Abstract
Klassevirus is a proposed new genus of picornavirus that has been associated with pediatric diarrhea. In this study, we used recombinant klassevirus 3C protease as the capture antigen for an indirect serological enzyme-linked immunosorbent assay (ELISA). Four of six klassevirus reverse transcription (RT)-PCR-positive individuals demonstrated seroconversion against the 3C protease, suggesting that klassevirus infection and replication occur in humans. Additional screening of 353 samples from an age-banded serological cohort from two St. Louis hospitals indicated a seroprevalence of 6.8%.
Klassevirus is the prototype member of a new picornavirus genus that has recently been discovered in human stool (3, 4, 7). Phylogenetic analysis shows klassevirus is most closely related to Aichi virus, a known cause of oyster-associated gastroenteritis in humans. Molecular detection studies have found klassevirus in pediatric stool and municipal sewage from the United States, Spain, Australia, Tunisia, and Nigeria (3, 4, 7). A small case control study found a statistically significant association between klassevirus and pediatric diarrhea (7). Furthermore, klassevirus RNA has been detected in high titer in pediatric stool (3). However, klassevirus has not yet been detected in any sterile sites in the human body, and thus it is still not clear whether the presence of the virus in stool represents a bona fide human infection.
In this study, we established a serological assay for klassevirus infection using recombinant klassevirus 3C protease to demonstrate seroconversion and human infection and to test for seroprevalence in an age-banded pediatric cohort from hospitals in the St. Louis area. Klassevirus 3C protease shares only 38% amino acid identity and only one potential 7-mer epitope with its closest relative, the 3C protease of Aichi virus. Previous studies of hepatitis A virus (HAV), a prototypic picornavirus, indicate that antibodies are made against the picornaviral 3C protease during bona fide picornavirus replication and thus can distinguish between vaccinated and actively infected chimpanzees, humans, and tamarins (5, 9).
MATERIALS AND METHODS
Expression and purification of 3C protease.
The klassevirus 3C protease gene (nucleotides [nt] 5825 to 6409 from strain 2394-01; NC_012986.1) flanked with NdeI and XhoI restriction sites was amplified from stool total RNA by reverse transcription (RT)-PCR with the following primers: klasse3C-NdeI (5′-CATATGGGTTTCGACCCTGCCGTCATGAAG-3′ and klasse3C-XhoI (5′-CTCGAGTCATCACTGAGGTGTGGCCAGGTTAGAGA-3′) (restriction sites italicized and stop codons underlined). The resulting product was sequence confirmed, digested with NdeI and XhoI, and subcloned into NdeI/XhoI-digested pET15b (Novagen), which contains a 6×His tag on the N terminus. The sequence-confirmed pET15b vector containing the klassevirus 3C gene was transformed into Escherichia coli BL21(DE3)LysS/pRIL and recombinant 6×His-klassevirus 3C protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C for 5 h. Cells were lysed in 150 mM NaCl, 25 mM Tris, pH 8.0, 5 mM imidazole, 2 mM β-mercaptoethanol, and 5% glycerol with 0.5 mg/ml lysozyme for 30 min in ice and 3 min of sonication (Branson Sonifier 250) at 60% power. 6×His-klassevirus 3C protein was purified from cell lysates via Ni-nitrilotriacetic acid (NTA) column chromatography. The resulting eluate was then concentrated using a 10-kDa Amicon Ultra (Millipore, Billerica, MA) and loaded on a 120-ml Superdex 200 gel filtration column (GE LifeSciences; Piscataway, NJ). Elution fractions were quantitated using 280-nm absorbance on a Nanodrop, and the 91-ml fraction was used for enzyme-linked immunosorbent assay (ELISA) experiments. SDS-PAGE was performed using 4 to 12% Bis-Tris NuPage gels (Invitrogen; Carlsbad, CA), and silver staining was performed using SilverXpress (Invitrogen). Two milligrams of purified 6×His-klassevirus 3C was used for polyclonal antibody generation in rabbits (Pacific Immunology; Ramona, CA).
RT-PCR screening of Indian samples.
Community- and hospital-based samples (n = 416) collected in Vellore, India, from 2005 to 2006 were screened for klassevirus. The community-based samples were from a birth cohort followed for 3 years. In this cohort, stool samples were collected every 2 weeks and with every episode of diarrhea. Additionally, serum samples were obtained every 3 months during the first 2 years and every 6 months during the third year of the study (8). The hospital-based samples were from a single-point collection of diarrheal stool for surveillance of children under the age of 5 years hospitalized for acute gastroenteritis (6).
RNA was extracted from 200 μl of an ∼20% fecal suspension by the Boom method and eluted in 40 μl of water (1). A Qiagen one-step RT-PCR kit (Qiagen, Valencia, CA) was used to screen 3 μl of extracted material from each sample, using primers LG0098 (5′-CGTCAGGGTGTTCGTGATTA-3′) and LG0093 (5′-AGAGAGAGCTGTGGAGTAATTAGTA-3′). These primers target the 2C region of klassevirus 1 and are expected to generate a 345-bp amplicon. RT-PCRs were performed using Qiagen one-step kit under the following conditions: 30 min for the RT step, with a 94°C hold for 10 min, followed by 40 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 60 s. Some amplicons were cloned into pCR4 (Invitrogen) and sequenced using standard Sanger sequencing technology (GenBank accession no. GU992865 to GU992870).
St. Louis serum cohort.
We analyzed 353 deidentified serum samples from patients 1 day to 79 years of age collected from St. Louis Children's Hospital and Barnes-Jewish Hospital in St. Louis, MO, from November 2007 through October 2008 for antibodies against the 3C protease from klassevirus. Serum samples were kindly provided by Greg Storch (St. Louis Children's Hospital) and Mitchell Scott (Barnes-Jewish Hospital). The patients were age stratified, and 30 samples were used for each pediatric age group, except for the groups 6 months to 1 year and 6 to 9 years, for which 29 samples were available. For the adult age groups, the following numbers of samples were available: 7 samples for 20 to 34 years, 15 samples for 35 to 49 years, 20 samples for 50 to 64 years, and 13 samples for 65 to 79 years. Collection of samples and clinical data were approved by the Human Research Protection Office of Washington University in St. Louis, School of Medicine.
ELISA.
Purified klassevirus 3C protein (0.15 μg/well) was coated overnight at 4°C in 50 μl phosphate-buffered saline (PBS) in Maxisorp 96-well microtiter plates (Nunc, Naperville, IL). Wells were washed twice with PBST (PBS containing 0.05% Tween 20) and blocked with blocking buffer (PBST containing 1% bovine serum albumin) for 2 h at room temperature. Wells were washed twice again with PBST and incubated with serum samples diluted 1:100 in blocking buffer in triplicate overnight at 4C. Serum samples for competition experiments were diluted 1:100 in blocking buffer and preincubated with 0.6 μg protein overnight at 4C. Plates were washed four times with PBST and incubated with horseradish peroxidase (HRP)-conjugated anti-human IgG antibody (Invitrogen) diluted 1:10,000 in 50 μl blocking buffer for 2 h at room temperature. Plates were then washed four times with PBST and incubated with 50 μl Ultra-TMB (Sigma; St. Louis, MO) for 30 min at room temperature. An equal volume of 2 M H2SO4 was added to stop the reaction, and the endpoint optical density values at 450 nm (OD450) were read immediately on a SpectraMax M2 plate reader. Statistical tests were done using a two-sample t test.
Western confirmation was performed for two matched serum samples from India by running 5 μg of purified klassevirus 3C protease on 4 to 12% Bis-Tris NuPage gels, transferring the samples using the XCell II blot module, staining them overnight at 4°C using 5 μl of human serum diluted 1:200 into 1 ml of 5% milk in PBST, and developing them with Western Lightning enhanced luminal reagent. Western blots were scanned on an Epson Perfection V350 photo scanner and quantitated using Licor Odyssey 2.1 software.
RESULTS
Purification of klassevirus 3C protease.
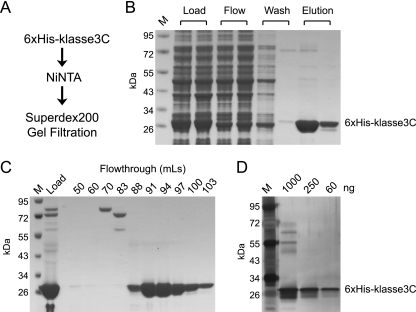
Klassevirus 3C protease was expressed as an N-terminal 6×His-tagged fusion protein, purified using Ni-NTA affinity chromatography, and followed by gel filtration fast protein liquid chromatography (FPLC). SDS-PAGE analysis with Coomassie stain and silver stain was used to confirm the purity of products at each stage (Fig. 1). The inclusion of reducing agents (at least 5 mM β-mercaptoethanol) was the most important factor in increasing the solubility of the product purified off the Ni-NTA column. The inclusion of 20% glycerol, 10 mM PMSF, 1× Roche proteinase inhibitor, or 1% Triton X did not prevent the elution from precipitating, as determined by visual inspection. The purified 6×His-klassevirus 3C protein was used as a capture antigen for an indirect ELISA for serological analysis of klassevirus infection.
FIG. 1.
Purification of klassevirus 3C protease. N-terminal His-tagged klassevirus 3C protease was expressed in E. coli and purified by Ni-NTA chromatography (A and B). Ni-NTA eluates were further purified by gel filtration chromatography using a Superdex 200 column (C). The 91-ml fraction was tested via silver stain to confirm purity of klassevirus 3C protease (D). Lane M, molecular mass markers.
Detection of klassevirus-positive stool samples in an Indian cohort.
Sixteen of the 203 community-based stool samples and 9 of the 213 hospital-based stool samples were positive by RT-PCR screening. Sequence was recovered from four of the community-based positive samples and two of the hospital-based positive samples.
Serological analysis of positive control samples and St. Louis cohort.
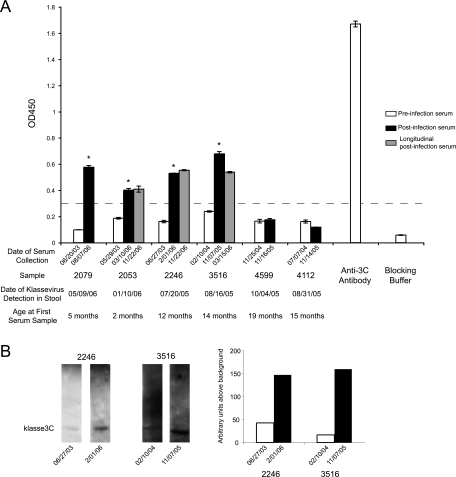
Matched preinfection and convalescent-phase human sera from klassevirus-positive Indian children who had PCR-positive klassevirus detected in stool were available for serological testing. A statistically significant (P < 0.005) increase in OD450 could be demonstrated in four of the six matched samples for as long as 11 months after klassevirus was detected in stool (Fig. 2A). Western blot analysis confirmed seroconversion between matched preinfection and postinfection serum samples 2246-5672 and 3516-5475, the only samples for which enough serum remained to attempt Western confirmation (Fig. 2B).
FIG. 2.
Klassevirus 3C ELISA for matched serum samples from klassevirus PCR-positive children from India. Endpoint OD450 values for matched preinfection serum samples (1:100 dilution; white) and postinfection serum samples (1:100 dilution; shaded) are grouped together, along with polyclonal rabbit antisera generated against klassevirus 3C and blocking buffer (A). The dotted line indicates an OD450 cutoff value of 0.30 that separates pre- and postinfection OD450 values. Asterisks indicate significant increases in OD450 (P < 0.005). Remaining serum was available from two matched serum samples (2246 and 3516) for Western confirmation, which showed a 4-fold and 8-fold increase in signal between pre- and post-klassevirus infection serum samples at a dilution of 1:200, respectively (B).
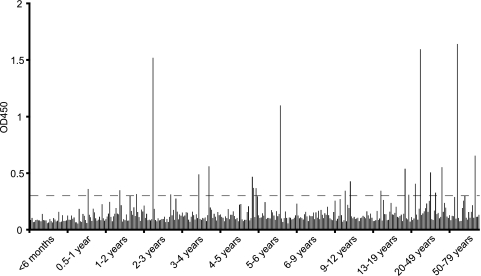
We next screened 353 age-banded serum samples from two St. Louis hospitals (Fig. 3). To control for plate-to-plate variation, a single control serum sample from a healthy donor was analyzed on every ELISA plate. The coefficient of variance for this sample was 8.8% across the 12 plates, indicating a high level of consistency for the assay. Incubation of the single control serum sample with recombinant Taq polymerase as the capture reagent resulted in a comparable OD450 with no plated antigen (OD450, <0.10). The mean OD450 of this serum sample in the St. Louis cohort was equivalent to its average OD450 when run with the matched Indian serum samples (coefficient of variation [CV], 3%). The median OD450 for the St. Louis children's hospital samples was 0.114, while the mean OD450 for the 0- to 6-month samples was 0.093 (standard deviation [SD], 0.023). Twenty-six (8.7%) of the pediatric St. Louis samples and 12 (21.8%) of the adult serum samples had an average OD450 that was greater than the highest OD450 for a preinfection Indian serum sample. Using a specific OD450 cutoff of 0.300 (a value that separates preinfection and postinfection OD450 values in the seroconverting Indian samples) yielded 16 (5.3%) seropositives in the pediatric St. Louis cohort and eight (14.5%) seropositives in the adult cohort. There was no significant association between klassevirus seropositivity and sex in either the pediatric or adult cohort.
FIG. 3.
Klassevirus 3C ELISA for 353 age-banded samples from St. Louis Children's Hospital and Barnes-Jewish Hospital. Approximately 30 samples for each age band were included. A specific OD450 cutoff value of 0.30 based on the matched Indian serum in Fig. 2 suggested 24 (6.8%) of the serum samples were seropositive for klassevirus.
DISCUSSION
The data in this study indicate that IgG antibodies are generated against the 3C protease after klassevirus infection. This represents the first demonstration of a human antibody response to a klassevirus antigen and suggests that bona fide human infection by klassevirus occurs. Based on the available convalescent-phase serum samples, IgG antibodies could be detected in some instances as long as 7 months after infection. It is unclear how long the antibody response to klassevirus 3C persists, and more extensive studies, using longitudinal serum samples are needed to address this issue.
Screening of an age-banded cohort of 353 serum samples from St. Louis Children's Hospital yielded only a handful of positive samples, using a cutoff value defined by the average of the preinfection samples from India. This observation suggests either that the klassevirus infection rate in the United States is quite low or that anti-klassevirus 3C antibodies may not be long lived. To assess this further, a larger sample size and/or serial samples taken from individuals with klassevirus confirmed stool samples would be required.
To date, klassevirus has only been detected via PCR of stool and sewage samples (3, 4, 7). Importantly, 3C protease is not present in picornaviral virions, and studies of hepatitis A experimental infections in chimpanzees indicate that anti-3C antibodies are only generated in an actively infected individual (5). Thus, our data are consistent with the notion that klassevirus replicates in humans and is not merely a virus of some human foodstuff.
No culture system for klassevirus has been described to date. Thus, recombinant viral proteins are the only currently viable capture reagents for determining seroprevalence. Attempts to use capsid VP1 protein for serological analysis, as has been used for other picornaviruses, were hindered by an inability to refold the purified denatured protein. Our experience with purification of VP1 is not atypical. Previous attempts have required purification from inclusion bodies and subsequent refolding on gel filtration columns (2, 10). Use of previous published protocols for the purification of VP1 were not successful, although this does not preclude future optimization to recover correctly folded protein (2, 10). Nonstructural proteins, such as 3C, have also been used to determine seroprevalence of picornaviruses (5). From a technical perspective, nonstructural proteins are more likely to be soluble than structural proteins and are easier to assay for activity and correct folding. Furthermore, detection of an immune response mediated against nonstructural proteins that are not present in the virion is suggestive of viral replication in vivo. Previous studies of hepatitis A virus (HAV) infection also suggested the severity of infection is correlated with the development of anti-3C antibodies (5, 9). However, these studies also suggest that there is heterogeneity in the development of anti-3C and anti-structural protein antibodies. Not every infected individual will develop anti-3C antibodies, even though anti-structural protein antibodies and evidence of clinical infection may develop. In this study, we note that two of six klassevirus-infected individuals showed no increase in anti-3C antibody levels.
Unambiguously ascertaining the seroprevalence of new agents such as klassevirus in a population is complicated by the lack of true positive and negative controls. For example, the infection history of the “preinfection” Indian serum samples and the 0- to 6-month-olds in the St. Louis cohort is not known. In this study, we made the assumption that the “preinfection” Indian serum samples represent seronegative individuals to create a specific cutoff value for seropositivity. Using these criteria, a seroprevalence of 6.8% was found in 353 serum samples from St. Louis. This seroprevalence is fairly low for a virus that has, to date, only been found in young children. This may be indicative of highly sporadic infection of klassevirus in St. Louis or may be suggestive of a short-lived antibody response to the 3C protease, as indicated by an inconsistent increase in seroprevalence with increasing age (Fig. 3). An alternative way to analyze the seroprevalence would be to assume that the 0- to 6-month samples are negative based on their low OD450 values and extremely young age and to create a common ELISA seropositivity cutoff 2 or 3 SDs above the mean 0- to 6-month OD450. The number of seropositive samples using this method would be 121 (34.3%) or 85 (24%) samples, respectively. However, using the 2-SD seropositive cutoff would count five of the six preinfection matched Indian serum samples as seropositive. This may be a reasonable conclusion given the higher detection of klassevirus by RT-PCR in stool from Indian children than has been reported in the United States (3, 4, 7). However, maternal antibodies would be unlikely to account for any seropositivity in the preinfection samples, as four of the six children were more than a year of age at the date of serum collection.
In this study, a small number of longitudinal serum samples (up to a year following the acute positive stool sample) from klassevirus-infected individuals were tested, demonstrating that high titers persist for at least 6 months. Ideally, a much longer longitudinal cohort should be examined to define the lifetime of an anti-klassevirus 3C response in a given individual. Studies on anti-3C antibodies from HAV suggest that anti-3C antibodies remain detectable in serum for up to 15 months after infection (5). The possibility of short-lived anti-3C antibody may explain why we failed to observe a clear upward trend in seropositivity across different age bands, although sample sizes for each age band were small and the adult population seemed to have a higher seroprevalence than the pediatric population. We also failed to observe seropositivity in children <6 months old, indicating a lack of maternally transmitted anti-3C antibodies in this population.
Future studies will be required to ascertain seroprevalence across different populations, environments, and age bands. Neutralization tests or virus-like particle-based ELISAs could provide an orthogonal test for klassevirus serology. We would also like to know how often klassevirus can infect an individual and whether neutralizing antibodies are generated. Of special interest is the use of klassevirus serology to determine potential associations with clinical disease in humans aside from diarrhea.
Acknowledgments
This work was supported by the Doris Duke Foundation, Howard Hughes Medical Institute, the Packard Foundation, the National Institutes of Health under the Ruth L. Kirschstein National Research Service Award (F32 DK083867-01) from the NIDDK, and P30 DK52574. D.W. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. J.L.D. and D.G. are supported by the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 25 August 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu, C. Y., A. L. Greninger, E. C. Chen, T. D. Haggerty, J. Parsonnet, E. Delwart, J. L. DeRisi, and D. Ganem. 2010. Cultivation and serological characterization of human Theiler's-like cardiovirus associated with diarrheal disease. J. Virol. 84:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greninger, A. L., C. Runckel, C. Y. Chiu, T. Haggerty, J. Parsonnet, D. Ganem, and J. L. DeRisi. 2009. The complete genome of klassevirus—a novel picornavirus in pediatric stool. Virol. J. 6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtz, L. R., S. R. Finkbeiner, G. Zhao, C. D. Kirkwood, R. Girones, J. M. Pipas, and D. Wang. 2009. Klassevirus 1, a previously undescribed member of the picornaviral family is globally widespread. Virol. J. 6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabrane-Lazizi, Y., S. U. Emerson, C. Herzog, and R. H. Purcell. 2001. Detection of antibodies to HAV 3C protease in experimentally infected chimpanzees and in naturally infected children. Vaccine 19:2878-2883. [DOI] [PubMed] [Google Scholar]
- 6.Kang, G., R. Arora, S. D. Chitambar, J. Deshpande, M. D. Gupte, M. Kulkarni, T. N. Naik, D. Mukherji, S. Venkatasubramaniam, J. R. Gentsch, R. I. Glass, and U. D. Parashar. 2009. Multicenter, hospital-based surveillance of rotavirus disease and strains among Indian children aged <5 years. J. Infect. Dis. 200:S147-S153. [DOI] [PubMed] [Google Scholar]
- 7.Li, L., J. Victoria, A. Kapoor, O. Blinkova, C. Wang, F. Babrzadeh, C. J. Mason, P. Pandey, H. Triki, O. Bahri, B. S. Oderinde, M. M. Baba, D. N. Bukbuk, J. M. Besser, J. M. Bartkus, and E. L. Delwart. 2009. A novel picornavirus associated with gastroenteritis. J. Virol. 83:12002-12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehman, A. M., B. P. Gladstone, V. P. Verghese, J. Muliyil, S. Jaffar, and G. Kang. 2009. Chronic growth faltering amongst a birth cohort of Indian children begins prior to weaning and is highly prevalent at three years of age. Nutr. J. 8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart, D. R., T. S. Morris, R. H. Purcell, and S. U. Emerson. 1997. Detection of antibodies to the nonstructural 3C proteinase of hepatitis A virus. J. Infect. Dis. 176:593-601. [DOI] [PubMed] [Google Scholar]
- 10.Wang, J. H., C. M. Liang, J. M. Peng, J. J. Shieh, M. H. Jong, Y. L. Lin, M. Sieber, and S. M. Liang. 2003. Induction of immunity in swine by purified recombinant VP1 of foot-and-mouth disease virus. Vaccine 21:3721-3729. [DOI] [PubMed] [Google Scholar]