Abstract
Human herpesvirus 8 (HHV-8) is the etiological agent of Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. It is postulated that CD8+ T cell responses play an important role in controlling HHV-8 infection and preventing development of disease. In this study, we investigated monofunctional and polyfunctional CD8+ T cell responses to HHV-8 lytic proteins gB (glycoprotein B) and K8.1 and latency proteins LANA-1 (latency-associated nuclear antigen-1) and K12. On the basis of our previous findings that dendritic cells (DC) reveal major histocompatibility complex (MHC) class I epitopes in gB, we used a DC-based system to identify 2 novel epitopes in gB, 2 in K8.1, 5 in LANA-1, and 1 in K12. These new HHV-8 epitopes activated monofunctional and polyfunctional CD8+ T cells that produced various combinations of gamma interferon, interleukin 2, tumor necrosis factor alpha, macrophage inhibitory protein 1β, and cytotoxic degranulation marker CD107a in healthy HHV-8-seropositive individuals. We were also able to detect HHV-8-specific CD8+ T cells in peripheral blood samples using HLA A*0201 pentamer complexes for one gB epitope, one K8.1 epitope, two LANA-1 epitopes, and one K12 epitope. These immunogenic regions of viral lytic and latency proteins could be important in T cell control of HHV-8 infection.
Human herpesvirus 8 (HHV-8), also referred to as Kaposi's sarcoma-associated herpesvirus, is a gammaherpesvirus that causes Kaposi's sarcoma (KS), primary effusion lymphoma, and multicentric Castleman's disease. The importance of developing effective prevention and treatment for HHV-8 infection is evident in that KS, a neoplasm of endothelial origin, continues to be the most common cancer among human immunodeficiency virus (HIV)-infected patients (8). KS is also the leading cause of cancer in children in sub-Saharan Africa (7). Although the incidence of KS in HIV-infected persons declined with the advent of antiretroviral therapy (ART) (10), KS can occur in persons on ART with suppressed HIV infection and high CD4+ T cell counts (25).
The immune responses responsible for controlling HHV-8 infection and preventing KS are not clear. CD8+ T cell immunity likely plays a significant role in HHV-8 infection, as these cells have been shown to be crucial in controlling infection caused by the other human gammaherpesvirus, i.e., Epstein-Barr virus (EBV) (11, 14). In support of this hypothesis, our laboratory (40-42) and others (4-6, 12, 19, 23, 26-28, 31, 32, 36, 37, 43, 44) have shown that CD8+ T cells produce gamma interferon (IFN-γ) in response to HHV-8 immunodominant epitopes presented by major histocompatibility complex class I (MHC-I) in HHV-8-seropositive individuals. Little is known whether T cells produce other immune mediators in response to HHV-8 infection. Indeed, polyfunctional T cells, i.e., single cells producing two or more immune mediators, have been linked to control of HIV and other persistent infections (1, 24, 29, 33) and could play a role in controlling HHV-8 infection. In one recent study, HHV-8 epitope-specific, polyfunctional T cells were detected in patients with multicentric Castleman's disease, but these cells did not differ in number from those in healthy controls (13). Another study has found that patients with controlled KS had HHV-8-specific CD8+ T cells that secreted IFN-γ and tumor necrosis factor alpha (TNF-α) but that patients with progressive disease had weaker and less polyfunctional CD8+ T cells (2).
HHV-8 epitope-specific monofunctional and polyfunctional T cell immunity could be important in development of HHV-8 vaccines that induce T cell responses that target these viral epitopes. In the present study, we therefore investigated CD8+ T cell responses to two HHV-8 lytic proteins, gB (glycoprotein B) and K8.1, and two latency proteins, LANA-1 (latency associated nuclear antigen-1) and K12. We previously showed that optimal induction of T cell reactivity to the HHV-8 protein gB required 1 week of stimulation with peptide-loaded, autologous, mature, monocyte-derived dendritic cells (DC) (40). Using this enhanced DC-T cell stimulation system, we now have revealed several new epitopes for these four lytic and latency HHV-8 proteins in healthy HHV-8-seropositive individuals, which induce both monofunctional and polyfunctional CD8+ T cells. These regions of HHV-8 could be critical in understanding HHV-8 immunopathogenesis and in vaccine development.
MATERIALS AND METHODS
Study subjects.
Healthy, HIV-1-negative subjects were selected based on their HHV-8 antibody status and MHC-I genotype, and written informed consent was obtained. Detection of HHV-8 serum antibody specific for viral lytic antigens was done using an indirect immunofluorescence assay (40). High-resolution HLA molecular typing was conducted by the University of Pittsburgh Medical Center Tissue Typing Laboratory. The donors were classified into HLA A*0201-positive HHV-8-seropositive and HLA A*0201-positive HHV-8-seronegative groups.
Synthetic peptides.
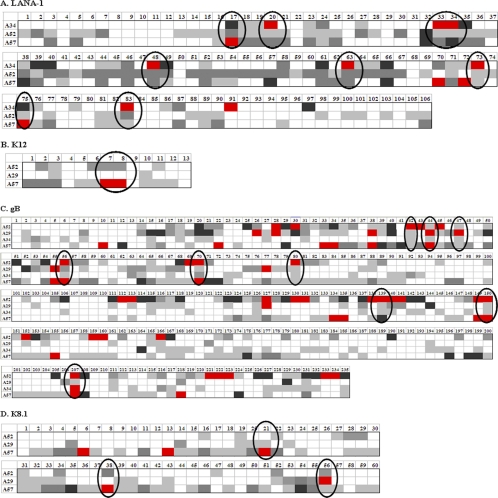
For the initial studies, libraries of 15-mer peptides overlapping by 11 amino acids (aa) derived from K12, gB, and K8.1 protein sequences were synthesized (PEPscreen; Sigma, St. Louis, MO). For the larger protein LANA-1, a library of 15- to 20-mer peptides overlapping by 11 aa was used. Protein sequences were obtained from the National Center for Biotechnology Information (NCBI) database, with accession number AAD46501 for LANA-1, accession number AAD46499 for K12, accession number ABD28851 for gB, and accession number ABD28902 for K8.1. As LANA-1 contains a large repeat region, we used one set of representative peptides to span this region (peptides 59 to 75) (Fig. 1A). For epitope mapping studies, putative optimal 9-mer peptides were synthesized based on anchor residues for HLA A*0201 (30) as well as peptides 1 N or C terminus amino acid shorter or longer than optimal (15). The following previously published 9-mer, HLA A*0201-restricted HHV-8 epitopes were also used: LANA-1238-246 (WATESPIYV) (12), LANA-11116-1124 (QMARLAWEA) (12), K1217-25 (LLNGWRWRL) (6), gB492-500 (LMWYELSKI) (40), and K8.1209-217 (LVLILYLCV) (5).
FIG. 1.
IFN-γ ELISPOT responses to four HHV-8 latency and lytic proteins. CD14− PBMC from healthy HLA A*0201-positive HHV-8-seropositive donors were stimulated with autologous, mature DC that were loaded with overlapping 15- to 20-mer peptides derived from each of the HHV-8 proteins LANA-1, K12, gB, and K8.1. IFN-γ production was measured by a DC-enhanced ELISPOT assay, and the number of spots produced by cells without peptide was subtracted from the number of spots produced by cells with peptide to give net values for spots. The donors (three or four donors used for each protein) are listed in each row, while the representative peptide numbers are listed in each column. The colors for the boxes represent the net number of IFN-γ spots per 106 cells as follows: white, <1 spot; light gray, 1 to 199 spots; dark gray, 200 to 399 spots; black, 400 to 599 spots; and red, ≥600 spots. Hot spots are circled for LANA-1 (A), K12 (B), gB (C), and K8.1 (D).
T cell stimulation.
CD14+ cells were isolated from donor peripheral blood mononuclear cells (PBMC) using anti-CD14 monoclonal antibody (MAb)-coated magnetic beads (Miltenyi Biotec, Auburn, CA). Immature DC were generated by culturing the CD14+ cells (1 × 106) for 7 days in AIM-V medium (GIBCO, Grand Island, NY) containing 1,000 U/ml each of recombinant interleukin-4 (IL-4) (R&D Systems, Minneapolis, MN) and recombinant granulocyte-monocyte colony-stimulating factor (GM-CSF) (Bayer Health Care, Seattle, WA) at 37°C in a 5% CO2 atmosphere. Fresh IL-4 and GM-CSF were added every 2 days. On day 5, CD40 ligand (CD40L) (1 μg/ml; R&D) was added to induce maturation. Mature DC were loaded with peptide (2 μg/ml) for 2 h at 37°C. To assess T cell reactivity, we used DC as antigen-presenting cells (APC) and autologous CD14− cells as responders (40). Antigen-loaded DC or DC without antigen (1 × 104 cells) were mixed with CD14− cells (1 × 105) and cocultured for 7 days in AIM-V at 37°C, with 100 U/ml IL-2 (Chiron, Emeryville, CA) added on day 2. T cell stimulation was assessed for HHV-8-seronegative donors as controls.
Single-cell IFN-γ production.
Following a 7-day coculture of peptide-loaded mature DC stimulators and CD14− cell responders, our DC-enhanced, single-cell IFN-γ enzyme-linked immunospot (ELISPOT) assay was performed based on our previously described method for assessing T cell reactivity (40). Briefly, nitrocellulose-bottomed, 96-well ELISPOT plates (Millipore, Bedford, MA) were precoated with an anti-IFN-γ monoclonal antibody (10 μg/ml; Mabtech, Mariemont, OH) overnight at 4°C and then blocked for 2 h at 37°C with RPMI 1640 medium (GIBCO) supplemented with 10% heat-inactivated human AB+ serum (Gemini Bio-Products, West Sacramento, CA). Autologous PBMC (1 × 104) were loaded with peptide (2 μg/ml) for 2 h at 37°C and then combined with week-long T cell cultures as described above to serve as APC. These samples were added to ELISPOT plates, cultured overnight at 37°C, and developed as previously described (9). Spots were counted using an ELISPOT reader (AID, Strassberg, Germany). The results were expressed as numbers of virus-specific spot-forming cells per 106 cells by subtracting the number of spots in T cell cultures stimulated by DC without peptide from the number of spots induced by DC with peptide. T cell responses were considered positive if the number of spots produced by cells stimulated with peptide was greater than the number of spots produced by cells not stimulated with peptide.
Generation of T cell lines.
HHV-8 epitope-specific T cell lines were generated to confirm the novel epitopes and their HLA A*0201 restriction. Frozen PBMC were thawed, washed, and resuspended in RPMI 1640 medium with 10% heat-inactivated fetal calf serum (Cellgro, Lawrence, KS). Cells were added to a 24-well plate at 2 × 106 cells per well and kept at 37°C. Peptides were added at 10 μg/ml. On day 3, IL-2 was added (100 U/ml), with fresh IL-2 added every 3 days for a total of 15 days. On day 15, a B cell line that expresses only one MHC-I haplotype, HLA A*0201, was used as APC; a B cell line that does not express HLA (null) was used as a control (34). The B cells were loaded with peptide (2 μg/ml) for 2 h at 37°C and then used as APC with the 15-day-cultured cells in an IFN-γ ELISPOT assay performed as described above.
MHC-I binding assay.
HLA A*0201 binding experiments were performed by ProImmune (Oxford, United Kingdom) with their class I REVEAL binding assay. Briefly, the assay determines the binding of a peptide to an HLA molecule by its ability to stabilize the MHC-peptide complex. Binding is scored relative to the level for a positive control involving a known T cell epitope with very strong binding properties. A score is reported quantitatively as a percentage of the signal generated by the test peptide versus the level for the positive-control peptide.
Pentamer staining.
Along with analysis of immune responses by the IFN-γ ELISPOT assay, MHC-I pentamers were also used to quantify epitope-specific CD8+ T cells. On the basis of the results for the ProImmune class I REVEAL binding assay described above, 5 high-affinity HLA-A*0201 binding peptides were used to generate MHC class I-peptide pentamer complexes (ProVE pentamers; ProImmune). Frozen PBMC (1 × 106 to 2 × 106 cells per well) were thawed, washed, and then incubated for 10 min with unlabeled pentamer (0.5 μg per test). The cells were washed and then stained for 20 min in the dark with phycoerythrin (PE)-conjugated Pro5 Fluorotag (8 μl per well; ProImmune), CD3-allophycocyanin-Cy7 clone SK7 (5 μl per well; BD), and CD8-peridinin chlorophyll protein (PerCP)-Cy5.5 clone SK1 (20 μl per well; BD). CD4-V450 clone RPA-T4, CD19-V450 clone HIB19, and CD14-V450 clone MOP9 (all from BD) were also added at 5 μl each per well for exclusion gating. Cells were washed and fixed with BD stabilizing solution and analyzed with an LSR II flow cytometer (BD Immunocytometry Systems, San Diego, CA).
ICS and polychromatic flow cytometry.
Intracellular staining (ICS) for various immune mediators was performed as described previously (1, 20), with minor modifications. On day 7 of the DC-enhanced T cell cultures described above, cells were collected and plated (1 × 106 to 2 × 106 cells per well). The peptide stimulus (10 μg/ml), costimulatory CD28/49d MAb (1 μg/ml; BD Biosciences, San Jose, CA), monensin (5 μg/ml; Sigma), brefeldin A (5 μg/ml; Sigma), and anti-CD107a-fluorescein isothiocyanate (anti-CD107a-FITC) (20 μl per well; BD) were added. Unstimulated cells were also included. The cells were incubated for 6 h at 37°C, and then EDTA (2 mM) (GIBCO) was added. The cells were washed and resuspended in 100 μl phosphate-buffered saline (PBS) (GIBCO) per well containing 1 μl/well aqua viability dye (Invitrogen) for 30 min in the dark. The cells were washed and resuspended in 1× fluorescence-activated cell sorting (FACS) lysis solution (BD) and kept at 4°C overnight. Cells were washed and permeabilized using 1× Perm solution (BD) and then washed and stained for 45 min in the dark with the directly conjugated antibodies CD3-allophycocyanin-Cy7, TNF-α-PE-Cy7, macrophage inhibitory protein 1β (MIP-1β)-PE, IL-2-allophycocyanin, and IFN-γ-AF700 at 5 μl each per well and CD8-PerCP-Cy5.5 (all from BD) at 20 μl per well. CD4-V450, CD19-V450, and CD14-V450 (all from BD) were also added at 5 μl each per well for exclusion gating. Cells were washed and fixed with BD stabilizing solution and analyzed with an LSR II flow cytometer (BD Immunocytometry Systems). Results were analyzed with Boolean gating to create the full array of possible cytokine combinations using BDFACS Diva (version 6.0) software. Polyfunctional bar and pie charts for each peptide were created using Simplified Presentation of Incredibly Complex Evaluations (SPICE) software (version 4.2.3; provided by Mario Roederer, VRC/NIAID/NIH).
Statistics.
We used analysis of variance (ANOVA) and the Student t test for comparisons between groups.
RESULTS
Novel T cell epitopes in HHV-8 lytic and latency proteins.
To define MHC-I epitopes that generate CD8+ T cell responses to HHV-8 proteins, a DC-enhanced IFN-γ ELISPOT assay was used. This was based on our previous finding that this method was necessary to reveal an epitope in HHV-8 gB due to the nonrobust nature of the immune response to HHV-8 in healthy HHV-8-seropositive persons (40). T cells were stimulated with autologous mature DC that were loaded with overlapping peptides derived from HHV-8 LANA-1, K12, gB, and K8.1, and single-cell IFN-γ production was measured by the ELISPOT assay.
The IFN-γ ELISPOT assay results for four HLA A*0201-positive HHV-8-seropositive donors (A29, A34, A52, and A57) to peptides from each viral protein are displayed in Fig. 1. For the larger proteins LANA-1 and gB, peptides containing possible epitopes, termed “hot spots” (Fig. 1, circled areas), were defined as peptides that generated a positive response above the background level of mock-stimulated cells for all donors tested, with at least one donor responding in the highest category (red boxes). For the smaller proteins K12 and K8.1, we considered hot spots to be those peptides that the majority of donors responded to above the background level, with at least one donor responding in the highest category. For LANA-1, out of 106 peptides tested, we found 8 hot spots (Fig. 1A). For K12, out of 13 peptides tested, we found 1 hot spot (Fig. 1B). For gB, out of 235 peptides tested, we found 9 hot spots, including 1 containing our previously described epitope (40) (Fig. 1C). For K8.1, out of 60 peptides tested, we found 3 hot spots, including 2 containing previously described epitopes (5, 43) (Fig. 1D). Two HLA A*0201-positive HHV-8-seronegative donors did not respond above the background level to any of these hot spots from the four viral proteins (data not shown). Thus, these hot spots represented regions of the four HHV-8 proteins that displayed positive reactivity associated with HHV-8 seropositivity.
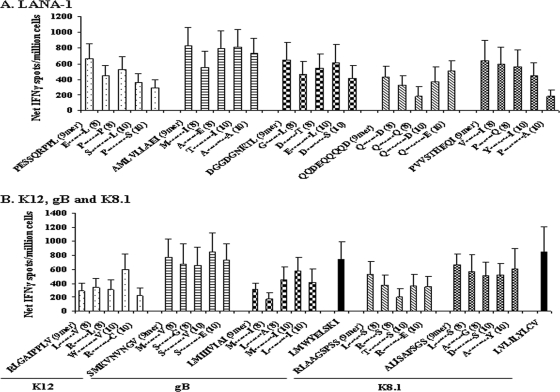
We next mapped minimal epitope sequences for 10 of these hot-spot peptides. To determine the minimal, optimal epitope, we used peptide families consisting of the putative optimal 9-mer based on anchor residues for HLA A*0201, along with peptides with 1 N or C terminus amino acid truncation (8-mers) or extension (10-mers). Initially, we adapted the conventional method to define optimal T cell epitopes, i.e., T cell response to different concentrations of N- and C-terminal extensions and truncations of peptide in a 16-h ELISPOT assay, to our 7-day extended assay. However, stimulating PBMC for 7 days with DC loaded with 5 different 10-fold concentrations of these peptide families did not differentiate a dominant peptide response (data not shown). Therefore, we tested the peptide families for the optimal epitopes by stimulating the cells for 7 days with a single concentration of peptide (2 μg/ml), followed by a conventional 16-h ELISPOT assay using the same peptide. We assessed five peptide families from LANA-1, one from K12, two from gB, and two from K8.1. One of the hot spots that we identified (K8.1 peptide 38 [Fig. 1D]) contained a published 15-mer epitope (43). Therefore, we included this peptide family to determine the minimal epitope sequence. Two of the hot spots (gB peptide 139 [Fig. 1C] and K8.1 peptide 56 [Fig. 1D]) contained previously published 9-mer epitopes (5, 40) that we included as controls. As shown in Fig. 2, the DC-enhanced assay revealed positive IFN-γ responses for the known HHV-8 epitopes as well as for the peptide families of each protein. We defined the optimal epitope as the peptide from each family that generated the highest level of IFN-γ production above the level for the background (i.e., mock-stimulated cell cultures). By these approaches, we were able to define five novel, minimal epitope sequences for LANA-1, one for K12, two for gB, and two for K8.1 (Table 1).
FIG. 2.
IFN-γ ELISPOT responses to known epitopes and peptide families derived from the HHV-8 protein hot spots. CD14− PBMC from healthy HLA A*0201-positive HHV-8-seropositive donors were stimulated for 1 week with autologous mature DC that were loaded with peptide. IFN-γ production was measured as described for Fig. 1. Mean + SE numbers of spot-forming cells from 10 experiments using three donors in response to DC loaded with peptide families are shown for LANA-1 (A) and for K12, gB, K8.1, and known gB492-500 (40) and K8.1209-217 (5) epitopes (B).
TABLE 1.
Novel HHV-8 HLA A*0201 epitopes identified in LANA-1, K12, gB, and K8.1
| Protein | Peptide(s)a | Amino acid positions | Sequence |
|---|---|---|---|
| LANA-1 | 17 | 140-148 | PESSQRPPL |
| LANA-1 | 33, 34 | 281-289 | AMLVLLAEI |
| LANA-1 | 48 | 417-425 | DGGDGNKTL |
| LANA-1 | 63 | 688-697 | QQDEQQQQDE |
| LANA-1 | 83 | 920-928 | PVVSTHEQI |
| K12 | 7, 8 | 23-32 | WRLGAIPPLV |
| gB | 44 | 159-168 | SSMKVNVNGV |
| gB | 207 | 736-745 | MLMIIIVIAI |
| K8.1 | 21 | 73-81 | RLAAGSPSS |
| K8.1 | 38 | 135-143 | ALISAFSGS |
Representative peptide numbers from Fig. 1.
MHC-I restriction and binding of novel HHV-8 epitopes.
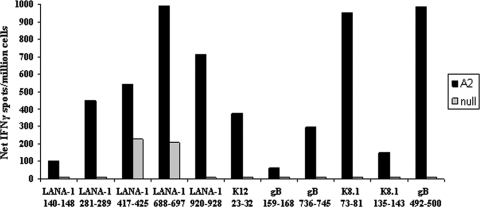
To verify that the novel epitopes were HLA A*0201 restricted, we generated epitope-specific T cell lines and used B cells that express only HLA A*0201 (A2 cells) or do not express any HLA (null cells) as APC in the standard ELISPOT assay. Positive IFN-γ responses with the use of A2 cells as APC were evident for our five novel LANA-1 epitopes, one novel K12 epitope, two novel gB epitopes, and two novel K8.1 epitopes (Fig. 3, black bars). IFN-γ responses were also detected with the use of A2 cells as APC for our previously determined HLA A*0201-restricted gB epitope (40) (Fig. 3, black bars). For all peptides tested, response levels were lower when the null B cells were used as APC (Fig. 3, gray bars). Taken together, these results support that our novel epitopes from the four HHV-8 proteins are HLA A*0201 restricted.
FIG. 3.
CD8+ T cell line IFN-γ ELISPOT responses obtained with the use of HLA A2 cells as APC. PBMC from healthy HLA A*0201-positive HHV-8-seropositive donors were stimulated with peptide, with IL-2 added every 3 days. On day 15, B cell lines expressing only HLA A*0201 (A2 cells) or no HLA molecules (null cells) were used as APC. IFN-γ production was measured by an ELISPOT assay as described for Fig. 1. Responses to novel epitopes from LANA-1, K12, gB, K8.1, and a known epitope from gB (gB492-500) (40) are shown for one representative donor out of two donors used.
Having demonstrated that these peptides are presented in the context of HLA A*0201, we then tested them in an MHC-I binding assay to confirm their specificity. To this end, ProImmune performed their class I REVEAL binding assay on our novel minimal epitopes as well as several known epitopes from the four HHV-8 proteins used in our study. The binding rates are reported as percentages relative to the binding of a known, strong HLA A*0201 T cell epitope. The peptides that showed high binding rates (i.e., above the intermediate control of a known, weaker HLA A*0201 T cell epitope) were three known epitopes (gB492-500, K1217-25, and LANA-11116-1124) as well as two of our novel epitopes (LANA-1281-289 and K8.1135-143), which supports that all of these peptides are HLA A*0201-restricted T cell epitopes (see Fig. S1 in the supplemental material). The remaining novel epitopes were classified as weak binders, i.e., below the intermediate control (Fig. S1). However, these binding scores are relative measures and general guidelines. Thus, peptides classified as weak binders are not necessarily poor T cell epitopes. Indeed, two known epitopes (LANA-1238-246 and K8.1209-217) were classified as weak binders in this assay (Fig. S1). Overall, we conclude that our peptides are novel HLA A*0201 epitopes.
Direct identification of circulating CD8+ T cells specific for HHV-8 epitopes.
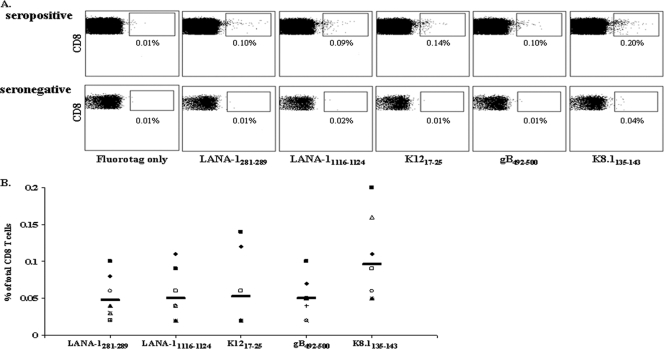
On the basis of the highest scores of our peptide-MHC class I binding results, we synthesized five HLA A*0201 pentamers for two novel epitopes (LANA281-289 and K8.1135-143) and three known epitopes (gB492-500, K1217-25, and LANA-11116-1124). As displayed in Fig. 4, the mean ± standard error (SE) percentages of epitope-specific CD8+ T cells in eight healthy HLA A*0201-positive HHV-8-seropositive individuals were 0.048% ± 0.010% for LANA281-289, 0.050% ± 0.012% for LANA-11116-1124, 0.053% ± 0.018% for K1217-25, 0.050% ± 0.009% for gB492-500, and 0.096% ± 0.020% for K8.1135-143. The mean levels of pentamer-positive CD8+ T cells were 0.020% ± 0.003% in three healthy HLA A*0201-positive HHV-8-seronegative individuals (Fig. 4). Overall, the level of pentamer-positive CD8+ T cells in the seropositive donors was significantly greater than that in the seronegative donors (P < 0.05).
FIG. 4.
HLA A*0201 pentamer staining in peripheral blood samples of HHV-8-seropositive donors. PBMC from HLA A*0201-positive HHV-8-seropositive donors were stained with fluorotag alone or MHC class I-peptide pentamer complexes specific for LANA-1281-289, LANA-11116-1124 (12), K1217-25 (6), gB492-500 (40), and K8.1135-143. Results are shown for one representative HHV-8-seropositive donor and one representative seronegative donor (A) and for all eight HHV-8-seropositive donors tested, with the average response indicated by black bar (B).
Monofunctional and polyfunctional CD8+ T cell responses to novel HHV-8 epitopes.
There are many cytokines in addition to IFN-γ, as well as chemokines and cytotoxic molecules, that are important in antiviral T cell responses. Moreover, polyfunctional T cells, which are defined as a single cell producing two or more such immune mediators, are an important immune correlate of protection against HIV-1 disease progression (1). As it is currently not clear whether monofunctional and polyfunctional CD8+ T cells play a role in controlling HHV-8 infection, we used a panel of markers for immune mediators to examine polyfunctional CD8+ T cell responses. This panel included the cytokines IFN-γ, TNF-α, and IL-2, the chemokine MIP-1β, and the degranulation mobilization marker CD107a. We used a modified procedure with DC-enhanced T cell cultures for the ICS assay developed in our laboratory (16) to improve detection of production of these immune mediators (for a representative analysis, see Fig. S2 in the supplemental material).
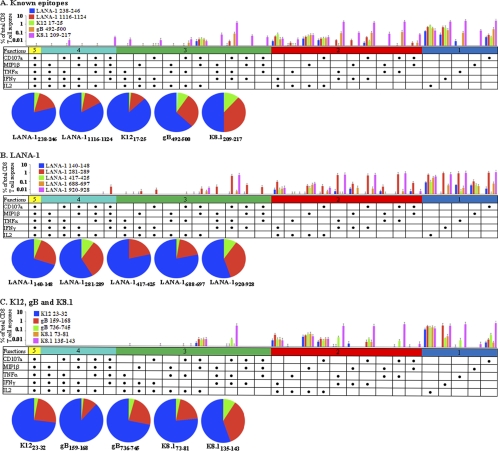
As shown in Fig. 5, positive responses for a variety of immune mediators were detected for our novel minimal epitopes and several known epitopes. The data are shown as bars representing the frequency of the listed mediator combination, with each color representing a different viral peptide. Each pie chart represents total responses to the listed peptide, with each color representing the number of cytokines produced. The ICS results revealed that in healthy HHV-8-seropositive individuals controlling infection, HHV-8 latency and lytic protein epitopes induced both monofunctional and polyfunctional CD8+ T cell responses (Fig. 5). Although all of the peptides displayed a trend of more monofunctional responses than polyfunctional responses, the differences were not significant. With all of the HHV-8 epitopes tested taken into consideration, all five immune mediators were produced by monofunctional T cells, with a predominance of IFN-γ, IL-2, and CD107a (Fig. 5). Polyfunctional T cells producing two immune mediators were notable for the combinations of IFN-γ and IL-2 and for MIP-1β and IL-2. Predominant patterns of polyfunctional responses for three immune mediators included IFN-γ, TNF-α, and CD107a or MIP-1β, IL-2, and CD107a. Polyfunctional T cells producing four immune mediators included IFN-γ, TNF-α, MIP-1β, and CD107a or IFN-γ, TNF-α, IL-2, and CD107a.
FIG. 5.
Polyfunctional CD8+ T cell responses to known and novel HHV-8 epitopes. CD14− PBMC from healthy HLA A*0201-positive HHV-8-seropositive donors were stimulated for 1 week with autologous mature DC loaded with known HHV-8 epitopes (5, 6, 12, 40) (A) or novel epitopes from LANA-1 (B) or K12, gB, or K8.1 (C). Responses were measured by polychromatic flow cytometry, and net response values were averaged for three donors (mean + SE). Diagrams were generated using SPICE.
Different HHV-8 epitopes induced different patterns of both monofunctional and polyfunctional responses. For example, the known epitope from gB (gB492-500) induced notable monofunctional responses consisting of IL-2 or CD107a and polyfunctional responses consisting of MIP-1β and IL-2 or IFN-γ, TNF-α, and CD107a (Fig. 5A). The known epitope K8.1 (K8.1209-217) displayed notable monofunctional responses consisting of IFN-γ or TNF-α and polyfunctional responses consisting of the combination of (i) CD107a and TNF-α, (ii) MIP-1β, TNF-α, and IFN-γ, or (iii) MIP-1β, TNF-α, IFN-γ, and CD107a (Fig. 5A).
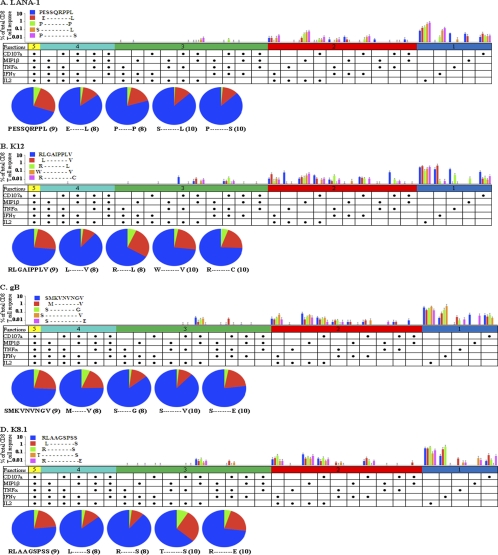
The traditional method for mapping epitopes is through an IFN-γ ELISPOT assay. However, while analysis using one marker allows for the determination of response magnitude, the inclusion of several markers provides insight regarding the quality of the response. Therefore, along with our novel and known HHV-8 epitopes, we also investigated polyfunctional responses to one peptide family from each of the four HHV-8 proteins included in our study (Fig. 6). While trends were evident, no single peptide had a significantly higher magnitude of responses or more polyfunctional responses than the other peptides within the peptide families examined.
FIG. 6.
Polyfunctional CD8+ T cell responses to HHV-8 peptide families. CD14− PBMC from healthy HLA A*0201-positive HHV-8-seropositive donors were stimulated for 1 week with autologous mature DC loaded with peptide. Peptide families are shown for LANA-1 17 (A), K12 7 (B), gB 44 (C), and K8.1 21 (D). Responses were measured by polychromatic flow cytometry, and net response values were averaged for three donors (mean + SE). Diagrams were generated using SPICE.
Taken together, these results show that the CD8+ T cell epitopes from the four HHV-8 proteins presented by DC induced a variety of both monofunctional and polyfunctional T cell responses in HHV-8-seropositive healthy donors. The specificity of this T cell response is less evident in polyfunctional T cell determinations.
DISCUSSION
It is postulated that CD8+ T cell responses play a significant role in controlling HHV-8 infection and preventing development of KS (2, 12). A central issue in understanding the CD8+ T cell immunity in HHV-8 infection is the identification of MHC-I epitopes for the more than 80 open reading frames (ORFs) of the virus. This has been difficult to do mainly because of relatively nonrobust CD8+ T cell responses to the virus (22). In this study, we employed a DC-based enhancement of CD8+ T cell responses to four HHV-8 lytic and latency proteins modified from our previous approach (40) to provide a greater insight into immunopathogenesis of the virus. Using this DC-enhanced IFN-γ ELISPOT assay, we have revealed five novel HLA A*0201-restricted CD8+ T cell epitopes in LANA-1, one in K12, two in gB, and two in K8.1. We were also able to detect epitope-specific CD8+ T cells directly in peripheral blood samples using HLA A*0201 pentamer complexes specific for one gB epitope, one K8.1 epitope, two LANA-1 epitopes, and one K12 epitope. These results were expanded and confirmed by using a DC-enhanced ICS assay with polychromatic flow cytometry to detect multiple immune mediators. By this approach, we revealed that HHV-8-seropositive healthy donors controlling infection have circulating monofunctional and polyfunctional CD8+ T cells specific for an array of HHV-8 lytic and latency protein epitopes.
The novel epitopes in our study did not all correspond to published motifs for the preferred HLA A*0201 anchor residues at positions 2 and 9. Such lack of correspondence to MHC-I allele-specific peptide motifs has been recognized for T cell epitopes of other viruses. Thus, the immunodominant HLA A*0201-restricted influenza A virus M158-66 epitope GILGFVFTL does not have the preferred HLA A*0201 anchor residue L or M at position 2 (35). Moreover, over half of the HLA A*0201-restricted epitopes for vaccinia virus do not fit the optimal peptide binding motif (35). Aside from binding to MHC-I molecules, there are other important factors that determine the T cell response to a peptide, such as the presence CD8+ T cell precursors, interactions with T cell receptors and peptide transporters, and generation of peptides by different protease cleavage pathways (3, 26). Furthermore, our epitopes were determined by two different functional assays (ELISPOT and ICS assays) and represent CD8+ T cell positive reactivity in HLA A*0201-positive HHV-8-seropositive individuals.
Given the large number of ORFs in HHV-8, we focused on ORFs that code for lytic and latency proteins that are considered important in HHV-8 pathogenesis and oncogenesis. We selected two latency cycle proteins, LANA-1 and K12, and two lytic cycle proteins, gB and K8.1. The lytic protein gB is a virion glycoprotein that binds cell surface heparan sulfate and induces signaling pathways, and the lytic protein K8.1 is a highly immunogenic (antibody-inducing) virion glycoprotein that also binds cell surface heparan sulfate (40, 43). During latency, LANA-1 is a cell cycle regulatory protein important in antiapoptotic functions and episome maintenance. The K12 (kaposin) latency protein has roles in B cell signaling, apoptosis, and cell transformation induction (12, 26). While we do acknowledge that a limitation of our study is the small sample size, we found T cell reactivity to peptides of these lytic and latency proteins, indicating that immunity to each could be important in control of HHV-8 infection. We are currently applying a battery of our newly identified and previously documented epitopes to assess T cell immunity to HHV-8 infection and progression to KS in the Multicenter AIDS Cohort Study. Remarkably, given the need to use DC to reveal this T cell function, direct staining of unstimulated PBMC with HLA A*0201 pentamer complexes for two lytic and three latency protein epitopes identified these antigen-specific CD8+ T cells. HHV-8 specific CD8+ T cells have previously been detected in blood samples of HHV-8-seropositive transplant recipients and patients with AIDS-related and classical KS (19). However, to our knowledge, this is the first direct evidence of HHV-8-specific CD8+ T cells in blood samples of healthy HHV-8-seropositive individuals. We are currently determining the memory phenotype of these circulating T cells and if there is a functional downregulation of these cells.
Previous studies have shown that the central repeat region of LANA-1 inhibits proteasomal degradation and slows protein synthesis in order to inhibit cellular surveillance for CD8+ T cell epitopes (17). This model predicts that host T cell reactivity to LANA-1 would be minimal. However, we detected CD8+ T cell reactivity to several regions of LANA-1 in these healthy HHV-8-seropositive subjects. Others have shown anti-LANA-1 CD8+ T cell responses in patients with KS (12). Similarly, CD8+ T cell recognition has been found for EBV-encoded nuclear antigen 1 (21, 39), which had previously been proposed to escape CD8+ T cell recognition through cis inhibition of synthesis or blockade of proteasomal degradation by its glycine-alanine repeat domain. Thus, it appears that CD8+ T cells can mount functional responses to certain regions of LANA-1 regardless of cis effects on its production.
Polyfunctional CD8+ T cells, i.e., cells producing more than one immune mediator, are associated with superior control of persistent viral infections such as those with HIV-1 (1, 24). A recent investigation found similar patterns of polyfunctional T cell responses in patients with multicentric Castleman's disease and healthy controls (13). Other investigators have reported that patients with nonprogressive KS have stronger and more frequent polyfunctional CD8+ T cell responses than those with progressive KS (2). In the present study, we established that polyfunctional T cells specific for known and novel epitopes are present in the PBMC of healthy HHV-8-seropositive individuals. We found that in these healthy, nonimmunosuppressed subjects controlling HHV-8 infection, there was a trend of more monofunctional CD8+ T cell responses than polyfunctional responses for all of the epitopes. Overall, our data suggest that the tight control of HHV-8 infection in healthy individuals may not require a predominance of circulating, virus-specific, polyfunctional CD8+ T cell reactivity. Furthermore, we found that different epitopes induced different patterns of monofunctional and polyfunctional responses. Similar results were found for T cell responses to influenza virus, where CD8+ T cells specific for some but not all viral proteins produced a wide range of cytokines (18). This could be explained by differences in T cell avidity for the peptide-MHC complex.
Traditionally, the standard in monitoring CD8+ T cell responses has been measuring single-cell IFN-γ production in response to antigenic stimuli by an overnight ELISPOT assay. While analysis using one marker allows for the determination of response magnitude, the inclusion of several markers provides insight regarding response quality. Therefore, we examined ICS responses to families of putative optimal epitopes consisting of peptides with N- and C-terminal extensions and truncations from each viral protein to further delineate the minimal epitope sequences. We found that there were different patterns of T cell reactivity revealed by ICS compared to those revealed by the ELISPOT assay. This stresses the need to move beyond IFN-γ ELISPOT assays in defining T cell immunity to viral infections (1, 38). The data also suggest that CD8+ T cells can mount diverse, functional reactivity to 8- to 10-mer variants of an MHC-I 9-mer epitope when presented by DC. This is supported by a recent finding in our laboratory that DC are able to generate efficient monofunctional and polyfunctional T cell responses against 8- to 10-mer N- and C-terminal variants of HIV Gag and Nef epitopes (15a). This enhancement of T cell responses by DC to peptide variations could also explain the broad spectrum of ELISPOT responses evident in our HHV-8 epitope mapping study.
In conclusion, we have revealed several novel HLA A*0201 minimal epitopes in both HHV-8 lytic and latency proteins. We have also shown the presence of antigen-specific CD8+ T cells in peripheral blood samples specific for two lytic and three latency protein epitopes. We have demonstrated that epitopes within these HHV-8 lytic and latency proteins induced both single and multiple immune mediators in CD8+ T cells from healthy HHV-8-seropositive individuals. The likely involvement of CD8+ T cells in the immune response to HHV-8 infection has implications for the prevention and treatment of HHV-8-associated cancers. These targeted regions of the virus that induce immune responses by CD8+ T cells could be critical in HHV-8 immunopathogenesis and the progression to KS.
Supplementary Material
Acknowledgments
We thank L. Borowski, K. Stojka, and J. Roper for technical assistance, W. Buchanan for clinical assistance, and M. Roederer (VRC/NIAID/NIH) for SPICE (version 4.2.3).
This work was supported by National Institutes of Health grants R01 CA 82053, U01 AI 35041, and T32 AI065380.
Footnotes
Published ahead of print on 18 August 2010.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bihl, F., C. Berger, J. V. Chisholm III, L. M. Henry, B. Bertisch, A. Trojan, D. Nadal, R. F. Speck, M. Flepp, C. Brander, and N. J. Mueller. 2009. Cellular immune responses and disease control in acute AIDS-associated Kaposi's sarcoma. AIDS 23:1918-1922. [DOI] [PubMed] [Google Scholar]
- 3.Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H. S. Hewitt, L. Henry, C. H. Linde, J. V. Chisholm III, T. M. Zaman, E. Pae, S. Mallal, B. D. Walker, A. Sette, B. T. Korber, D. Heckerman, and C. Brander. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094-4101. [DOI] [PubMed] [Google Scholar]
- 4.Bihl, F., M. Narayan, J. V. Chisholm III, L. M. Henry, T. J. Suscovich, E. E. Brown, T. M. Welzel, D. E. Kaufmann, T. M. Zaman, S. Dollard, J. N. Martin, F. Wang, D. T. Scadden, K. M. Kaye, and C. Brander. 2007. Lytic and latent antigens of the human gammaherpesviruses Kaposi's sarcoma-associated herpesvirus and Epstein-Barr virus induce T-cell responses with similar functional properties and memory phenotypes. J. Virol. 81:4904-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourboulia, D., D. Aldam, D. Lagos, E. Allen, I. Williams, D. Cornforth, A. Copas, and C. Boshoff. 2004. Short- and long-term effects of highly active antiretroviral therapy on Kaposi sarcoma-associated herpesvirus immune responses and viraemia. AIDS 18:485-493. [DOI] [PubMed] [Google Scholar]
- 6.Brander, C., P. O'Connor, T. Suscovich, N. G. Jones, Y. Lee, D. Kedes, D. Ganem, J. Martin, D. Osmond, S. Southwood, A. Sette, B. D. Walker, and D. T. Scadden. 2001. Definition of an optimal cytotoxic T lymphocyte epitope in the latently expressed Kaposi's sarcoma-associated herpesvirus kaposin protein. J. Infect. Dis. 184:119-126. [DOI] [PubMed] [Google Scholar]
- 7.Butler, L. M., G. Dorsey, W. Hladik, P. J. Rosenthal, C. Brander, T. B. Neilands, G. Mbisa, D. Whitby, P. Kiepiela, A. Mosam, S. Mzolo, S. C. Dollard, and J. N. Martin. 2009. Kaposi sarcoma-associated herpesvirus (KSHV) seroprevalence in population-based samples of African children: evidence for at least 2 patterns of KSHV transmission. J. Infect. Dis. 200:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, A., D. G. Wolf, E. Guttman-Yassky, and R. Sarid. 2005. Kaposi's sarcoma-associated herpesvirus: clinical, diagnostic, and epidemiological aspects. Crit. Rev. Clin. Lab. Sci. 42:101-153. [DOI] [PubMed] [Google Scholar]
- 9.Colleton, B. A., X. L. Huang, N. M. Melhem, Z. Fan, L. Borowski, G. Rappocciolo, and C. R. Rinaldo. 2009. Primary human immunodeficiency virus type 1-specific CD8+ T-cell responses induced by myeloid dendritic cells. J. Virol. 83:6288-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallafent, J. H., S. E. Buskin, P. B. De Turk, and D. M. Aboulafia. 2005. Profile of patients with Kaposi's sarcoma in the era of highly active antiretroviral therapy. J. Clin. Oncol. 23:1253-1260. [DOI] [PubMed] [Google Scholar]
- 11.Gottschalk, S., H. E. Heslop, and C. M. Rooney. 2005. Adoptive immunotherapy for EBV-associated malignancies. Leuk. Lymphoma 46:1-10. [DOI] [PubMed] [Google Scholar]
- 12.Guihot, A., N. Dupin, A. G. Marcelin, I. Gorin, A. S. Bedin, P. Bossi, L. Galicier, E. Oksenhendler, B. Autran, and G. Carcelain. 2006. Low T cell responses to human herpesvirus 8 in patients with AIDS-related and classic Kaposi sarcoma. J. Infect. Dis. 194:1078-1088. [DOI] [PubMed] [Google Scholar]
- 13.Guihot, A., E. Oksenhendler, L. Galicier, A. G. Marcelin, L. Papagno, A. S. Bedin, F. Agbalika, N. Dupin, J. Cadranel, B. Autran, and G. Carcelain. 2008. Multicentric Castleman disease is associated with polyfunctional effector memory HHV-8-specific CD8+ T cells. Blood 111:1387-1395. [DOI] [PubMed] [Google Scholar]
- 14.Hislop, A. D., and S. Sabbah. 2008. CD8+ T cell immunity to Epstein-Barr virus and Kaposi's sarcoma-associated herpes virus. Semin. Cancer Biol. 18:416-422. [DOI] [PubMed] [Google Scholar]
- 15.Honeyborne, I., A. Rathod, R. Buchli, D. Ramduth, E. Moodley, P. Rathnavalu, S. Chetty, C. Day, C. Brander, W. Hildebrand, B. D. Walker, P. Kiepiela, and P. J. Goulder. 2006. Motif inference reveals optimal CTL epitopes presented by HLA class I alleles highly prevalent in southern Africa. J. Immunol. 176:4699-4705. [DOI] [PubMed] [Google Scholar]
- 15a.Huang, X. L., Z. Fan, L. Borowski, R. B. Mailliard, M. Roland, J. I. Mullins, R. D. Day, and C. R. Rinaldo. Dendritic cells reveal a broad range of MHC class I epitopes for HIV-1 in persons with suppressed viral load on antiretroviral therapy. PlosOne, in press. [DOI] [PMC free article] [PubMed]
- 16.Huang, X. L., Z. Fan, L. Borowski, and C. R. Rinaldo. 2009. Multiple T-cell responses to human immunodeficiency virus type 1 are enhanced by dendritic cells. Clin. Vaccine Immunol. 16:1504-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwun, H. J., S. R. da Silva, I. M. Shah, N. Blake, P. S. Moore, and Y. Chang. 2007. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J. Virol. 81:8225-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Gruta, N. L., S. J. Turner, and P. C. Doherty. 2004. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J. Immunol. 172:5553-5560. [DOI] [PubMed] [Google Scholar]
- 19.Lambert, M., M. Gannage, A. Karras, M. Abel, C. Legendre, D. Kerob, F. Agbalika, P. M. Girard, C. Lebbe, and S. Caillat-Zucman. 2006. Differences in the frequency and function of HHV8-specific CD8 T cells between asymptomatic HHV8 infection and Kaposi sarcoma. Blood 108:3871-3880. [DOI] [PubMed] [Google Scholar]
- 20.Lamoreaux, L., M. Roederer, and R. Koup. 2006. Intracellular cytokine optimization and standard operating procedure. Nat. Protoc. 1:1507-1516. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. P., J. M. Brooks, H. Al-Jarrah, W. A. Thomas, T. A. Haigh, G. S. Taylor, S. Humme, A. Schepers, W. Hammerschmidt, J. L. Yates, A. B. Rickinson, and N. W. Blake. 2004. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J. Exp. Med. 199:1409-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little, R. F., and R. Yarchoan. 2006. Poor specific T cell responses to human herpesvirus 8: a key to unleashing Kaposi sarcoma? J. Infect. Dis. 194:1030-1031. [DOI] [PubMed] [Google Scholar]
- 23.Lonard, B. M., M. Sester, U. Sester, H. W. Pees, N. Mueller-Lantzsch, H. Kohler, and B. C. Gartner. 2007. Estimation of human herpesvirus 8 prevalence in high-risk patients by analysis of humoral and cellular immunity. Transplantation 84:40-45. [DOI] [PubMed] [Google Scholar]
- 24.Makedonas, G., and M. R. Betts. 2006. Polyfunctional analysis of human T cell responses: importance in vaccine immunogenicity and natural infection. Springer Semin. Immunopathol. 28:209-219. [DOI] [PubMed] [Google Scholar]
- 25.Maurer, T., M. Ponte, and K. Leslie. 2007. HIV-associated Kaposi's sarcoma with a high CD4 count and a low viral load. N. Engl. J. Med. 357:1352-1353. [DOI] [PubMed] [Google Scholar]
- 26.Micheletti, F., P. Monini, C. Fortini, P. Rimessi, M. Bazzaro, M. Andreoni, M. Giuliani, S. Traniello, B. Ensoli, and R. Gavioli. 2002. Identification of cytotoxic T lymphocyte epitopes of human herpesvirus 8. Immunology 106:395-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen, S. J., R. Sarid, Y. Chang, and P. S. Moore. 2000. Evaluation of the latency-associated nuclear antigen (ORF73) of Kaposi's sarcoma-associated herpesvirus by peptide mapping and bacterially expressed recombinant Western blot assay. J. Infect. Dis. 182:306-310. [DOI] [PubMed] [Google Scholar]
- 28.Osman, M., T. Kubo, J. Gill, F. Neipel, M. Becker, G. Smith, R. Weiss, B. Gazzard, C. Boshoff, and F. Gotch. 1999. Identification of human herpesvirus 8-specific cytotoxic T-cell responses. J. Virol. 73:6136-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Precopio, M. L., M. R. Betts, J. Parrino, D. A. Price, E. Gostick, D. R. Ambrozak, T. E. Asher, D. C. Douek, A. Harari, G. Pantaleo, R. Bailer, B. S. Graham, M. Roederer, and R. A. Koup. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J. Exp. Med. 204:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rammensee, H. G., T. Friede, and S. Stevanoviic. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41:178-228. [DOI] [PubMed] [Google Scholar]
- 31.Ribechini, E., C. Fortini, M. Marastoni, S. Traniello, S. Spisani, P. Monini, and R. Gavioli. 2006. Identification of CD8+ T cell epitopes within lytic antigens of human herpes virus 8. J. Immunol. 176:923-930. [DOI] [PubMed] [Google Scholar]
- 32.Robey, R. C., D. Lagos, F. Gratrix, S. Henderson, N. C. Matthews, R. J. Vart, M. Bower, C. Boshoff, and F. M. Gotch. 2009. The CD8 and CD4 T-cell response against Kaposi's sarcoma-associated herpesvirus is skewed towards early and late lytic antigens. PLoS One 4:e5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seder, R. A., P. A. Darrah, and M. Roederer. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 8:247-258. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu, Y., and R. DeMars. 1989. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J. Immunol. 142:3320-3328. [PubMed] [Google Scholar]
- 35.Sidney, J., E. Assarsson, C. Moore, S. Ngo, C. Pinilla, A. Sette, and B. Peters. 2008. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stebbing, J., D. Bourboulia, M. Johnson, S. Henderson, I. Williams, N. Wilder, M. Tyrer, M. Youle, N. Imami, T. Kobu, W. Kuon, J. Sieper, F. Gotch, and C. Boshoff. 2003. Kaposi's sarcoma-associated herpesvirus cytotoxic T lymphocytes recognize and target Darwinian positively selected autologous K1 epitopes. J. Virol. 77:4306-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strickler, H. D., J. J. Goedert, F. R. Bethke, C. M. Trubey, T. R. O'Brien, J. Palefsky, J. E. Whitman, D. Ablashi, S. Zeichner, and G. M. Shearer. 1999. Human herpesvirus 8 cellular immune responses in homosexual men. J. Infect. Dis. 180:1682-1685. [DOI] [PubMed] [Google Scholar]
- 38.Sun, Y., E. Iglesias, A. Samri, G. Kamkamidze, T. Decoville, G. Carcelain, and B. Autran. 2003. A systematic comparison of methods to measure HIV-1 specific CD8 T cells. J. Immunol. Methods 272:23-34. [DOI] [PubMed] [Google Scholar]
- 39.Tellam, J., G. Connolly, K. J. Green, J. J. Miles, D. J. Moss, S. R. Burrows, and R. Khanna. 2004. Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1. J. Exp. Med. 199:1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, Q. J., X. L. Huang, G. Rappocciolo, F. J. Jenkins, W. H. Hildebrand, Z. Fan, E. K. Thomas, and C. R. Rinaldo, Jr. 2002. Identification of an HLA A*0201-restricted CD8(+) T-cell epitope for the glycoprotein B homolog of human herpesvirus 8. Blood 99:3360-3366. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Q. J., F. J. Jenkins, L. P. Jacobson, L. A. Kingsley, R. D. Day, Z. W. Zhang, Y. X. Meng, P. E. Pellett, K. G. Kousoulas, A. Baghian, and C. R. Rinaldo, Jr. 2001. Primary human herpesvirus 8 infection generates a broadly specific CD8(+) T-cell response to viral lytic cycle proteins. Blood 97:2366-2373. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Q. J., F. J. Jenkins, L. P. Jacobson, Y. X. Meng, P. E. Pellett, L. A. Kingsley, K. G. Kousoulas, A. Baghian, and C. R. Rinaldo, Jr. 2000. CD8+ cytotoxic T lymphocyte responses to lytic proteins of human herpes virus 8 in human immunodeficiency virus type 1-infected and -uninfected individuals. J. Infect. Dis. 182:928-932. [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson, J., A. Cope, J. Gill, D. Bourboulia, P. Hayes, N. Imami, T. Kubo, A. Marcelin, V. Calvez, R. Weiss, B. Gazzard, C. Boshoff, and F. Gotch. 2002. Identification of Kaposi's sarcoma-associated herpesvirus (KSHV)-specific cytotoxic T-lymphocyte epitopes and evaluation of reconstitution of KSHV-specific responses in human immunodeficiency virus type 1-infected patients receiving highly active antiretroviral therapy. J. Virol. 76:2634-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodberry, T., T. J. Suscovich, L. M. Henry, J. N. Martin, S. Dollard, P. G. O'Connor, J. K. Davis, D. Osmond, T. H. Lee, D. H. Kedes, A. Khatri, J. Lee, B. D. Walker, D. T. Scadden, and C. Brander. 2005. Impact of Kaposi sarcoma-associated herpesvirus (KSHV) burden and HIV coinfection on the detection of T cell responses to KSHV ORF73 and ORF65 proteins. J. Infect. Dis. 192:622-629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.