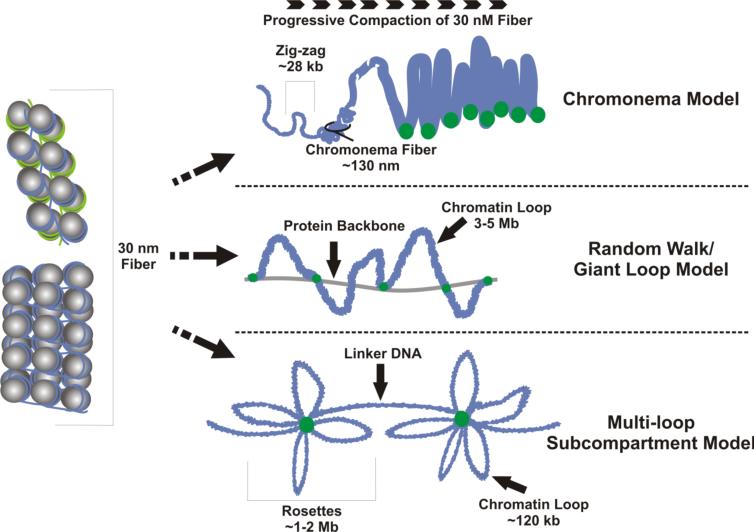
It is now well-accepted that individual interphase chromosomes are organized into discrete territories [reviewed in (Cremer and Cremer 2001)] but the structural organization within the chromosome territories remains to be discerned. Figure 2 summarizes several models including the chromonema model (Belmont and Bruce 1994), the random walk/giant loop model (Sachs et al. 1995) and the multi-loop sub compartment model (Munkel et al. 1999). The chromonema model of organization was derived by following the various states of condensation and intermediates of interphase chromatin through G1. As G1 proceeded, folded structures of chromonema fibers of approximately 100-130 nm in diameter decondensed to 60-80 nm. It was suggested that condensed chromonema fibers created looped domains that are stabilized by attaching to either specific non-histone nuclear scaffold proteins or to condensed chromatin within adjacent chromonema fibers. The random walk/giant loop model considered the large-scale aggregation of chromatin into DNA foci. It was developed by assessing the distances between defined genomic sequences visualized by fluorescence in situ hybridization (Sachs et al. 1995). This mathematical model assumed that a series of flexible 3 million base-pair chromatin domains were randomly attached to a nonchromatin nuclear substructure. However, this did not consider the presence of smaller loops addressed in the multi-loop sub compartment model of chromosome territories (Munkel et al. 1999). Among the random walks/giant loops, the multi-120 kb loop sub compartment model considered the intertwining of fibers. This would promote long-range interactions that were constrained by requiring that individual sub compartments provide a high level of specification within each chromosome territory.
Figure 2. Models of Chromosomal Looping.
Several models have been proposed for the looping of the 30 nm fibers. The chromonema model proposes the formation of a higher order structure through the progressive compaction of the 30 nm fiber into ~130 nm chromonema fibers. Each chromonema fiber is composed of condensed ~28 kb zig-zags of DNA. The chromonema fibers are then coiled to create looped domains that may be attached to the nuclear scaffold/matrix (dots). The random walk/giant loop model suggests that large condensed chromatin loops of 3-5 Mb are randomly attached to a non-DNA backbone (green dots) within the nucleus. The multi-loop sub compartment model is based on individual ~120 kb chromatin loops anchored to the nuclear scaffold/matrix (green dots). They are organized into rosettes that contain a total of 1-2 Mb of DNA connected to one another by linker DNA
III. Nucleosome remodeling, modification and transcription
It is well established that post-translational modifications of the amino terminal tails of the core histone proteins that are typified by, but certainly not limited to, acetylation, methylation and phosphorylation, can alter DNA structure. These individual modifications serve to mark regions for active transcription or silencing. It appears that a dynamic balance between multiple histone modifications maintains a chromatin environment that is either permissive or restrictive to transcription (for a review, see Jenuwein and Allis 2001). For example, a histone modification module consisting of a combination of 17 modifications has been defined. This module tends to be overrepresented within the promoters of the highly expressed genes in human CD4+ T cells (Wang et al. 2008). However, these histone modifications do not uniquely determine expression, as other modifications are required to coordinately regulate structure and poise a gene for transcription.
The regions immediately surrounding active genes can be completely devoid of nucleosomes. Several studies have shown that nucleosome presence may not be conducive to transcription (Wasylyk et al. 1979; Lorch et al. 1987) requiring their removal for transcriptional activation (Han and Grunstein 1988). For example, chromatin-remodeling complexes such as the multiprotein SWI/SNF complex can alter nucleosome positioning to reduce steric hinderence to promote recognition of specific sequences by transcription factors (Richmond and Davey 2003). This can be achieved through several mechanisms including sliding/moving the histone octamer to a new position to expose the DNA (Hamiche et al. 1999; Langst et al. 1999; Whitehouse et al. 1999), ejecting the octamer (Lorch et al. 1999; Boeger et al. 2003; Reinke and Horz 2003; Boeger et al. 2004), removing the H2A-H2B dimer (Bruno et al. 2003; Yang et al. 2007) and/or replacing dimers with histone variants e.g., exchanging H2A-H2B with H2B-H2A.Z (Mizuguchi et al. 2004).
Transcriptional regulation by structure must also be considered. For example, regions of Z DNA upstream of yeast promoters demarcate the boundaries of neighboring nucleosome rich regions (Wong et al. 2007). These alternative conformations provide a means to absorb torsional stress thereby relieving local super coiling induced by active transcription. This highlights one specific mechanism for altering DNA structure. Additional mechanisms have been identified including the binding of high mobility group proteins (Javaherian et al. 1978) as well as the action of enzymes such as topoisomerase II to stabilize and/or relax the DNA [reviewed in (Giaever et al. 1988)].
IV. Higher order levels of organization and transcription
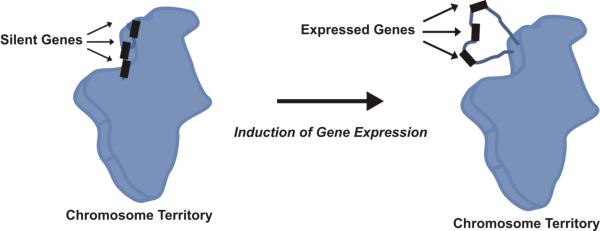
The regulation of gene expression by higher order structure is not well understood. Perhaps this reflects the poor understanding of chromatin organization beyond the level of the 30 nm fiber. As summarized in Figure 3, it is well established that chromosomes occupy distinct territories within the nucleus (Cremer and Cremer 2001) and genes undergoing active transcription can loop out away from their respective chromosome territories into structurally distinct interchromatin compartments (Albiez et al. 2006). On one hand, looping has been shown to coincide with the activation of the major histocompatibility locus (Volpi et al. 2000) and the HoxB gene cluster (Chambeyron and Bickmore 2004). Perhaps chromosomal looping may be a result of the local density of expressed genes (Mahy et al. 2002) reflecting the aggregation of transcriptionally active genes into transcription factories [reviewed in (Marenduzzo et al. 2007)]. On the other hand, X inactivation activates multiple domains of higher order looping. These may form separate chromatin hubs at the noncoding Xite and Tsix loci as part of the sex-specific X inactivation center induced developmental chromosomal looping (Tsai et al. 2008).
Figure 3. Induction of Gene Expression Coordinates with Looping.
Groups of genes that are poised for expression are located at the periphery of the chromosome territory, but not necessarily expressed (‘silent genes’). Upon induction of gene expression, the domain loops out away from the chromosome territory, perhaps into a transcription factory (‘expressed genes’).
There is a large body of evidence for transcription factories [reviewed in (Jackson 2005)], i.e., complex centers of transcription where active genes from distant locations on the same or different chromosomes are clustered together (Osborne et al. 2007). Joining non- linear regions of the genome within the same transcription factory suggests that chromatin may physically relocate to the factory. One of the most well studied examples of looping is that of the β-globin locus. Activation of this gene cluster relies on the developmentally timed interaction of the locus control region with specific promoters by looping of the intervening DNA (Tolhuis et al. 2002). Looping appears to be mediated by the attachment of at least one hypersensitive site within the LCR to the nuclear matrix (Ostermeier et al. 2003). Similar to the concept of a transcription factory, where distant genomic regions are brought together within one nuclear complex, the β-globin locus provided a key developmental example of an active chromatin hub. Active chromatin hubs that are part of transcription factories are assembled through looping to promote the interactions of distant elements to facilitate tissue specific expression [reviewed in (de Laat and Grosveld 2003)].
The positioning of specific genes and or chromosomes relative to the boundaries of the nucleus may also affect transcription. For example, gene dense chromosomes are often located towards the nuclear interior while gene poor chromosomes are located peripherally, just interior to the nuclear lamina (Bridger et al. 2000; Tanabe et al. 2002). Interestingly, genes targeted to the inner nuclear membrane and nuclear laminas are silenced (Reddy et al. 2008). Tethering of genomic regions to the nuclear lamina may serve as a silencing mechanism as lamin B1 associated domains (LADs) appear preferentially bound in gene poor regions (Guelen et al. 2008). Silencing may also be achieved by specific promoter region binding to type A lamins (Lee et al. 2009). While recruitment of genes to the nuclear periphery can alter expression, its effect is not always measurable (Finlan et al. 2008).
The seemingly conflicting data of gene silencing at the nuclear periphery suggests that other factors may play a role. For example, Ikaros regulates lymphocyte cell expression by developing interphase heterochromatinization foci (Brown et al. 1997). As expected, inactive genes were associated with heterochromatic foci whereas active genes were located elsewhere. This is consistent with the view that genes are selectively recruited to repressive foci of heterochromatization.
V. The Nuclear scaffold, matrix and nucleoskeleton
The proteinacious nuclear matrix was first observed when interphase nuclei were treated with high salt solutions (Zbarskii and Debov 1951). The protein components of the residual body of the interphase nuclear matrix were subsequently characterized from rat liver nuclei (Berezney and Coffey 1974). This clearly showed that the protein constituents of the high salt extracted nuclear matrix could be differentiated from that of untreated nuclei. Looping constrained by a residual protein body was subsequently verified (Cook and Brazell 1976). This structure was shown to persist in metaphase nuclei after histones were depleted by extraction with 2 M NaCl (Paulson and Laemmli 1977; McCready et al. 1979). Subsequent removal of the DNA component revealed a protein scaffold (Adolphs et al. 1977) suggesting attachment to a static structure. Similarly, isolation of the interphase nuclear matrix was visualized as loops of super coiled DNA that appeared as a halo of DNA constrained by the nuclear matrix (Vogelstein 1980). This respectively defined the nuclear scaffold and nuclear matrix. As expected, their protein constituents differed and their nuclear constituents were classified into three components: the peripheral nuclear lamina, an internal network of proteins and residual nucleoli upon which the nuclear scaffold composed primarily SC1, i.e., Topoisomerase II (Earnshaw et al. 1985; Gasser et al. 1986) and SC2 (Lewis and Laemmli 1982) was overlaid.
It was suggested that the ability to resolve sites of attachment may be compromised by sliding or the rearrangement of the sites of DNA attachment to the nuclear matrix when prepared with high salt. To address this limitation, extraction with LIS, or 25 mM lithium diiodosalicylate, was introduced as a “milder” means to extricate the non-histone nuclear protein fraction. When applied, this procedure also revealed a nonrandom interphase loop structure organized by the residual extracted proteins (Mirkovitch et al. 1984). However, lamins along with their corresponding sites of attachment were absent from LIS scaffold preparations (Mirkovitch et al. 1988). Nevertheless other protein complexes (4 of the 6 sites remained attached to both metaphase and interphase scaffolds) appeared constant throughout the cell cycle (Mirkovitch et al. 1988). The terms nuclear scaffold referring to LIS extraction and nuclear matrix referring to extraction with 2M NaCl have since been adopted.
The methodology of nuclear matrix isolation continues to develop (Fey et al. 1986; Wan et al. 1999) as part of the toolset to study both replication (Vogelstein 1980) and transcription (Jackson et al. 1981). This includes ‘physiological’ methods that encapsulate cells in agarose beads prior to hypotonic lysis followed by the gentle salt extraction of histones that resolves the nucleoskeleton (Jackson et al. 1988). Studies comparing some of these methods have shown method dependent resolution of sites of attachment (Donev 2000) suggesting that the various extraction methods may isolate different subgroups of proteins. However, throughout the literature the various methods are often interchangeably used without addressing the implications. This has fueled the debate of the existence of a matrix/scaffold within the nucleus (Pederson 2000). It has recently been resolved (Heng et al. 2004) with the aid of a comprehensive description of the biological role of the sites of nuclear matrix and scaffold attachment (Linnemann and Krawetz 2009; Linnemann et al. 2009).
VI. Specific constituents of the nuclear scaffold/matrix
Isolation of the metaphase scaffold by different methods of extraction does not appear to affect the types of proteins that are isolated. Initial characterization of the scaffold proteins revealed that the major constituents are the protein scaffold component 1 (SC1), i.e., topoisomerase II (Earnshaw et al. 1985; Gasser et al. 1986) and scaffold component 2 (SC2) (Lewis and Laemmli 1982). Although at a reduced level to metaphase cells, topoisomerase II persists in interphase cells as a part of the nuclear scaffold/matrix (Berrios et al. 1985; Berrios and Fisher 1988; Mirkovitch et al. 1988). Multiple variants of topoisomerase II have been shown to differentially mark potentiation of the protamine gene domain during mouse spermatogenesis (Martins and Krawetz 2007). This includes sites of matrix attachment that position the locus within a small looped region coincident with expression.
Some, but not all, of the non-histone proteins identified from interphase nuclei differ based on the type of isolation method used. Visualization of the interphase nuclear matrix and nucleoskeleton has revealed proteins that appear to have a structural role in the nucleus. Electron microscopy of thick resin less sections of HeLa cells revealed that residual chromatin loops were associated with a network of proteins that appeared as intermediate filaments (Jackson and Cook 1988; He et al. 1990). Interestingly, cytoplasmic type intermediate filaments also interact with SARs and are isolated as a part of the nuclear scaffold (Tolstonog et al. 2002). The presence of nuclear lamins supporting the nuclear envelope as well as their dispersion throughout the nucleus suggests that these proteins may play a significant structural role as a part of the nuclear scaffold/matrix [reviewed in (Goldman et al. 2002)]. The lamins that dominate and define the nuclear lamina are resistant to high salt solubilization while the more diffusely organized lamins throughout the nucleus are less resistant. For example, lamin B1 is widely distributed throughout the nucleus as a part of the nucleoskeleton. It was recently shown to be necessary for RNA synthesis (Tang et al. 2008). The relative ease that the lamins can be extracted with solutions of high salt suggests that this method can be used to differentiate proteins that appear fixed and static from those serving a more dynamic role. It is likely that these intermittently attached proteins specifically function as needed during dynamic processes like transcription. Thus their differential isolation may provide insight into the direct role of these nuclear scaffold/matrix components of transcription.
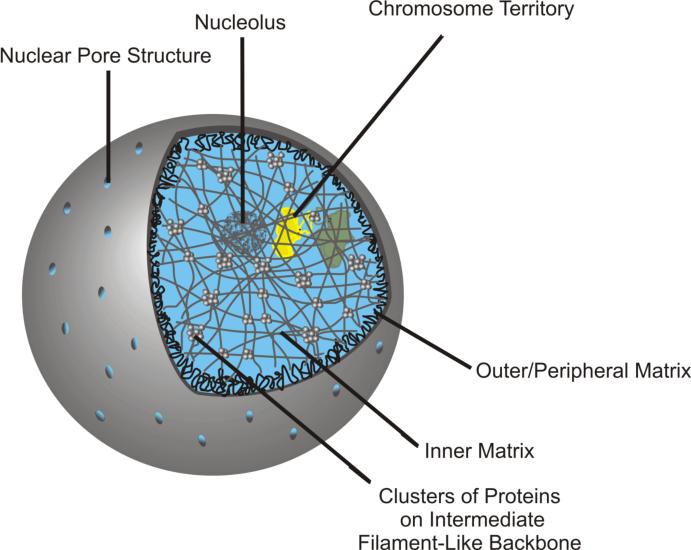
As depicted in Figure 4, the inner and peripheral networks within the nucleus appear physically continuous but remain functionally distinct. Their differential isolation by high salt extraction with or without cation/heat stabilization has led to their description as either the Type I or Type II nuclear matrix respectively (Lebkowski and Laemmli 1982a; Lebkowski and Laemmli 1982b). The isolation of the type II nuclear matrix without stabilization yielded peripheral nuclear matrix components including proteins of the lamina and nuclear pore complexes. Similarly, it has been shown that MARs do specifically bind to type II matrix components of the nuclear lamina (Luderus et al. 1992). In comparison the protein constituents of the type I or inner nuclear matrix have been characterized with the use of cation/heat stabilization. The array of proteins characteristic of type I nuclear matrices is similar to those isolated as part of the LIS nuclear scaffold.
Figure 4. The Nuclear Matrix.
Within the nucleus, the nuclear matrix can be subdivided into the outer/peripheral, or type II nuclear matrix. This can be isolated without the use of heat or cation stabilization. The type II nuclear matrix includes proteins of the nuclear lamina as well as proteins of the nuclear pore complexes. The inner, or type I nuclear matrix can be isolated after nuclei stabilization with heat or the addition of cations such as Cu2+. This portion of the nuclear matrix is complex, comprised of hundreds of different types of proteins. These appear to form clusters on a backbone of filaments of similar diameter as the intermediate filaments.
Electron microscopy showed intermediate like filaments that span clusters of globular type proteins (Jackson and Cook 1988) suggesting clustered foci of additional functional non-histone proteins. At least 200 proteins have been detected by 2D gel electrophoresis (Capco et al. 1982) and over 300 by tandem mass spectrometry (Ishii et al. 2008). The wide range of proteins that have been identified as constituents of the nuclear scaffold/matrix attests to its complex nature. However, the distribution of proteins present in relatively small proportions begs the question: does this merely represent a snapshot of dynamic interactions such as the induction of expression of a group of functionally related genes?
One of the most thoroughly investigated nuclear matrix proteins with functional impact is SATB1. This protein is expressed predominantly in the cells of the thymus, where it binds to MARs at the bases of chromatin loops to regulate global gene expression during T cell development (Dickinson et al. 1992; de Belle et al. 1998; Alvarez et al. 2000). SATB1 has recently been shown to direct a significant increase in the number of loops present at the cytokine locus upon T-helper type 2-cell activation and is necessary for the activation of the locus (Cai et al. 2006). Another protein that has gained interest as a potential nuclear matrix interacting protein is CTCF. Interestingly, the binding of CTCF insulates discrete regions of a genome (Antes et al. 2001; Magdinier et al 2004; Yusufzai and Felsenfeld 2004). However, CTCF may not be a nuclear matrix protein (Goetze et al. 2005) requiring reconciliation. Additional proteins such as scaffold attachment factors A and B [SAF-A (Romig et al. 1992) and SAF-B (Renz and Fackelmayer 1996)] also bind both SARs and MARs in vitro, but are only extracted as a part of the nuclear scaffold. SAF-A is analogous to hn-RNPU and analysis of the two protein isoforms has indicated that they both bind to SAR elements with similar affinity (Fackelmayer and Richter 1994). Several transcription factors, such as the pit-1 and the histone acetyltransferase/ transcriptional coactivator CBP, have also been isolated as part of the cadre of nuclear matrix proteins [reviewed in (Hendzel et al. 2001)].
Characterization of the nuclear substructure has established a link between the local physical packing of DNA and the global reorganization of chromosomes during cellular differentiation (Mayer et al. 2005; Martins and Krawetz 2007). It has been suggested that homeotic proteins (Boulikas 1992) like the homeobox domain containing protein Oct-1, may anchor the genome to the nuclear lamina (Imai et al. 1997). RUNX, or Runt-related transcription factors, are also associated with the nuclear matrix. Their targeting to discrete foci within the nucleus essential to regulate RUNX dependent genes and in turn tissue specific differentiation during embryonic development (Choi et al. 2001). This intranuclear targeting to transcriptionally active foci occurs in partnership with Smad, which attaches to the nuclear matrix as a RUNX- Smad protein complex (Zaidi et al. 2002). Recently it was suggested that the sperm nuclear matrix is necessary for zygotic development (Shaman et al. 2007). Curiously, it is enriched in RNAs (Lalancette et al. 2008).
VII. Attachment of the genome to the nuclear scaffold/matrix
The human genome is organized into loop domains attached at their bases to the nuclear scaffold/matrix by S/MARs that exhibit a range of sizes depending on extraction method (Jackson et al. 1990). Considering an average loop size of approximately 100 kb, it can be estimated that there are 32,000 looped domains throughout the human genome. If each looped domain is demarcated by an attachment site on either side, this would correspond to approximately 64,000 MARs throughout the human genome. In mature spermatozoa, toroid loops are estimated to be approximately 50 kb in length (Brewer et al. 1999), or half that of somatic cells. If every toroid loop were attached to the nuclear matrix in the intervening linker DNA, this would also correspond to ~64,000 sites of attachment in the sperm genome.
Several studies have suggested that MARs are not evenly distributed throughout the mammalian genome. For example, loops as small as 1 kb have been observed at telomeres (Luderus et al. 1996). Similarly, the imprinted region on mouse distal chromosome 7 resolves 52 MARs in a 1 Mb domain (Purbowasito et al. 2004). Although the average MAR spacing was ~20 kb, the majority of attachments clustered into almost equal groups of 58 kb and 169 kb regions flanked by MARs. These observations have now been reconciled following the recent genome- wide analysis (Linnemann and Krawetz 2009; Linnemann et al. 2009). This has shown that S/MARs segment the genome into a wide range of cell-type dependent differentially sized loops. Taken together, the estimates of the number of S/MARs are merely predictions. It is important to note that both cellular environment and cell type likely influence the number of attachments observed at any given point in time.
VIII. Sequence specificity and in-silico prediction of nuclear scaffold / matrix attachment
Although many S/MARs that have been identified in vitro and in vivo have been studied in depth by sequence analysis, the identification of an obvious consensus sequence for scaffold/matrix attachment has proven difficult. Perhaps this reflects the many different protein interactions that appear to contribute to the operational definition of an S/MAR. While to date no consensus has been derived, S/MARs have displayed several reiterative characteristics. Initial observations based on a small subset of all genomic sites suggest that S/MARs are approximately 300 to 1000 bp in length, AT enriched, containing both oligo d[A] and d[T] tracks (Gasser and Laemmli 1986) as well as ATATAT boxes (Cockerill and Garrard 1986). As S/MARs are preferentially bound and cleaved by topoisomerase II both in vitro (Adachi et al. 1989; Razin et al. 1991) and in vivo (Kas and Laemmli 1992), many are expected to contain topoisomerase II binding sites.
While S/MARs have often been generally characterized as AT rich regions or sequences containing topoisomerase II (Platts et al. 2006), there is evidence that these two characteristics alone are not sufficient for nuclear matrix binding (Das et al. 1993; Bode et al. 2006). Perhaps this simply reflects the lack of examples or a conformational dependence (Yamamura and Nomura 2001). For example, there is direct evidence of an intrinsically curved structure in the tobacco S/M II MAR where the nuclear proteins appear to bind to the MAR because of structure and not sequence (Fukuda 2000). A body of evidence is also accumulating to suggest that mammalian MARs coincide with Base Unpairing Regions (BURs) that alleviate the torsional strain induced by transcription of nearby regions (Bode et al. 2006).
These general characteristics have been used in silico to independently and in combination predict the locations of S/MARs [reviewed in (Platts et al. 2006)]. The application of these algorithms often yields weak and/or seemingly over predictive models of nuclear scaffold/matrix attachment. However, because S/MARs are known to display cell type specificity, what appears as an over prediction may represent potential attachment in other cell types. Additionally the sequence characteristics identified thus far may work in combination with structural features such as BURs and perhaps additional unidentified sequences or local chromatin modifications to direct attachment to the nuclear scaffold/matrix are required. Recent evidence suggests that in some cases the inclusion of structural elements can lead to stronger prediction algorithms (Girod et al. 2007).
IX. The multifactorial nature of MARs
S/MARs can be classified as constitutive or facultative, perhaps possessing either structural or functional characteristics respectively. In this respect, the human epsilon globin gene has been shown to have both facultative and constitutive MARs (Yan and Qian 1998). Additionally, several MARs within a 1 Mb region of human chromosome 19 have been identified and classified as either structural or regulatory (Chernov et al. 2002). What is perhaps of note is that the S/MARs are merely a snapshot of a dynamic environment. Differences in conditions or the time of measurement could yield varying levels of attachment for an individual S/MAR. Thus their observed roles, be that constitutive or facultative, may not be easily subdivided through the study of a single point in time, environment or cell type.
As a function of both the cell cycle and cell type specificity, only a subset of potential S/MARs are expected to be observed at any given time. In specific cases S/MARs have been shown to regulate conformation in both a cell type specific and cell cycle context specific manner (Britanova et al. 2005; Koina and Piper 2005; Martins and Krawetz 2007). Evidence for cell type specific nuclear scaffold/matrix attachment suggests facultative and constitutive attachment can be differentiated. For example, MARs of the osteocalcin gene are tissue specific and transiently associated with the nuclear matrix (Bidwell et al. 1993) while the tyrosine hydroxylase MARs bind the nuclear matrix independent of tissue type (Lenartowski et al. 2003). Removal of the chicken lysozyme MARs increases ectopic expression (Bonifer et al. 1994) confirming their role in tissue specific gene expression. Recent genome wide surveys have shown that SARs and MARs provide complementary activities that either facilitate or suppress gene expression (Linnemann et al. 2009). In this manner cell type specific attachment mediates identity (Linnemann and Krawetz 2009).
X. The role of MARs in disease pathogenesis
Epigenetic contributions to disease have been long suspected in studies of monozygotic and dizygotic twins (Wong et al. 2005). The linkage of the epigenetic control of gene expression, often via aberrant methylation and acetylation, to disease is well appreciated (Kalebic 2003). While little is known about the direct effect of S/MARs on tissue specific genes in a normal or diseased state, changes in nuclear scaffold/matrix attachment by either the loss of association or through binding to a previously cryptic site have been implicated in the onset of several genetic disorders. Even synonymous mutations that might not affect protein sequence could result in changes in scaffold/matrix binding resulting in disease if the local structure is perturbed. However, in this case they would perhaps be better characterized as non-synonymous. For example, the loss of a MAR may arrest expression of a male factor and present as infertility (Kramer et al. 1997). Similarly, in vitro loss of a MAR has been correlated with the onset of FSHD, Fascioscapulohumeral muscular dystrophy (Petrov et al. 2006). Deletions in conserved noncoding sequences of heritable diseases (Kleinjan and van Heyningen 2005; Sjakste and Sjakste 2005) are often underscored by regions containing MARs and insulators. Although the nuclear lamins bind to S/MARs, the key laminopathies resulting from defects in the A/C type lamins, most often muscular dystrophy and cardiomyopathy, are not necessarily directly linked to the function of the nuclear matrix. The primary phenotypic effect of these mutations is an inability of cells with contractile behavior to survive the stress associated with cellular function. The result is that the nuclei are damaged and thus destroyed so that the muscle fibers harbor reduced numbers of nuclei and cannot function. Other laminopathies induced through lamin A mutations including Hutchinson-Gilford progeria syndrome that also presents artherosclerosis (McClintock et al. 2006), and dilated cardiomyopathy (Taylor et al. 2003) may act by directly affecting nuclear scaffold/matrix attachment to destabilize the gene expression program.
The role of MARs in cancer is well established. For example, sites of chromosomal fragmentation preferentially localize to MARs (Schoenlein et al. 1999; Bode et al. 2000a; Bode et al. 2000b). In addition, progression to a malignant phenotype may be promoted by the binding of mutant p53 to the nuclear matrix thereby dysregulating the cell-cycle (Deppert 1996). Similarly, aberrant binding to the nuclear matrix may destabilize the expression of a host of genes that maintain cells in a non- cancerous state (Linnemann and Krawetz 2009). The most direct evidence for cancer by perturbed nuclear scaffold/matrix attachment is that induced by SATB1. As a genomic organizer, SATB1 regulates transcription during T cell development (Dickinson et al. 1992; de Belle et al. 1998; Alvarez et al. 2000; Cai et al. 2006). SATB1 is not dominant in other cell types but its expression in breast cancer cells is indicative of an aggressive phenotype (Han et al. 2008). Interestingly, ectopic expression of SATB1 in non-invasive cancer cells induces an invasive phenotype whereas removal of SATB1 from metastatic cells reverses the invasive phenotype and inhibits tumor growth.
S/MARs continue to reveal a multitude of varied roles in development and disease. Continued analyses should provide a window to their underlying mechanism(s) of action reflective of their complex nature. This is essential as we begin to take these elements from the bench to the bedside for use as effective gene therapeutics.
Acknowledgements
This work was supported in part by National Institutes of Health grant HD36512, the Wayne State University Research Enhancement Program in Computational Biology and the Charlotte B. Failing Professorship if Fetal Therapy and Diagnosis to SAK.
Biography
The Krawetz Laboratory: Front row from left to right, Amelia Linnemann, Kathryn Drennan, Claudia Lalancette, Back row from left to right, Robert Goodrich, Adrian Platts, Stephen A. Krawetz, Seema Mahadev, Graham Johnson
References
- Adachi Y, Kas E, Laemmli UK. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. Embo J. 1989;8(13):3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolphs KW, Cheng SM, Paulson JR, Laemmli UK. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc Natl Acad Sci U S A. 1977;74(11):4937–4941. doi: 10.1073/pnas.74.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14(7):707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14(5):521–535. [PMC free article] [PubMed] [Google Scholar]
- Antes TJ, Namciu SJ, Fournier RE, Levy-Wilson B. The 5' boundary of the human apolipoprotein B chromatin domain in intestinal cells. Biochemistry. 2001;40(23):6731–6742. doi: 10.1021/bi0100743. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Bruce K. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol. 1994;127(2):287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C, Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Berrios M, Osheroff N, Fisher PA. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc Natl Acad Sci U S A. 1985;82(12):4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrios S, Fisher PA. Thermal stabilization of putative karyoskeletal protein-enriched fractions from Saccharomyces cerevisiae. Mol Cell Biol. 1988;8(10):4573–4575. doi: 10.1128/mcb.8.10.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwell JP, Van Wijnen AJ, Fey EG, Dworetzky S, Penman S, Stein JL, Lian JB, Stein GS. Osteocalcin gene promoter-binding factors are tissue-specific nuclear matrix components. Proc Natl Acad Sci U S A. 1993;90(8):3162–3166. doi: 10.1073/pnas.90.8.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J, Benham C, Ernst E, Knopp A, Marschalek R, Strick R, Strissel P. Fatal connections: when DNA ends meet on the nuclear matrix. J Cell Biochem Suppl Suppl. 2000a;35:3–22. doi: 10.1002/1097-4644(2000)79:35+<3::aid-jcb1121>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Bode J, Benham C, Knopp A, Mielke C. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Crit Rev Eukaryot Gene Expr. 2000b;10(1):73–90. [PubMed] [Google Scholar]
- Bode J, Winkelmann S, Gotze S, Spiker S, Tsutsui K, Bi C, A KP, Benham C. Correlations between scaffold/matrix attachment region (S/MAR) binding activity and DNA duplex destabilization energy. J Mol Biol. 2006;358(2):597–613. doi: 10.1016/j.jmb.2005.11.073. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11(6):1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14(5):667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Bonifer C, Yannoutsos N, Kruger G, Grosveld F, Sippel AE. Dissection of the locus control function located on the chicken lysozyme gene domain in transgenic mice. Nucleic Acids Res. 1994;22(20):4202–4210. doi: 10.1093/nar/22.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. Homeotic protein binding sites, origins of replication, and nuclear matrix anchorage sites share the ATTA and ATTTA motifs. J Cell Biochem. 1992;50(2):111–123. doi: 10.1002/jcb.240500202. [DOI] [PubMed] [Google Scholar]
- Brewer LR, Corzett M, Balhorn R. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 1999;286(5437):120–123. doi: 10.1126/science.286.5437.120. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Boyle S, Kill IR, Bickmore WA. Remodelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol. 2000;10(3):149–152. doi: 10.1016/s0960-9822(00)00312-2. [DOI] [PubMed] [Google Scholar]
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21(3):658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91(6):845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell. 2003;12(6):1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38(11):1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Capco DG, Wan KM, Penman S. The nuclear matrix: three-dimensional architecture and protein composition. Cell. 1982;29(3):847–858. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18(10):1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov IP, Akopov SB, Nikolaev LG, Sverdlov ED. Identification and mapping of nuclear matrix-attachment regions in a one megabase locus of human chromosome 19q13.12: long-range correlation of S/MARs and gene positions. J Cell Biochem. 2002;84(3):590–600. [PubMed] [Google Scholar]
- Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, et al. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci U S A. 2001;98(15):8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cook PR, Brazell IA. Conformational constraints in nuclear DNA. J Cell Sci. 1976;22(2):287–302. doi: 10.1242/jcs.22.2.287. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2(4):292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Das AT, Luderus ME, Lamers WH. Identification and analysis of a matrix-attachment region 5' of the rat glutamate-dehydrogenase-encoding gene. Eur J Biochem. 1993;215(3):777–785. doi: 10.1111/j.1432-1033.1993.tb18092.x. [DOI] [PubMed] [Google Scholar]
- de Belle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol. 1998;141(2):335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11(5):447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- Deppert W. Binding of MAR-DNA elements by mutant p53: possible implications for its oncogenic functions. J Cell Biochem. 1996;62(2):172–180. doi: 10.1002/(sici)1097-4644(199608)62:2<172::aid-jcb5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70(4):631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Donev RM. The type of DNA attachment sites recovered from nuclear matrix depends on isolation procedure used. Mol Cell Biochem. 2000;214(1-2):103–110. doi: 10.1023/a:1007159421204. [DOI] [PubMed] [Google Scholar]
- Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Halligan B, Cooke CA, Heck MM, Liu LF. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J Cell Biol. 1985;100(5):1706–1715. doi: 10.1083/jcb.100.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackelmayer FO, Richter A. Purification of two isoforms of hnRNP-U and characterization of their nucleic acid binding activity. Biochemistry. 1994;33(34):10416–10422. doi: 10.1021/bi00200a024. [DOI] [PubMed] [Google Scholar]
- Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102(5):1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4(3):e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y. Interaction of nuclear proteins with intrinsically curved DNA in a matrix attachment region of a tobacco gene. Plant Mol Biol. 2000;44(1):91–98. doi: 10.1023/a:1006416929665. [DOI] [PubMed] [Google Scholar]
- Gasser SM, Laemmli UK. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986;188(4):613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Giaever GN, Snyder L, Wang JC. DNA supercoiling in vivo. Biophys Chem. 1988;29(1-2):7–15. doi: 10.1016/0301-4622(88)87020-0. [DOI] [PubMed] [Google Scholar]
- Girod PA, Nguyen DQ, Calabrese D, Puttini S, Grandjean M, Martinet D, Regamey A, Saugy D, Beckmann JS, Bucher P, et al. Genome-wide prediction of matrix attachment regions that increase gene expression in mammalian cells. Nat Methods. 2007;4(9):747–753. doi: 10.1038/nmeth1076. [DOI] [PubMed] [Google Scholar]
- Goetze S, Baer A, Winkelmann S, Nehlsen K, Seibler J, Maass K, Bode J. Performance of genomic bordering elements at predefined genomic loci. Mol Cell Biol. 2005;25(6):2260–2272. doi: 10.1128/MCB.25.6.2260-2272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16(5):533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97(7):833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452(7184):187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55(6):1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110(3):569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Kruhlak MJ, MacLean NA, Boisvert F, Lever MA, Bazett-Jones DP. Compartmentalization of regulatory proteins in the cell nucleus. J Steroid Biochem Mol Biol. 2001;76(1-5):9–21. doi: 10.1016/s0960-0760(00)00153-9. [DOI] [PubMed] [Google Scholar]
- Heng HH, Goetze S, Ye CJ, Liu G, Stevens JB, Bremer SW, Wykes SM, Bode J, Krawetz SA. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J Cell Sci. 2004;117(Pt 7):999–1008. doi: 10.1242/jcs.00976. [DOI] [PubMed] [Google Scholar]
- Imai S, Nishibayashi S, Takao K, Tomifuji M, Fujino T, Hasegawa M, Takano T. Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression. Mol Biol Cell. 1997;8(12):2407–2419. doi: 10.1091/mbc.8.12.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Hirano Y, Araki N, Oda T, Kumeta M, Takeyasu K, Furukawa K, Horigome T. Nuclear matrix contains novel WD-repeat and disordered-region-rich proteins. FEBS Lett. 2008 doi: 10.1016/j.febslet.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Jackson DA. The amazing complexity of transcription factories. Brief Funct Genomic Proteomic. 2005;4(2):143–157. doi: 10.1093/bfgp/4.2.143. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. Visualization of a filamentous nucleoskeleton with a 23 nm axial repeat. Embo J. 1988;7(12):3667–3677. doi: 10.1002/j.1460-2075.1988.tb03248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Dickinson P, Cook PR. The size of chromatin loops in HeLa cells. Embo J. 1990;9(2):567–571. doi: 10.1002/j.1460-2075.1990.tb08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, McCready SJ, Cook PR. RNA is synthesized at the nuclear cage. Nature. 1981;292(5823):552–555. doi: 10.1038/292552a0. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Yuan J, Cook PR. A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J Cell Sci. 1988;90(Pt 3):365–378. doi: 10.1242/jcs.90.3.365. [DOI] [PubMed] [Google Scholar]
- Javaherian K, Liu JF, Wang JC. Nonhistone proteins HMG1 and HMG2 change the DNA helical structure. Science. 1978;199(4335):1345–1346. doi: 10.1126/science.628842. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kalebic T. Epigenetic changes: potential therapeutic targets. Ann N Y Acad Sci. 2003;983:278–285. doi: 10.1111/j.1749-6632.2003.tb05982.x. [DOI] [PubMed] [Google Scholar]
- Kas E, Laemmli UK. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. Embo J. 1992;11(2):705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koina E, Piper A. An inactive X specific replication origin associated with a matrix attachment region in the human X linked HPRT gene. J Cell Biochem. 2005;95(2):391–402. doi: 10.1002/jcb.20425. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184(139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kramer JA, Zhang S, Yaron Y, Zhao Y, Krawetz SA. Genetic testing for male infertility: a postulated role for mutations in sperm nuclear matrix attachment regions. Genet Test. 1997;1(2):125–129. doi: 10.1089/gte.1997.1.125. [DOI] [PubMed] [Google Scholar]
- Lalancette C, Miller D, Li Y, Krawetz SA. Paternal contributions: new functional insights for spermatozoal RNA. J Cell Biochem. 2008;104(5):1570–1579. doi: 10.1002/jcb.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97(7):843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Lebkowski JS, Laemmli UK. Evidence for two levels of DNA folding in histone-depleted HeLa interphase nuclei. J Mol Biol. 1982a;156(2):309–324. doi: 10.1016/0022-2836(82)90331-x. [DOI] [PubMed] [Google Scholar]
- Lebkowski HS, Laemmli UK. Non-histone proteins and long-range organization of HeLa interphase DNA. J Mol Biol. 1982b;156(2):325–344. doi: 10.1016/0022-2836(82)90332-1. [DOI] [PubMed] [Google Scholar]
- Lee DC, Welton KL, Smith ED, Kennedy BK. A-type nuclear lamins act as transcriptional repressors when targeted to promoters. Exp Cell Res. 2009;315(6):996–1007. doi: 10.1016/j.yexcr.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartowski R, Grzybowski T, Miscicka-Sliwka D, Wojciechowski W, Goc A. The bovine tyrosine hydroxylase gene associates in vitro with the nuclear matrix by its first intron sequence. Acta Biochim Pol. 2003;50(3):865–873. [PubMed] [Google Scholar]
- Lewis CD, Laemmli UK. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982;29(1):171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- Linnemann AK, Krawetz SA. Silencing by Nuclear Matrix Attachment Distinguishes Cell-type Specificity: Association with Increased Proliferation Capacity. Nucleic Acids Research. 2009;37(9):2779–88. doi: 10.1093/nar/gkp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann AK, Platts AE, Krawetz SA. Differential nuclear scaffold/matrix attachment marks expressed genes. Hum Mol Genet. 2009;18(4):645–654. doi: 10.1093/hmg/ddn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96(3):389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- Luderus ME, de Graaf A, Mattia E, den Blaauwen JL, Grande MA, de Jong L, van Driel R. Binding of matrix attachment regions to lamin B1. Cell. 1992;70(6):949–959. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- Luderus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135(4):867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Magdinier F, Yusufzai TM, Felsenfeld G. Both CTCF-dependent and -independent insulators are found between the mouse T cell receptor alpha and Dad1 genes. J Biol Chem. 2004;279(24):25381–25389. doi: 10.1074/jbc.M403121200. [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159(5):753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23(3):126–133. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17(4):849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- Martins RP, Krawetz SA. Decondensing the protamine domain for transcription. Proc Natl Acad Sci U S A. 2007;104(20):8340–8345. doi: 10.1073/pnas.0700076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 2005;6:44. doi: 10.1186/1471-2121-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock D, Gordon LB, Djabali K. Hutchinson-Gilford progeria mutant lamin A primarily targets human vascular cells as detected by an anti-Lamin A G608G antibody. Proc Natl Acad Sci U S A. 2006;103(7):2154–2159. doi: 10.1073/pnas.0511133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready SJ, Akrigg A, Cook PR. Electron-microscopy of intact nuclear DNA from human cells. J Cell Sci. 1979;39:53–62. doi: 10.1242/jcs.39.1.53. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J, Gasser SM, Laemmli UK. Scaffold attachment of DNA loops in metaphase chromosomes. J Mol Biol. 1988;200(1):101–109. doi: 10.1016/0022-2836(88)90336-1. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J, Mirault ME, Laemmli UK. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303(5656):343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Munkel C, Eils R, Dietzel S, Zink D, Mehring C, Wedemann G, Cremer T, Langowski J. Compartmentalization of interphase chromosomes observed in simulation and experiment. J Mol Biol. 1999;285(3):1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J Mol Biol. 2007;368(4):1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5(8):e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Liu Z, Martins RP, Bharadwaj RR, Ellis J, Draghici S, Krawetz SA. Nuclear matrix association of the human beta-globin locus utilizing a novel approach to quantitative real-time PCR. Nucleic Acids Res. 2003;31(12):3257–3266. doi: 10.1093/nar/gkg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Pederson T. Half a century of “the nuclear matrix”. Mol Biol Cell. 2000;11(3):799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Pirozhkova I, Carnac G, Laoudj D, Lipinski M, Vassetzky YS. Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc Natl Acad Sci U S A. 2006;103(18):6982–6987. doi: 10.1073/pnas.0511235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienta KJ, Coffey DS. A structural analysis of the role of the nuclear matrix and DNA loops in the organization of the nucleus and chromosome. J Cell Sci Suppl. 1984;1:123–135. doi: 10.1242/jcs.1984.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- Platts AE, Quayle AK, Krawetz SA. In-silico prediction and observations of nuclear matrix attachment. Cell Mol Biol Lett. 2006;11(2):191–213. doi: 10.2478/s11658-006-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purbowasito W, Suda C, Yokomine T, Zubair M, Sado T, Tsutsui K, Sasaki H. Large-scale identification and mapping of nuclear matrix-attachment regions in the distal imprinted domain of mouse chromosome 7. DNA Res. 2004;11(6):391–407. doi: 10.1093/dnares/11.6.391. [DOI] [PubMed] [Google Scholar]
- Razin SV, Vassetzky YS, Hancock R. Nuclear matrix attachment regions and topoisomerase II binding and reaction sites in the vicinity of a chicken DNA replication origin. Biochem Biophys Res Commun. 1991;177(1):265–270. doi: 10.1016/0006-291x(91)91977-k. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452(7184):243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11(6):1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Renz A, Fackelmayer FO. Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Res. 1996;24(5):843–849. doi: 10.1093/nar/24.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423(6936):145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci U S A. 2006;103(17):6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romig H, Fackelmayer FO, Renz A, Ramsperger U, Richter A. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. Embo J. 1992;11(9):3431–3440. doi: 10.1002/j.1460-2075.1992.tb05422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs RK, van den Engh G, Trask B, Yokota H, Hearst JE. A random-walk/giant-loop model for interphase chromosomes. Proc Natl Acad Sci U S A. 1995;92(7):2710–2714. doi: 10.1073/pnas.92.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenlein PV, Barrett JT, Welter D. The degradation profile of extrachromosomal circular DNA during cisplatin-induced apoptosis is consistent with preferential cleavage at matrix attachment regions. Chromosoma. 1999;108(2):121–131. doi: 10.1007/s004120050359. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. The sperm nuclear matrix is required for paternal DNA replication. J Cell Biochem. 2007;102(3):680–688. doi: 10.1002/jcb.21321. [DOI] [PubMed] [Google Scholar]
- Shen X, Yu L, Weir JW, Gorovsky MA. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82(1):47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- Sjakste N, Sjakste T. Nuclear matrix proteins and hereditary diseases. Genetika. 2005;41(3):293–298. [PubMed] [Google Scholar]
- Smith MF, Athey BD, Williams SP, Langmore JP. Radial density distribution of chromatin: evidence that chromatin fibers have solid centers. J Cell Biol. 1990;110(2):245–254. doi: 10.1083/jcb.110.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H, Muller S, Neusser M, von Hase J, Calcagno E, Cremer M, Solovei I, Cremer C, Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci U S A. 2002;99(7):4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CW, Maya-Mendoza A, Martin C, Zeng K, Chen S, Feret D, Wilson SA, Jackson DA. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J Cell Sci. 2008;121(Pt 7):1014–1024. doi: 10.1242/jcs.020982. [DOI] [PubMed] [Google Scholar]
- Taylor MR, Fain PR, Sinagra G, Robinson ML, Robertson AD, Carniel E, Di Lenarda A, Bohlmeyer TJ, Ferguson DA, Brodsky GL, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41(5):771–780. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10(6):1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Tolstonog GV, Sabasch M, Traub P. Cytoplasmic intermediate filaments are stably associated with nuclear matrices and potentially modulate their DNA-binding function. DNA Cell Biol. 2002;21(3):213–239. doi: 10.1089/10445490252925459. [DOI] [PubMed] [Google Scholar]
- Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128(4):651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Tsai CL, Rowntree RK, Cohen DE, Lee JT. Higher order chromatin structure at the X-inactivation center via looping DNA. Dev Biol. 2008;319(2):416–425. doi: 10.1016/j.ydbio.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008 doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Pardoll DM, Coffey DS. Supercoiled loops and eukaryotic DNA replication. Cell. 1980;22(1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wan KM, Nickerson JA, Krockmalnic G, Penman S. The nuclear matrix prepared by amine modification. Proc Natl Acad Sci U S A. 1999;96(3):933–938. doi: 10.1073/pnas.96.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B, Thevenin G, Oudet P, Chambon P. Transcription of in vitro assembled chromatin by Escherichia coli RNA polymerase. J Mol Biol. 1979;128(3):411–440. doi: 10.1016/0022-2836(79)90095-0. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400(6746):784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- Williams SP, Athey BD, Muglia LJ, Schappe RS, Gough AH, Langmore JP. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J. 1986;49(1):233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AH, Gottesman II, Petronis A. Phenotypic differences in genetically identical organisms: the epigenetic perspective. Hum Mol Genet. 2005;14:R11–18. doi: 10.1093/hmg/ddi116. Spec No 1. [DOI] [PubMed] [Google Scholar]
- Wong B, Chen S, Kwon JA, Rich A. Characterization of Z-DNA as a nucleosome-boundary element in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104(7):2229–2234. doi: 10.1073/pnas.0611447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Frado LL, Rattner JB. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984;99(1 Pt 1):42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A, Strogatz S, Riley D. Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci U S A. 1981;78(3):1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Bassett A, Travers A. A variable topology for the 30-nm chromatin fibre. EMBO Rep. 2007;8(12):1129–1134. doi: 10.1038/sj.embor.7401115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura J, Nomura K. Analysis of sequence-dependent curvature in matrix attachment regions. FEBS Lett. 2001;489(2-3):166–170. doi: 10.1016/s0014-5793(01)02098-1. [DOI] [PubMed] [Google Scholar]
- Yan ZJ, Qian RL. The 5'-flanking cis-acting elements of the human epsilon-globin gene associates with the nuclear matrix and binds to the nuclear matrix proteins. Cell Res. 1998;8(3):209–218. doi: 10.1038/cr.1998.21. [DOI] [PubMed] [Google Scholar]
- Yaneva J, Schroth GP, van Holde KE, Zlatanova J. High-affinity binding sites for histone H1 in plasmid DNA. Proc Natl Acad Sci U S A. 1995;92(15):7060–7064. doi: 10.1073/pnas.92.15.7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat Struct Mol Biol. 2007;14(6):540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Felsenfeld G. The 5'-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc Natl Acad Sci U S A. 2004;101(23):8620–8624. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proc Natl Acad Sci U S A. 2002;99(12):8048–8053. doi: 10.1073/pnas.112664499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbarskii IB, Debov SS. [Protein fractions in the cell nuclei.]. Biokhimiia. 1951;16(5):390–395. [PubMed] [Google Scholar]