Abstract
Objectives
Prenatal exposure to cocaine causes cytoarchitectural alterations in the developing neocortex. Previously, we reported that cocaine inhibits neural progenitor cell proliferation through oxidative endoplasmic reticulum stress and consequent down-regulation of cyclin A, whereas cyclin A expression was increased in astrocytes. In the present study, cell type-specific responses to cocaine were further explored.
Methods
Gene expression profiles were examined in five types of cells obtained from the human fetal cerebral cortex at 20 weeks gestation. Cells were treated with 100 µM cocaine in vitro for 24 hr, followed by gene expression analysis using a human neural/stem cell/drug abuse-focused cDNA array, with verification by quantitative real-time RT-PCR.
Results
Cocaine influenced transcription of distinct categories of genes in a cell type-specific manner. Cocaine down-regulated cytoskeleton-related genes including ezrin, γ2 actin, α3d tubulin and α8 tubulin in neural and/or A2B5+ progenitor cells. In contrast, cocaine modulated immune and cell death-related genes in microglia and astrocytes. In microglia, cocaine up-regulated the immunoregulatory and pro-apoptotic genes IL-1β and BAX. In astrocytes, cocaine down-regulated the immune response gene glucocorticoid receptor and up-regulated the anti-apoptotic genes 14-3-3 ε and HVEM. Therefore, cell types comprising the developing neocortex show differential responses to cocaine.
Conclusions
These data suggest that cocaine causes cytoskeletal abnormalities leading to disturbances in neural differentiation and migration in progenitor cells, while altering immune and apoptotic responses in glia. Understanding the mechanisms of cocaine’s effects on human CNS cells may help in the development of therapeutic strategies to prevent or ameliorate cocaine-induced impairments in fetal brain development.
Keywords: cocaine, microarray, gene expression profiling, human fetal CNS cells, brain development
Understanding the mechanisms through which cocaine causes adverse effects on the developing brain has received considerable attention, because several hundred thousand fetuses per year are exposed to cocaine in the United States alone (Singer et al., 2002). Clinical studies have correlated neocortical architecture modifications and behavioral disturbances with in utero cocaine exposure (Chiriboga et al., 1999; Bellini et al., 2000; Singer et al., 2008). In sub-human primates, administration of cocaine at the time of neocortical neurogenesis (the second trimester) disturbs development of cerebral cortex, causing reduced neocortex volume, disturbed lamination, altered positioning of cerebral cortical neurons, and reduced density and number of cortical neurons (Lidow, 1995; Lidow and Song, 2001). Notably, these anatomical abnormalities result in neurobehavioral impairments in infant and adolescent sub-human primates similar to those seen in humans, including deficits in attention, orientation, state control, and motor maturity (He et al., 2004). In addition to the action of cocaine on neurons, interference with astroglial development in developing cerebral cortex has been reported in animal fetuses prenatally exposed to cocaine (Akbari et al., 1994; Lidow, 1995; Nassogne et al., 1998).
In vitro, cocaine has been shown to selectively induce neuronal death without affecting the survival of glial cells (Nassogne et al., 1995). We have also found that cocaine specifically inhibits proliferation of neural progenitor cells by inducing down-regulation of cyclin A2 triggered by oxidative endoplasmic reticulum stress, an effect which is not seen in neurons or microglia (Lee et al., 2008). In contrast, cocaine increases the cyclin A2 transcript in human astrocytes (Lee et al., 2008). These data suggested that prenatal cocaine exposure may cause abnormalities in the developing cerebral cortex through multiple and divergent pathways in different types of cells.
By using a human neural/stem cell/drug abuse-focused cDNA array (hNSDA array) designed for analysis of development/maintenance of central nervous system (CNS) cell types as well as for effects of drugs of abuse, we profiled cocaine-induced gene expression changes in five types of CNS cells obtained from the developing human fetal cerebral cortex. The effects of cocaine on brain development are largely based on studies in rodent model systems, and although rodents and humans have generally similar patterns of brain development, there are some differences (Kornack and Rakic, 1998; Letinic et al., 2002). Characterization of gene expression changes induced by cocaine in primary human CNS cells in vitro may help to extend the understanding of cocaine’s effects on brain development to the human brain. The results suggest that cocaine down-regulates cytoskeleton-related genes in progenitor cells, while modulating expression of immune and cell death-related genes in glial cells. Cocaine-induced cell-type specific alterations in gene expression may cooperate to cause overall cytoarchitectural abnormalities, by interrupting morphological development of cortical progenitor cells while altering immune and apoptotic functions of glial cells.
MATERIALS AND METHODS
Cell Culture and Treatments
Primary human fetal CNS cells (ScienCell Research Laboratories, San Diego, CA), including neural progenitor cells, A2B5+ progenitor cells, neurons, astrocytes and microglia, were obtained from ~20-week human fetal cerebral cortexes, in accordance with principles embodied in the Declaration of Helsinki (Code of Ethics of the World Medical Association). Primary human cells were cultured as previously described (Lee et al., 2008). Human fetal CNS cultures were maintained for 8–21 days in vitro and treated with 100 µM cocaine hydrochloride (National Institute on Drug Abuse, Baltimore, MD) for 24 hr. The concentration of 100 µM is frequently used for in vitro studies of cocaine (Nassogne et al., 1995; Lee et al., 2008). Although relatively high, this concentration can be reached in the brain of chronic human cocaine users (Kalasinsky et al., 2000). Additionally, there is a dose-response relationship between the effects of cocaine and its concentration; exposure to high levels of cocaine in utero is correlated with the greatest impairments in human prenatal brain growth (Delaney-Black et al., 1996; Chiriboga et al., 1999). We employed this dose to produce maximal effects, and thus to avoid finding differences between cell types based on variable sensitivity to lower doses of cocaine.
Human Neural/Stem Cell/Drug Abuse-Focused cDNA Array (hNSDA Array)
Two independent samples from individual brains were used for each group analyzed by microarray. Total RNA was extracted from human fetal CNS cultures using RNA STAT-60 (TEL-TEST, Friendswood, TX). Microarray analysis was performed using a hNSDA array containing triplicate repeats of 2400 clones, of which 56% were from the human mammalian gene collection (Strausberg et al., 1999), 33% from Research Genetics, and 11% from Invitrogen. The hNSDA array includes 2400 genes related to nervous system function, neurogenesis and neural stem cell differentiation. In addition, genes related to drug metabolism, stress signaling, cell survival and toxicity are included.
Complete materials and methods including immunocytochemistry, microarray procedure, microarray data analysis, quantitative real-time RT-PCR, and associated references are available as Supplementary methods.
RESULTS
Cell Characterization
The human neural progenitor cells used in this study were nestin-positive and A2B5-negative (> 95% of cells, data not shown), and A2B5+ progenitor cells were A2B5-positive and nestin-positive (> 90% of cells, data not shown) (Fig. 1A). A2B5+ progenitor cells can only differentiate to glial cells, oligodendrocytes and astrocytes, while neural progenitor cells can become both glial cells and neurons. The human neurons, which were MAP-2 immunopositive (Fig. 1A), were immunoreactive for glutamate (44 ± 3%), and GABA (69 ± 5%), but not for tyrosine hydroxylase (TH), choline acetyltransferase (ChAT), or 5-hydroxytryptamine (5-HT) (data not shown). In addition, the human astrocytes and microglia were glial fibrillary acidic protein (GFAP)-positive and OX42 (CD11b/c)-positive, respectively (> 99% of cells) (Fig. 1A).
FIGURE 1.
Clustering of transcriptional profiles for five distinct types of human fetal CNS cells. (A) Phase contrast and immunostaining images of human fetal CNS cells. Five different types of human primary cells from cerebral cortex (8–21 days in vitro) were characterized by the CNS cell type-specific markers including nestin (green) for neural progenitor cells, A2B5 (green) for A2B5+ progenitor cells, MAP2 (green) for neurons, GFAP (green) for astrocytes and OX42 (green) for microglia. DAPI nuclear staining is shown in blue. All scale bars are 25 µm, except marker expression of neural progenitor cells and morphology of A2B5+ progenitor cells are 15 µm, and marker expression of A2B5+ progenitor cells, morphology and marker expression of astrocytes are 35 µm. (B) Hierarchical clustering result, showing that the two progenitor cell types, neural progenitor cells and A2B5+ progenitor cells, shared a high degree of similarity in transcriptional expression patterns (sublevel A). Two differentiated CNS cell types, neurons and astrocytes, also shared a high degree of similarity (sublevel B). Microglia showed a pattern of transcription different from the other four types of CNS cells. (C) Principal components analysis differentiating the five cell types.
Human Neural/Stem Cell/Drug Abuse-Focused cDNA Array (hNSDA Array)
To test the ability of the hNSDA array to discriminate CNS cell types, gene expression profiles in five types of CNS cells (Fig. 1A) obtained from two independent 20-week human fetal cerebral cortexes and maintained in vitro were analyzed. Hierarchical clustering was employed to examine overall similarities and differences in gene expression for the five types of CNS cells. The clustering program was supplied with the averages of all individual gene z-scores, which were obtained from replicate microarray data of each cell type. A two-way clustering analysis separated the five cell types into three distinct groups (Fig. 1B). The two types of human fetal progenitor cells (neural and A2B5+ progenitors) shared a high degree of similarity in transcriptional profiles, but diverged from those of differentiated CNS neurons and astrocytes, and from hematopoetic lineage-derived microglia. The similarity between neural and A2B5+ progenitor cells, and between neurons and astrocytes was also reflected by the principal components analysis (PCA) results (Fig. 1C), in a manner similar to that seen by hierarchical clustering. The six points for each cell type represent triplicates of all 2400 genes for two independent samples. These data suggest that the hNSDA array successfully distinguished differences between CNS cell populations by gene expression profiles.
Profiling of Cocaine-Regulated Genes in Primary Human CNS Cells
The five different types of primary human CNS cells described above were treated with 100 µM cocaine for 24 hr, and RNA from two independent paired experiments was subjected to transcriptional profiling with a hNSDA array. Z test analysis identified cocaine-responsive genes that fulfilled a stringent cut-off of p<0.05 and |z ratio|≥ 2.5. By these criteria, numbers of genes responsive to cocaine were 15 in neural progenitor cells, 29 in A2B5+ progenitor cells, 27 in neurons, 23 in microglia, and 15 in astrocytes (Table 1).
Table 1.
Cocaine-induced gene expression in human CNS cells.
| A. Neural progenitor cells | |||
|---|---|---|---|
| Gene Name | Gene Symbol | GenBank | Z ratio |
| amyloid beta (A4) precursor-like protein 2 | APLP2 | BC000373 | 12.9 |
| chloride channel Ka | CLCNKA | BC035373 | 8.7 |
| unc-51-like kinase 1 | ULK1 | NM003565 | 7.9 |
| neurogranin (protein kinase C substrate, RC3) | NRGN | BC002835 | 6.1 |
| insulin-like growth factor 1 receptor | IGF-1R | NM000875 | 2.9 |
| syndecan binding protein (syntenin) | SDCBP | BC013254 | 2.8 |
| Kruppel-like factor 8 | KLF8 | BC031355 | 2.6 |
| phospholipase C, beta 2 | PLCB2 | BC009009 | −2.6 |
| actin, gamma 2, smooth muscle, enteric (γ2 actin) | ACTG2 | BC012617 | −2.7 |
| NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | NDUFV1 | BC008146 | −2.7 |
| dopamine beta-hydroxylase (dopamine beta-monooxygenase) | DBH | BC017174 | −3.0 |
| villin 2 (ezrin) | VIL2 | BC013903 | −3.1 |
| protein kinase, Y-linked | PRKY | BC074851 | −6.5 |
| general transcription factor IIIC, polypeptide 3, 102kDa | GTF3C3 | BC015995 | −10.1 |
| nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | NFATC1 | BC033848 | −11.4 |
| B. A2B5+ progenitor cells | |||
|---|---|---|---|
| Gene Name | Gene Symbol | Genbank | Z ratio |
| fibroblast growth factor 13 | FGF13 | BC034340 | 8.1 |
| nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | NR3C1 | BC015610 | 3.6 |
| homer homolog 3 | HOMER3 | BC012113 | 3.5 |
| destrin (actin depolymerizing factor) | DSTN | BC009477 | 3.2 |
| filamin A, alpha (actin binding protein 280) | FLNA | BC014654 | 2.8 |
| insulin-like growth factor binding protein 4 | IGFBP4 | BC016041 | 2.7 |
| opioid growth factor receptor | OGFR | BC014137 | 2.5 |
| growth arrest and DNA-damage-inducible, alpha | GADD45A | BC011757 | 2.5 |
| galactosidase, alpha | GLA | BC002689 | −2.5 |
| DR1-associated protein 1 (negative cofactor 2 alpha) | DRAP1 | BC010025 | −2.6 |
| bone morphogenetic protein 8b (osteogenic protein 2) | BMP8B | BC023526 | −2.6 |
| glutamate receptor, ionotropic, N-methyl D-asparate-associated protein 1 (glutamate binding) | GRINA | BC041788 | −2.6 |
| sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G | SEMA3G | BC098104 | −2.6 |
| protein phosphatase 1, catalytic subunit, beta isoform | PPP1CB | BC002697 | −2.6 |
| enolase 2 (gamma, neuronal) | ENO2 | BC002745 | −2.8 |
| GNAS complex locus | GNAS | BC002722 | −2.9 |
| homeobox B5 | HOXB5 | BC008940 | −2.9 |
| v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | ERBB3 | BC002706 | −2.9 |
| SRY (sex determining region Y)-box 10 | SOX10 | BC007595 | −3.0 |
| chloride channel 6 | CLCN6 | BC050457 | −3.1 |
| dopamine beta-hydroxylase (dopamine beta-monooxygenase) | DBH | BC017174 | −3.2 |
| guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type | GNAL | BC050021 | −3.3 |
| natriuretic peptide precursor A | NPPA | BC005893 | −3.9 |
| tubulin, alpha 8 (α8 tubulin) | TUBA8 | BC074827 | −4.1 |
| villin 2 (ezrin) | VIL2 | BC013903 | −4.7 |
| Fas apoptotic inhibitory molecule 3 | FAIM3 | BC006401 | −5.5 |
| tubulin, alpha 3d (α3d tubulin) | TUBA3D | BC057810 | −5.6 |
| E2F transcription factor 4, p 107/p130-binding | E2F4 | BC033180 | −6.0 |
| S100 calcium binding protein G | S100G | BC112174 | −6.8 |
| C. Neurons | |||
|---|---|---|---|
| Gene Name | Gene Symbol | Genbank | Z ratio |
| cadherin 3, type 1, P-cadherin (placental) | CDH3 | BC014462 | 2.7 |
| signal sequence receptor, alpha (translocon-associated protein alpha) | SSR1 | BC007710 | 2.6 |
| sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A | SEMA7A | BC101643 | 2.6 |
| choline kinase beta | CHKB | BC024286 | −2.5 |
| solute carrier family 9 (sodium/hydrogen exchanger), member 1 (antiporter, Na+/H+, amiloride sensitive) | SLC9A1 | BC012121 | −2.5 |
| paired box gene 5 (B-cell lineage specific activator) | PAX5 | NM016734 | −2.5 |
| G protein-coupled receptor 132 | GPR132 | BC004555 | −2.6 |
| activin A receptor, type IIB | ACVR2B | BC096243 | −2.7 |
| protein tyrosine phosphatase, receptor type, B | PTPRB | BC051329 | −2.7 |
| latent transforming growth factor beta binding protein 2 | LTBP2 | BC032434 | −2.8 |
| G protein-coupled receptor kinase 5 | GRK5 | BC018116 | −2.8 |
| fibroblast growth factor 5 | FGF5 | BC071227 | −2.8 |
| v-raf murine sarcoma viral oncogene homolog B1 | BRAF | BC038966 | −2.9 |
| SMAD specific E3 ubiquitin protein ligase 1 | SMURF1 | BC008574 | −2.9 |
| POU domain, class 4, transcription factor 1 (BRN-3A) | POU4F1 | BC041859 | −2.9 |
| CD34 antigen | CD34 | BC03146 | −2.9 |
| COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | COPS4 | BC004302 | −3.0 |
| netrin G1 | NTNG1 | BC030220 | −3.0 |
| activator of basal transcription 1 | ABTI | BC048812 | −3.1 |
| distal-less homeobox 6 | DLX6 | BC069363 | −3.1 |
| EPH receptor B4 | EPHB4 | BC004264 | −3.1 |
| Kruppel-like factor 5 (intestinal) | KLF5 | BC042131 | −3.1 |
| gap junction protein, beta 5 (connexin 31.1) | GJB5 | BC004379 | −3.2 |
| ataxin 1 | ATXN1 | BC011026 | −3.2 |
| ubinuclein 1 | UBN1 | NM016936 | −3.2 |
| ring finger protein 150 | RNF150 | BC101992 | −3.6 |
| calcium-sensing receptor (hypocalciuric hypercalcemia 1, severe neonatal hyperparathyroidism) | CASR | BC104999 | −3.8 |
| D. Microglia | |||
|---|---|---|---|
| Gene Name | Gene Symbol | Genbank | Z ratio |
| BCL2-associated X protein | BAX | BC014175 | 7.8 |
| prominin 1 | PROM1 | BC012089 | 4.7 |
| integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | ITGA3 | BC015344 | 4.5 |
| GATA binding protein 1 (globin transcription factor 1) | GATA1 | BC009797 | 4.1 |
| Rho-associated, coiled-coil containing protein kinase 1 | ROCK1 | BC041849 | 3.4 |
| histone deacetylase 1 | HDAC1 | BC000301 | 3.3 |
| POU domain, class 4, transcription factor 2 | POU4F2 | NM004575 | 3.1 |
| ELK1, member of ETS oncogene family | ELK1 | BC048296 | 3.1 |
| interleukin 1, beta (IL-1β) | IL1B | BC008678 | 3.1 |
| synaptotagmin XI | SYT11 | BC004291 | 3.1 |
| RNA terminal phosphate cyclase-like 1 | RCL1 | BC001025 | 3.0 |
| guanylate binding protein 1, interferon-inducible, 67kDa | GBP1 | BC002666 | 2.9 |
| phosphoilpase D1, phophatidylcholine-specific | PLD1 | BC068976 | 2.8 |
| dual specificity phosphatase 13 | DUSP13 | BC009778 | 2.8 |
| mitogen-activated protein kinase 12 | MAPK12 | BC015741 | 2.7 |
| heparan sulfate proteoglycan 2 (perlecan) | HSPG2 | BC033152 | 2.5 |
| pyruvate dehydrogenase kinase, isozyme 3 | PDK3 | BC015948 | 2.5 |
| mitogen-activated protein kinase kinase kinase kinase 4 | MAP4K4 | BC010909 | −2.8 |
| transient receptor potential cation channel, subfamily A, member 1 | TRPA1 | NM007332 | −3.0 |
| gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) | GREM1 | BC069525 | −3.2 |
| collapsin response mediator protein 1 | CRMP1 | BC007898 | −3.2 |
| FK506 binding protein 12-rapamycin associated protein 1 | FRAP1 | NM004958 | −3.7 |
| S100 calcium binding protein G | S100G | BC112174 | −11.6 |
| E. Astrocytes | |||
|---|---|---|---|
| Gene Name | Gene Symbol | Genbank | Z ratio |
| chromobox homolog 3 (HP1 gamma homolog) | CBX3 | BC000954 | 2.9 |
| activin A receptor, type IB | ACVR1B | BC000254 | 2.8 |
| solute carrier family 1 (neutral amino acid transporter), member 5 | SLC1A5 | BC000062 | 2.8 |
| gamma-aminobutyric acid (GABA) A receptor, gamma 2 | GABRG2 | BC036030 | 2.7 |
| cytochrome P450, family 2, subfamily B, polypeptide 6 | CYP2B6 | BC067430 | 2.6 |
| tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (14-3-3 ε) | YWHAE | BC001440 | 2.6 |
| phosphoilpase C-like 1 | PLCL1 | BC101531 | 2.6 |
| catenin (cadherin-associated protein), alpha 2 | CTNNA2 | BC040458 | 2.5 |
| RAP1B, member of RAS oncogene family | RAP1B | BC000176 | 2.5 |
| tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator, HVEM) | TNFRSF14 | BC029848 | 2.5 |
| collagen, type VII, alpha 1 (epidermolysis bullosa, dystrophic, dominant and recessive) | COL7A1 | NM000094 | −2.8 |
| anti-Mullerian hormone receptor, type II | AMHR2 | NM020547 | −3.1 |
| glutamate receptor, ionotropic, kainate 5 | GRIKS | BC037358 | −4.2 |
| nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | NR3C1 | BC015610 | −5.7 |
| Fas apoptotic inhibitory molecule 3 | FAIM3 | BC006401 | −7.5 |
The FatiGO web tool, which extracts gene ontology terms that are significantly represented by groups of genes (Al-Shahrour et al., 2004), revealed that cocaine influenced distinct categories of genes in a cell-type specific manner (Supplementary Fig. 1). Supplementary figure 1 represents the ten most highly-represented categories influenced by cocaine for each type of cell. Although there were some categories that were altered across all or most of the five cell types (e.g., transcription, cellular protein metabolism, and phosphate metabolism), some ontology terms appeared to unique to certain cell types. Cocaine tended to down-regulate cytoskeleton-related genes including ezrin (villin 2), γ2 actin (actin, gamma 2, smooth muscle, enteric), α3d tubulin (tubulin, alpha 3d), and α8 tubulin (tubulin, alpha 8) in neural and/or A2B5+ progenitor cells (Tables 1A and 1B). Among these, ezrin was strongly down-regulated by cocaine in both neural (z ratio = − 3.1) and A2B5+ progenitor cells (z ratio = −4.7). In addition, cocaine down-regulated several transcription factors related to nervous system development such as DLX6 (distal-less homeobox 6), BRN-3A (POU domain, class 4, transcription factor 1), and PAX5 (paired box gene 5) in neurons (Table 1C). On the other hand, cocaine modified immunomodulatory and cell survival-related genes in microglia and astrocytes (Supplementary Fig. 1). Cocaine up-regulated the immunocytokine and pro-apoptotic genes IL-1β (interleukin 1, beta) and BAX (BCL2-associated X protein) in microglia (Table 1D), but down-regulated the immune response gene glucocorticoid receptor (nuclear receptor subfamily 3, group C, member 1), and up-regulated the anti-apoptotic genes 14-3-3 ε (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide) and HVEM (tumor necrosis factor receptor superfamily, member 14) in astrocytes (Table 1E). Thus, the effect of cocaine varies for different types of CNS cells that are involved in cerebral cortex development.
Chromatin remodeling has been suggested to be involved in cocaine-induced gene expression changes in neurons (Kumar, 2005). Exploration of cocaine-regulated genes using WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/), which aligns groups of genes along chromosomes, did not show a significant clustering of genes up-or down-regulated by cocaine.
The oPOSSUM web tool (http://burgundy.cmmt.ubc.ca/oPOSSUM/) can be used to identify transcription factor binding sites that are over-represented in the promoters of sets of genes. oPOSSUM was used to search for transcription factor binding sites common to multiple genes up-or down-regulated by cocaine. Six transcription factors, MZF1_5–13, SRF, Arnt, SP1, USF1, and Mycn, were identified as cocaine-induced transcriptional changes in the various cell types (Supplementary Table 2). Further analysis would be required to determine if these transcription factors are involved in the modulation of expression of these genes by cocaine.
Quantitative Real-Time RT-PCR
Quantitative real-time RT-PCR analyses of the ezrin, γ2 actin, α8 tubulin, DLX6, IL-1β , BAX, glucocorticoid receptor, and 14-3-3 ε gene products were performed for all five cell types. The quantitative real-time RT-PCR results were very similar to the results of the microarray (Table 2). Cocaine decreased expression of ezrin, γ2 actin and α8 tubulin, in neural progenitor cells (0.16 ± 0.05, 0.27 ± 0.02, and 0.32 ± 0.12, respectively) and A2B5+ progenitor cells (0.38 ± 0.07, 0.52 ± 0.05, and 0.23 ± 0.05, respectively), decreased expression of DLX6 (0.23 ± 0.11) in neurons, and increased expression of IL-1β (3.70 ± 0.19) and BAX (8.97 ± 2.63) in microglia. In astrocytes, decreased expression of glucocorticoid receptor (0.39 ± 0.03) and increased expression of 14-3-3 ε (2.10 ± 0.33) were confirmed.
Table 2.
Comparison between quantified values from the Micorarray and quantitative real-time RT-PCR
| Gene Name | Methods | Neural progenitor cells |
A2B5+ progenitor cells |
Neurons | Microglia | Astrocytes |
|---|---|---|---|---|---|---|
| Ezrin | Z ratio by Microarray | −3.09 | −4.75 | 0.14 | 0.36 | −0.79 |
| Relative expression by RT-PCR | 0.16±0.05*** | 0.38±0.07*** | 1.09±0.12 | 1.19±0.26 | 1.00±0.16 | |
| γ2 actin | Z ratio by Microarray | −2.68 | −1.99 | −0.98 | 1.01 | −1.29 |
| Relative expression by RT-PCR | 0.27±0.02** | 0.52±0.05** | 1.10±0.26 | 1.06±0.34 | 1.15±0.26 | |
| α8 tubulin | Z ratio by Microarray | −2.40 | −4.12 | 0.37 | −0.40 | −0.36 |
| Relative expression by RT-PCR | 0.32±0.12** | 0.23±0.05*** | 1.29±0.20 | 1.33±0.18 | 1.19±0.34 | |
| DLX6 | Z ratio by Microarray | −0.07 | −0.96 | −3.09 | 0.30 | −0.18 |
| Relative expression by RT-PCR | 0.80±0.12 | 0.89±0.08 | 0.23±0.11* | 0.74±0.15 | 0.95±0.18 | |
| IL-1β | Z ratio by Microarray | −0.05 | 0.77 | 1.95 | 3.08 | 1.12 |
| Relative expression by RT-PCR | 1.09±0.22 | 0.87±0.08 | 1.28±0.06 | 3.70±0.19** | 1.01±0.12 | |
| BAX | Z ratio by Microarray | −0.36 | −0.32 | −0.11 | 7.80 | 1.71 |
| Relative expression by RT-PCR | 1.09±0.30 | 0.94±0.09 | 1.02±0.11 | 8.97±2.63* | 0.87±0.19 | |
| Glucocorticoid receptor | Z ratio by Microarray | 0.92 | 3.62 | 3.31 | 0.75 | −5.72 |
| Relative expression by RT-PCR | 0.88±0.08 | 2.34±0.45* | 1.28±0.48 | 1.14±0.20 | 0.39±0.03*** | |
| 14-3-3 ε | Z ratio by Microarray | 0.75 | −0.78 | 1.29 | −0.03 | 2.57 |
| Relative expression by RT-PCR | 1.01±0.10 | 0.94±0.30 | 1.11±0.22 | 0.95±0.16 | 2.10±0.33* |
Data in red indicate significant differential expression compared to each respective control.
See “Methods” for detailed calculations of “Z ratio by Microarray”, “Relative expression by RT-PCR” represents ratios of cocaine-treated samples to respective controls.
p<0.05
p<0.01
p<0.001 compared to each control.
Although cocaine-induced down-regulation of α8 tubulin and γ2 actin (in neural and A2B5+ progenitor cells, respectively) did not fulfill our threshold criteria for positive transcripts from the microarray data (both genes p<0.001 but |z ratio|<2.5), quantitative real-time RT-PCR confirmed that both were significantly down-regulated by cocaine. Similarly, we found that cocaine significantly decreased cyclin A2 in neural and A2B5+ progenitor cells, and increased cyclin A2 in astrocytes (z ratio: neural, −1.0; A2B5, −1.1; astrocytes, 2.0). Although these changes did not meet the 2.5 z ratio criterion used in the present study, they were nonetheless statistically significant and consistent with similar changes found previously by quantitative RT-PCR (Lee et al., 2008). While it was necessary to employ the very restrictive 2.5 z ratio criterion due to the small sample size, this also would have minimized false-positives. However, only the transcripts most markedly affected by cocaine would have been detected in the present study.
DISCUSSION
Neonatal exposure to cocaine causes both morphological and neurobehavioral alterations in the developing human brain (Chiriboga et al., 1999; Bellini et al., 2000; Singer et al., 2008). The mechanisms involved in the adverse developmental effects of cocaine are, however, not fully known. Novikova and coworkers identified Wnt/cadherin and multiple apoptosis-signaling pathways as being involved in cocaine’s actions on brain development from gene expression profiling of the cortex of cocaine-treated mouse fetuses (Novikova et al., 2005a, 2005b). Nonetheless, the cell type or types and cell-type specific gene alterations related to developmental effects of cocaine are unknown.
Administration of cocaine during the second trimester only, the period when neocortical neurogenesis is most active, is capable of disturbing neocortical cytoarchitecture in non-human primates (Lidow et al., 2001). Cocaine has previously been shown to induce distinct effects on different types of CNS cells (Nassogne et al., 1995; Lee et al., 2008). We identified several cell type-specific effects of cocaine, including modulation of cytoskeleton-related transcripts in progenitor cells; CNS development-related transcription factors in neurons; and immune and cell survival-related transcripts in glial cells.
Cytoskeleton Organization and Biogenesis
Transcripts involved in cytoskeletal organization and biogenesis were down-regulated by cocaine in both neural and A2B5+ progenitor cells. One of these cytoskeleton-related transcripts is ezrin, a member of the ERM (ezrin-radixin-moesin) family of membrane-cytoskeleton linker proteins regionally expressed in the developing neuroepithelium (Gimeno et al., 2004). In cultured neurospheres, ezrin was specifically localized in filopodia of adherent neuronal progenitor cells (Gronholm et al., 2005). Ezrin, in addition to its role in the regulation of cell shape, adhesion, and migration, is also involved in cellular signaling pathways, like Rho and PI3-kinase (Louvet-Vallee, 2000). Ezrin may also be involved in early embryonic development (Dard et al., 2001).
A number of transcripts encoding subunits of actin and tubulin filaments, such as γ2 actin, α3d tubulin, and α8 tubulin, were down-regulated by cocaine in neural and/or A2B5+ progenitors. In eukaryotic cells, microtubule and actin filaments interact to form the cytoskeletal network involved in determination of cell architecture, mitosis, motility, and differentiation. Increased rates of cytoskeletal protein synthesis occur during chick brain development (Bryan et al., 1981). In contrast, cytoskeletal components are reduced in the frontal cortex of patients with neurodegenerative diseases, including Alzheimer’s disease, Down’s syndrome, and Pick’s disease (Pollack et al., 2003). Down-regulation of cytoskeletal genes in progenitor cells by cocaine might lead to disturbances in neural differentiation and migration.
Transcription Factors Related to CNS Development
Genes encoding transcription factors related to nervous system development, such as DLX6, BRN-3A, and PAX5, were down-regulated in neurons by cocaine. Among these, DLX6, a member of a homeobox transcription factor gene family homologous to the distal-less (Dll) gene of Drosophila, is primarily expressed in subpopulations of neurons in the developing forebrain with roles in forebrain development (Merlo et al., 2000; Panganiban and Rubenstein, 2002). Down-regulation of these transcription factors in neurons by cocaine might be a consequence of cocaine-induced cytoskeleton derangement in neural progenitors, which may impact ensuing cortical neurogenesis.
Immune Response and Apoptosis
Genes involved in immune response and apoptosis were specifically changed by cocaine in glial cells. Cocaine up-regulated the proinflammatory cytokine IL-1β and the pro-apoptotic regulator BAX in microglia. IL-1β, produced by activated microglia, exerts a number of effects on cell proliferation, differentiation, and apoptosis in the brain (Gibson et al., 2004). BAX, a member of BCL2 family, functions as an apoptotic activator by permeabilizing the outer mitochondrial membrane, leading to the release of apoptogenic proteins such as cytochrome c (Er et al., 2006). In fact, cocaine-induced up-regulation of Bcl2 family genes such as Bcl-x(L) in the brains of cocaine self-administering rats has been reported (Ahmed et al., 2005), and could reflect apoptotic pressure in the brain. It is also interesting to note that increased expression of Bcl-x(L) has been shown to stimulate synaptic transmission (Jonas et al., 2003).
Microglia are detected in the developing CNS during the period of neurogenesis (Ashwell, 1991). One role of microglia in the developing CNS is thought to be to eliminate dead cells and debris by phagocytosis. A developmental role of microglia, in directing migration and differentiation of embryonic neural progenitor cells, has also been suggested (Aarum et al., 2003). Therefore, abnormal activation and/or apoptosis of microglia by cocaine might interrupt microglial orchestration of neurogenesis during development.
In contrast to microglia, cocaine down-regulated the immunoreactive gene glucocorticoid receptor, while up-regulating the anti-apoptotic genes 14-3-3 ε and HVEM in astrocytes. The glucocorticoid receptor, a ligand-activated transcription factor, has previously been reported to be down-regulated by cocaine in human astrocytoma cells (Malaplate-Armand et al., 2005). Activation of glucocorticoid receptors causes anti-inflammatory and immunosuppressive actions through inhibition of the pro-inflammatory transcription factor NF-kappa B activity, by induction of I kappa B alpha (Auphan et al., 1995). 14-3-3 ε mediates an anti-apoptotic signal through interaction with Bad, resulting in retention of Bad in the cytoplasm, preventing subsequent cytotoxic interaction with Bcl-x(L) in mitochondria (Won et al., 2003). HVEM, a member of the TNF-receptor superfamily, protects cells against TNF-α-mediated apoptosis by inhibiting caspase-3 and caspase-8 activation (Matsui et al., 2002). Astrocytes, the guiding structures for migrating neurons during brain development, provide maintenance, support, and protection to neurons (Kirchhoff et al., 2001). Changes of immunological and cell survival signaling of astrocytes by cocaine might affect their structural and trophic functions.
Monoaminergic System Transcripts
Cocaine induces its primary effects in the brain through monoaminergic systems. Nevertheless, the expression of dopamine receptors (D1-5), adrenergic receptors (α-1A, 1B, 1D, 2A, 2B, β-1, 3 ), serotonin receptors (1E, 1F, 2A-C, 3A, 3B, 4, 5A, 6,7), and all three types of monoamine transporters including dopamine, norepinephrine, and serotonin transporters were not changed by cocaine in our CNS cell cultures, at least at the transcriptional level (Supplementary Table 3). The only neurotransmitter-related transcript which was changed by cocaine was dopamine beta-hydroxylase (DBH), which catalyzes the synthesis of norepinephrine from dopamine in noradrenergic neurons. DBH was decreased (z ratios = −3.0 and −3.2) in both neural and A2B5+ progenitor cells (Tables 1A, 1B and Supplementary Table 3). Cocaine also tended to increase DBH transcript expression in microglia (z ratio = 3.4), although this difference was not significant by our criteria. The functional significance of DBH in neural and A2B5+ progenitor cells is, however, unknown.
Clinical Relevance
Cocaine exposure in utero is associated with multiple and prolonged behavioral and neurological changes. In addition to slower intrauterine growth, these babies are born with a smaller head circumference and neurological abnormalities such as hypertonia and tremor (Chiriboga et al., 1999). Persistent developmental effects of fetal cocaine exposure have also been found (Singer et al., 2008). Despite neurological indications, relatively little is known about the direct effect of cocaine in the developing brain, and the mechanisms by which prenatal cocaine exposure causes CNS injury are not clear. We have studied cocaine-induced changes in gene expression patterns in order to obtain a better understanding of the mechanisms involved in the adverse effects of in utero cocaine exposure.
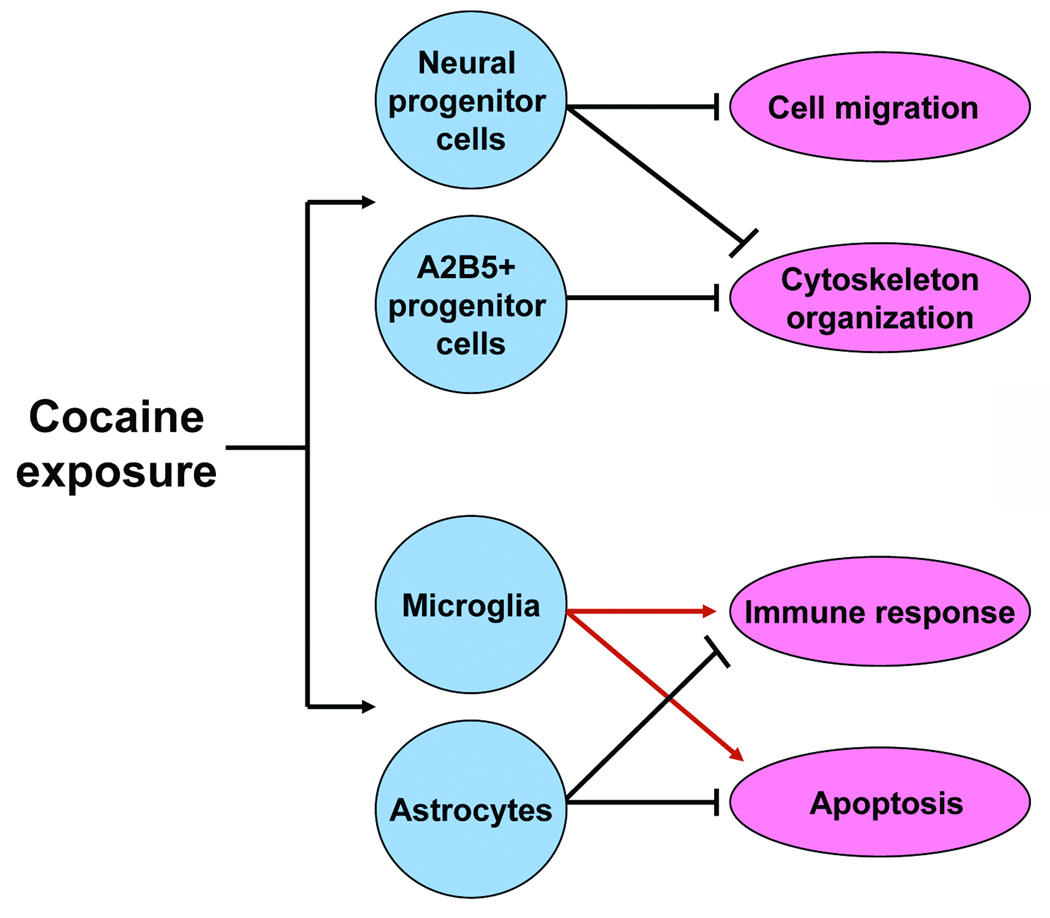
CONCLUSION
The present study indicates that cocaine can directly alter gene expression profiles in a cell-type specific manner, even in non-dopaminergic, non-serotonergic CNS cells. We found that cocaine down-regulates cytoskeleton-related genes in neural and A2B5+ progenitor cells, but modulates immune response and cell death-related genes in glial cells (Fig. 2). These data suggest that exposure of infants to cocaine could have adverse effects on the development of both neural progenitor cells and glia.
FIGURE 2.
Schematic diagram illustrating the cell type-specific action of cocaine on gene transcription. Alterations in cytoskeleton-related gene expression in multipotent cortical progenitor cells by cocaine may interrupt morphological development and migration of progenitors. Microglia provide neuronal immunomodulatory functions and play a developmental role during neurogenesis, and astrocytes provide trophic, metabolic and structural support for neurons. Cocaine may derange immunoregulatory functions and survival of glial cells. Activation of each specific response is denoted by →, inhibition of each specific response is denoted by  .
.
We previously found that cocaine inhibits neural progenitor cell proliferation, and that this effect was due to oxidative endoplasmic reticulum stress consequent to P450 metabolism of cocaine, leading to phosphorylation of EIF2α, up-regulation of ATF4, and down-regulation of cylin A (Lee et al., 2008). Although this effect of cocaine is likely to contribute to developmental brain injury, it does not necessarily comprise the entire reason for cocaine’s adverse developmental effects. It is necessary to understand the effects of cocaine on brain cell types other than neural progenitors, as well as obtaining a more comprehensive survey of the molecular effects of cocaine.
This study provides a general survey of the molecular effects of cocaine on the major cell types which are present in the developing brain, using primary human cells in vitro. At the present time, which of these molecular changes contribute to the adverse effects of cocaine on brain development is unknown. Future studies will focus on determining which if any of these effects are due to P450 metabolism of cocaine and consequent oxidative stress, how these various molecular events are interrelated, and which of these effects contribute to the adverse effects of cocaine on brain development. If the molecular events which lead to cocaine-induced developmental brain injury can be determined, it may be possible to develop drugs which can be used to prevent this form of developmental damage.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank William H. Wood III (Gene Expression and Genomics Unit, IRP, NIA IRP) for his expert assistance. We also thank Cindy Ambriz for preparing the manuscript.
Supported by the IRPs of NIDA and NIA, NIH, DHHS. U.S. Provisional Application No. 60/893,218 filed 06 Mar 2007: "Cytochrome P450 Inhibitors for Treatment of Cocaine-Induced Fetal Brain Injury" (CTL, WJF)
REFERENCES
- 1.Aarum J, Sandberg K, Haeberlein SL, et al. Migration and differentiation of neural precursor cells can be directed by microglia. Proc Natl Acad Sci USA. 2003;100:15983–15988. doi: 10.1073/pnas.2237050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed SH, Lutjens R, van der Stap L, et al. Gene expression evidence for remodeling of lateral hypothalamic circuitry in cocaine addiction. Proc Natl Acad Sci USA. 2005;102:11533–11538. doi: 10.1073/pnas.0504438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akbari HM, Whitaker-Azmitia PM, Azmitia EC. Prenatal cocaine decreases the trophic factor S-100 beta and induced microcephaly: reversal by postnatal 5-HT1A receptor agonist. Neurosci Lett. 1994;170:141–144. doi: 10.1016/0304-3940(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 4.Al-Shahrour F, Diaz-Uriate R, Dopazo J. FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 5.Ashwell K. The distribution of microglia and cell death in the fetal rat forebrain. Brain Res Dev Brain Res. 1991;58:1–12. doi: 10.1016/0165-3806(91)90231-7. [DOI] [PubMed] [Google Scholar]
- 6.Auphan N, DiDonato JA, Rosette C, et al. Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 7.Bellini C, Massocco D, Serra G. Prenatal cocaine exposure and the expanding spectrum of brain malformations. Arch Intern Med. 2000;160:2393. doi: 10.1001/archinte.160.15.2393. [DOI] [PubMed] [Google Scholar]
- 8.Bryan RN, Bossinger J, Hayashi M. Tubulin and actin synthesis during brain development. Dev Biol. 1981;81:349–355. doi: 10.1016/0012-1606(81)90299-2. [DOI] [PubMed] [Google Scholar]
- 9.Chiriboga CA, Brust JC, Bateman D, et al. Dose-response effect of fetal cocaine exposure on newborn neurologic function. Pediatrics. 1999;103:79–85. doi: 10.1542/peds.103.1.79. [DOI] [PubMed] [Google Scholar]
- 10.Dard N, Louvet S, Santa-Maria A, et al. In vivo functional analysis of ezrin during mouse blastocyst formation. Dev Biol. 2001;233:161–173. doi: 10.1006/dbio.2001.0192. [DOI] [PubMed] [Google Scholar]
- 11.Delaney-Black V, Covington C, Ostrea E, Jr, et al. Prenatal cocaine and neonatal outcome: Evaluation of dose-response relationship. Pediatrics. 1996;98:735–740. [PubMed] [Google Scholar]
- 12.Er E, Oliver L, Cartron PF, et al. Mitochondria as the target of the pro-apoptotic protein Bax. Biochim Biophys Acta. 2006;1757:1301–1311. doi: 10.1016/j.bbabio.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 13.Gibson RM, Rothwell NJ, Le Feuvre RA. CNS injury: the role of the cytokine IL-1. Vet J. 2004;168:230–237. doi: 10.1016/j.tvjl.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Gimeno L, Corradi A, Cobos I, et al. Ezrin gene, coding for a membrane-cytoskeleton linker protein, is regionally expressed in the developing mouse neuroepithelium. Gene Expr Patterns. 2004;4:749–754. doi: 10.1016/j.modgep.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Gronholm M, Teesalu T, Tyynela J, et al. Characterization of the NF2 protein merlin and the ERM protein ezrin in human, rat, and mouse central nervous system. Mol Cell Neurosci. 2005;28:683–693. doi: 10.1016/j.mcn.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 16.He N, Bai J, Champoux M, et al. Neurobehavioral deficits in neonatal rhesus monkeys exposed to cocaine in utero. Neurotoxicol Teratol. 2004;26:13–21. doi: 10.1016/j.ntt.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Jonas EA, Hoit D, Hickman JA, et al. Modulation of synaptic tranmission by the BCL-2 family proteína BCL-xL. J Neurosci. 2003;23:8423–8431. doi: 10.1523/JNEUROSCI.23-23-08423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalasinsky KS, Bosy TZ, Schmunk GA, et al. Regional distribution of cocaine in postmortem brain of chronic human cocaine users. J Forensic Sci. 2000;45:1041–1048. [PubMed] [Google Scholar]
- 19.Kirchhoff F, Dringen R, Giaume C. Pathways of neuron-astrocyte interactions and their possible role in neuroprotection. Eur Arch Psychiatry Clin Neurosci. 2001;251:159–169. doi: 10.1007/s004060170036. [DOI] [PubMed] [Google Scholar]
- 20.Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci USA. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A, Choi KH, Renthal W, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48(2):303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Lee CT, Chen J, Hayashi T, et al. A mechanism for the inhibition of neural progenitor cell proliferation by cocaine. PLoS Med. 2008;5:e117. doi: 10.1371/journal.pmed.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- 24.Lidow MS. Prenatal cocaine exposure adversely affects development of the primate cerebral cortex. Synapse. 1995;21:332–341. doi: 10.1002/syn.890210408. [DOI] [PubMed] [Google Scholar]
- 25.Lidow MS, Bozian D, Song ZM. Cocaine affects cerebral neocortical cytoarchitecture in primates only if administered during neocortical neuronogenesis. Brain Res Dev Brain Res. 2001;128:45–52. doi: 10.1016/s0165-3806(01)00139-0. [DOI] [PubMed] [Google Scholar]
- 26.Lidow MS, Song ZM. Primates exposed to cocaine in utero display reduced density and number of cerebral cortical neurons. J Comp Neurol. 2001;435:263–275. doi: 10.1002/cne.1028. [DOI] [PubMed] [Google Scholar]
- 27.Louvet-Vallee S. ERM proteins: from cellular architecture to cell signaling. Biol Cell. 2000;92:305–316. doi: 10.1016/s0248-4900(00)01078-9. [DOI] [PubMed] [Google Scholar]
- 28.Malaplate-Armand C, Ferrari L, Masson C, et al. Down-regulation of astroglial CYP2C, glucocorticoid receptor and constitutive androstane receptor genes in response to cocaine in human U373 MG astrocytoma cells. Toxicol Lett. 2005;159:203–211. doi: 10.1016/j.toxlet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Matsui H, Hikichi Y, Tsuji I, et al. LIGHT, a member of the tumor necrosis factor ligand superfamily, prevents tumor necrosis factor-alpha-mediated human primary hepatocyte apoptosis, but not Fas-mediated apoptosis. J Biol Chem. 2002;277(51):50054–50061. doi: 10.1074/jbc.M206562200. [DOI] [PubMed] [Google Scholar]
- 30.Merlo GR, Zerega B, Paleari L, et al. Multiple functions of Dlx genes. Int J Dev Biol. 2000;44:619–626. [PubMed] [Google Scholar]
- 31.Nassogne MC, Evrard P, Courtoy PJ. Selective neuronal toxicity of cocaine in embryonic mouse brain cocultures. Proc Natl Acad Sci USA. 1995;92:11029–11033. doi: 10.1073/pnas.92.24.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassogne MC, Gressens P, Evrard P, et al. In contrast to cocaine, prenatal exposure to methadone does not produce detectable alterations in the developing mouse brain. Brain Res Dev Brain Res. 1998;110:61–67. doi: 10.1016/s0165-3806(98)00094-7. [DOI] [PubMed] [Google Scholar]
- 33.Novikova SI, He F, Bai J, et al. Cocaine-induced changes in the expression of apoptosis-related genes in the fetal mouse cerebral wall. Neurotoxicol Teratol. 2005a;27:3–14. doi: 10.1016/j.ntt.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Novikova SI, He F, Bai J, et al. Neuropathology of the cerebral cortex observed in a range of animal models of prenatal cocaine exposure may reflect alterations in genes involved in the Wnt and cadherin systems. Synapse. 2005b;56:105–116. doi: 10.1002/syn.20134. [DOI] [PubMed] [Google Scholar]
- 35.Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 36.Pollak D, Cairns N, Lubec G. Cytoskeleton derangement in brain of patients with Down syndrome, Alzheimer's disease and Pick's disease. J Neural Transm Suppl. 2003:149–158. doi: 10.1007/978-3-7091-6721-2_13. [DOI] [PubMed] [Google Scholar]
- 37.Singer LT, Salvator A, Arendt R, et al. Effects of cocaine/polydrug exposure and maternal psychological distress on infant birth outcomes. Neurotoxicol Teratol. 2002;24:127–135. doi: 10.1016/s0892-0362(01)00208-2. [DOI] [PubMed] [Google Scholar]
- 38.Singer LT, Nelson S, Short E, et al. Prenatal cocaine exposure: drug and environmental effects at 9 years. J Pediatr. 2008;153(1):105–111. doi: 10.1016/j.jpeds.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strausberg RL, Feingold EA, Klausner RD, et al. The mammalian gene collection. Science. 1999;286:455–457. doi: 10.1126/science.286.5439.455. [DOI] [PubMed] [Google Scholar]
- 40.Won J, Kim DY, La M, et al. Cleavage of 14-3-3 protein by caspase-3 facilitates bad interaction with Bcl-x(L) during apoptosis. J Biol Chem. 2003;278:19347–19351. doi: 10.1074/jbc.M213098200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.