Abstract
The bacterial phylum Acidobacteria has a widespread distribution and is one of the most common and diverse phyla in soil habitats. However, members of this phylum have often been recalcitrant to cultivation methods, hampering the study of this presumably important bacterial group. In this study, we used a cultivation-independent metagenomic approach to recover genomic information from soilborne members of this phylum. A soil metagenomic fosmid library was screened by PCR targeting acidobacterial 16S rRNA genes, facilitating the recovery of 17 positive clones. Recovered inserts appeared to originate from a range of Acidobacteria subdivisions, with dominance of subdivision 6 (10 clones). Upon full-length insert sequencing, gene annotation identified a total of 350 open reading frames (ORFs), representing a broad range of functions. Remarkably, six inserts from subdivision 6 contained a region of gene synteny, containing genes involved in purine de novo biosynthesis and encoding tRNA synthetase and conserved hypothetical proteins. Similar genomic regions had previously been observed in several environmental clones recovered from soil and marine sediments, facilitating comparisons with respect to gene organization and evolution. Comparative analyses revealed a general dichotomy between marine and terrestrial genes in both phylogeny and G+C content. Although the significance of this homologous gene cluster across subdivision 6 members is not known, it appears to be a common feature within a large percentage of all acidobacterial genomic fragments recovered from both of these environments.
The genetic structure and functioning of the soilborne microorganisms are still poorly understood in part due to the huge complexity of these communities. Molecular methods targeting 16S rRNA genes have indeed revealed that soil microbial communities may be comprised of many thousands of different species in a single gram of soil (52). Moreover, the fraction of bacterial populations in soil that can be accessed through standard cultivation techniques is very low (>5%) (3, 10, 52), limiting our ability to obtain functional information about many of the microbial populations that dominate soil. Although novel methodologies for culturing soil microbes have been successful in culturing many soil bacteria (27), limitations remain with respect to biases and failure to detect essential microbial interactions and processes (49).
As phylogenetic inventories of soil microbial communities have accrued, it has become clear that numerous bacterial lineages appear to be quite common in the environment but are underrepresented in bacterial culture collections (19). One of the best examples of such a lineage is that of the Acidobacteria. This phylum is one of the most commonly detected phyla in soil habitats by molecular methods, suggesting that members of this phylum may be important drivers of key ecosystem processes in terrestrial ecosystems. However, physiological information on isolates is essentially lacking, and only three complete genome sequences of isolates of this phylum have been determined to date, including those for “Solibacter usitatus” (subdivision 3), “Korebacter versatilis” Ellin345 (subdivision 1), and Acidobacterium capsulatum (subdivision 1) (58).
The designation Acidobacteria was coined after the first described species of this phylum, Acidobacterium capsulatum, which was isolated in the late 1990s from an acidic mine drainage (36). The phylum name is somewhat misleading since Acidobacteria are not restricted to acidic environments, as evidenced by their common detection in other habitats, including ocean water, sediments, hot springs, peat bogs, and soil (5, 8, 11, 18, 34, 42, 43, 48). Members of the Acidobacteria seem to be particularly dominant in the soil, representing up to 52% of 16S rRNA gene sequences from clone libraries (46) and typically accounting for 20 to 30% of all bacterial 16S rRNA sequences amplified by PCR from soil DNA (19, 26).
By means of phylogenetic analyses, Acidobacteria were originally described as having 4 to 5 subdivisions (31), with subsequent expansion to having between 8 and 11 subdivisions (22, 59). With larger sequencing efforts, it has recently been proposed that the Acidobacteria may be composed of 26 or more subdivisions (4), the majority of which still lack culture representatives. Given their prevalence in soil environments, and the lack of described cultures for most subdivisions, the Acidobacteria have been proposed as ideal targets for study via large-insert metagenomic approaches (30).
The first description of acidobacterial genome fragments obtained from a metagenome library was provided by Liles et al. (33). By screening 24,400 bacterial artificial chromosome (BAC) clones from a soil metagenome library for 16S rRNA genes, those authors detected 12 clones putatively derived from members of the phylum Acidobacteria (9 clones from subdivision 6, 2 from subdivision 4, and 1 from subdivision 5). A single BAC insert (from subdivision 5) was selected for full-length sequencing, giving the first glimpse into the genome of an organism from this mysterious bacterial phylum. In the first study specifically targeting the Acidobacteria, Quaiser et al. (45) recovered 6 acidobacterial fosmid clone fragments (4 out of 6 related to subdivision 6) from a sandy ecosystem, followed by the recovery of an additional 11 clones from deep-sea environments (44). This allowed for a preliminary comparison of the genomic organizations in the chromosomal regions adjacent to rRNA operons, revealing a number of clones affiliated with subdivision 6, which contained a colinear region encoding genes involved in purine biosynthesis.
In this study, we sought to recover additional genomic information from Acidobacteria within soil environments, with the goal of providing greater comparative insight into the function and evolution of members of this phylum. To this end, a large-insert fosmid library (28,800 clones) was constructed from high-molecular-weight DNA isolated from the rhizosphere of Festuca ovina L. in a former agricultural soil and screened for inserts of acidobacterial origin. The resulting 17 positive clones were subsequently subjected to full-length DNA sequence analysis, followed by identification and annotation of open reading frames (ORFs). General genomic properties were determined for all sequenced inserts and compared across the Acidobacteria subdivisions represented in the library. A number of clones contained regions of synteny with previously described acidobacterial gene fragments, facilitating the phylogenetic analysis of genes contained within multiple clones and comparisons across soil and marine habitats.
MATERIALS AND METHODS
Field site and sampling.
Soil samples were collected from a former arable field site located near Ede, Netherlands (52o4′N, 5o45′E), where an ecosystem restoration experiment was initiated after the 1995 harvest season within the European project titled “Changing Land Usage: Enhancement of Biodiversity and Ecosystem Development” (6, 54). The soil is described as loamy sand and had a pH (KCl) of 5.8 and organic matter of 4.5%. Detailed soil characteristics can be found in the work of Van der Putten et al. (54). Rhizosphere soil samples were collected in August 2005 from the rhizosphere of Festuca ovina L. plants from within plots sown with a monoculture of this species, adjacent to the main plant diversity treatment experiment (28). For purposes of this study, rhizosphere was defined as a combined sample of rhizosphere and root surface (rhizoplane), where pieces of roots and adhering soil after shaking were collected. Soil samples were frozen at −20°C for approximately 2 weeks prior to DNA extraction.
Metagenome library construction and screening.
For large-insert metagenomic library construction, high-molecular-weight DNA was extracted from 10 g of soil (wet weight) as previously described (29, 55). The construction of the metagenome library utilized the CopyControl fosmid library production kit (Epicentre, Madison, WI) and was preformed according to the manufacturer's protocol. This protocol yielded 5′-phosphorylated blunt-end DNA, which was subsequently ligated into the pCC1FOS vector and transformed into Escherichia coli EPI300 (Epicentre, Madison, WI). The final metagenome library contained 28,800 chloramphenicol resistance clones selected on LB medium plates, with addition of 12.5 μg ml−1 of chloramphenicol.
All 28,800 clones in the metagenome library were subjected to a PCR-based screening strategy using a seminested clone pooling strategy (28, 29) and the primers pA/1378R (13) and Acd31F/1378R (5). Briefly, clones were cultured overnight in 96-well plates, with the contents of 4 plates (384 clones) combined in a single plasmid extraction (QIAprep Spin Miniprep kit; Qiagen, Venlo, Netherlands). The resulting mixed template was used for PCR with the pAF/1378R primer pair, followed by amplification with primers Acd31F/1378R, as described by Kielak et al. (28, 29). In the cases where PCR product was detected, clone pooling was reduced to a single plate. Pooling was further reduced to a single row of a culture plate and eventually to a single clone for subsequent reactions that produced positive product. After partial 16S rRNA gene sequencing and preliminary phylogenetic analysis, 17 clones affiliated to the Acidobacteria phylum were selected for full-length sequencing. Sequencing was performed by Macrogen (Seoul, South Korea). Contig assembly was carried out using Lasergene SeqMan (DNAStar). Remaining sequence gaps were closed by primer walking with sequence-derived oligonucleotides. A few gaps could not be closed due to the technical difficulties (Table 1).
TABLE 1.
General characteristics of acidobacterial fosmid clones
| Description | Value(s) for fosmid no.: |
Value(s) for fosmid no.: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 246 | 162 | 282 | 89 | 148 | 164 | 253 | 59 | 66 | 70 | 92 | 98 | 126 | 213 | 259 | 270 | 293 | |
| Subdivision | —c | 1 | 1 | 3 | 4 | 4 | 4 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
| Length | 32,608 | 36,502 | 29,311 | 31,620 | 30,784 | 33,664 | 33,004a | 37,659 | 29,899 | 33,759 | 33,281 | 30,831a | 28,124 | 35,443 | 31,287a | 38,585a | 36,563a |
| No. of gaps | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 3 | 4 |
| % GC | 57.7 | 58.7 | 55.5 | 59.3 | 70.3 | 54.4 | 53.8 | 64.5 | 63.4 | 62.7 | 62.7 | 65.5 | 66.3 | 65.2 | 65.9 | 65.0 | 69.2 |
| % GC in ORF | 57.4 | 60.8 | 59.9 | 71.1 | 55.2 | 53.6 | 65.9 | 64.8 | 63.9 | 63.8 | 67.6 | 67.9 | 67.1 | 68.3 | |||
| No. of ORF | 28 | 30 | 26 | 28 | 26 | 29 | 23 | 26 | 20 | 27 | 22 | 26 | 19 | 21 | 27 | 30 | 36 |
| No. ORFs matching nonhypothetical proteins | 13 | 20 | 17 | 14 | 10 | 14 | 13 | 13 | 9 | 12 | 5 | 20 | 5 | 10 | 13 | 15 | 24 |
| No. of ORFs matching hypothetical proteinsb | 3/12 | 3/7 | 3/6 | 10/4 | 3/13 | 3/12 | 4/6 | 6/7 | 6/5 | 10/5 | 7/10 | 4/2 | 7/7 | 5/6 | 5/9 | 11/4 | 3/9 |
| No. of tRNAs | 4 | 2 | 3 | 2 | 1 | 4 | 3 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 1 |
| Type(s) of rRNA | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 16S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S | 5S, 16S, 23S |
Length of recovered sequences without gap length estimation.
Number of ORFs with best matches to hypothetical proteins or proteins of unknown function/number of ORFs without any close match in the database.
—, subdivision designation unclear.
Annotation and sequence properties.
Open reading frames (ORFs) were assigned using the GLIMMER (12) and FGENESB (http://linux1.softberry.com) software tools. Annotation of putative ORFs was accomplished based on a similarity search against nonredundant protein databases using BLASTX and BLASTP algorithms, using an E value threshold of <10−15. ORFs shorter than 150 bp were discarded. tRNA genes were identified using the tRNAscan-SE program version 1.21 (35). ORFs were classified into functional categories as clusters of orthologous groups (COGs) by using COGNITOR (51). Searches for GC islands were performed using CpGFinder (http://linux1.softberry.com). For identification of potential horizontal gene transfer regions, the method described by Tamames and Moya (50) was applied to examine tetranucleotide frequencies. For comparative analysis, full-length fosmid inserts sequences were compared using polydot (http://emboss.bioinformatics.nl/cgi-bin/emboss/polydot). This was followed by reciprocal BLASTN and TBLASTX searches among selected fosmids using the BLAST version 2.2.18 package (1; ftp://ftp.ncbi.nih.gov). Genomic fragment comparisons were visualized using the Artemis Comparison Tool version 7 (7).
Phylogenetic analyses.
All obtained 16S rRNA gene sequences were aligned using the ARB software package (37) and manually edited, considering the secondary structure of the rRNA molecule. Gaps and ambiguously aligned positions were excluded. The following two different methods were used for phylogenetic tree construction based upon small-subunit (SSU) rRNA gene sequences: neighbor joining (NJ) and maximum likelihood (ML). MrModeltest2.2 (41) was used to determine the most appropriate nucleotide substitution model. The likelihood ratio test, as well as the Akaike information criterion, identified the general time-reversible (GTR) model with invariable sites and Γ-shaped distribution of substitution rates as the best-fitting model. Both trees were constructed using the PHYLIP 3.67 package (15), with the model parameters calculated by MrModeltest. The NJ tree was bootstrapped using 1,000 replicates, and the ML tree was bootstrapped using 100 replicates. For protein phylogeny reconstructions, closest orthologues were identified with BLASTP from NCBI (2; http://www.ncbi.nlm.nih.gov/BLAST). Phylogenetic reconstructions were performed with the PHYLIP 3.67 package (15), using NJ and ML.
Testing for potential sites of recombination.
To help identify potential sites of recombination, a multiple nucleotide sequence alignment was made for the overlapping regions of fosmids 270, 59, and 98 and fosmids with GenBank accession numbers AY281356, AY281352, EF597689, EU686586, EF597694, EU686638, EU686603, EU686608, and EU795230 (44, 45). This alignment was examined using several different statistical algorithms for predicting recombination probability, namely, RDP, GENECONV, MaxChi, Chimera, and SiScan. Analyses were carried out with the RDP software package, version 3.34 (38).
Nucleotide sequence accession numbers.
The sequences described in this study have been deposited in the GenBank database under accession numbers GU260698 to GU260714.
RESULTS AND DISCUSSION
General characteristics and annotation of acidobacterial genomic fragments recovered from a rhizosphere metagenomic library.
In total, 17 putatively acidobacterial genomic fragments were detected in the library (28,800 fosmid clones) based upon PCR detection of 16S rRNA genes affiliated with this phylum. Insert sizes ranged from approximately 30 to 38 kb. The majority of clones appeared to be derived from members of Acidobacteria subdivision 6 (10 out of 17 clones), and fragments from subdivisions 4, 1, and 3 were also recovered (3, 2, and 1 fosmid clones, respectively) (Table 1). This prevalence of subdivision 6 is in agreement with previous studies targeting soil habitats (5, 14, 20, 26, 31). It should be noted that the screening strategy used here to identify target inserts utilized an acidobacterium-specific primer, which has recently been demonstrated to miss some lineages within this phylum (4, 29). However, based upon general bacterial clone analyses of the soil used in this study (28), these lineages probably represent only a very small fraction of the total acidobacterial community.
The G+C contents of inserts ranged from 54 to 70%, and G+C contents were generally rather consistent within each subdivision. Subdivision 6 sequences had the highest average overall G+C content at 65.2%, ranging from 62.7% in clones 70 and 92 to 69.2% in clone 293. The G+C contents observed in this study were comparable to those obtained previously for soil fosmids by Quaiser et al. (45). However, these G+C contents differed sharply from the values observed in fosmids recovered from aquatic environments (44), where nine subdivision 6 inserts averaged only 54.1% G+C. It has previously been suggested that environment factors can influence G+C content and potentially the specific amino acid composition of proteins (16). Similar contrasts in average G+C content have been observed in the comparison of metagenomic data obtained from shotgun cloning and sequencing approaches. For instance, the G+C contents of soil metagenomic sequences versus surface water metagenomic sequences differ by almost 30% (61% versus 34%, respectively) (16, 53, 56). Interestingly, this dichotomy across habitats appears to hold for organisms that are phylogenetically rather closely related. However, the reasons and mechanisms in which this occurs are not yet known. The effects of genome size (21) and responses to shifts in optimal growth temperature have been suggested as potential factors involved in shaping G+C content (39, 40), although some evidence also speaks against the latter explanation (17, 23, 57).
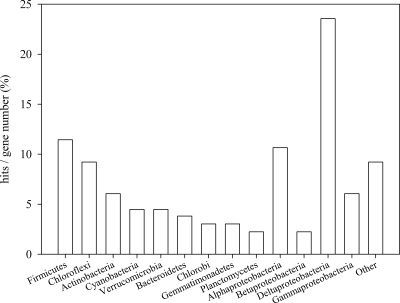
Full-length fosmid insert sequences were subjected to gene annotation, revealing a range of predicted gene functions (see Tables S1 to S17 in the supplemental material). A large fraction of predicted ORFs encoded housekeeping functions or were assigned to hypothetical protein groups. The majority of best BLAST hits were with genes from one of the three available Acidobacteria genome sequences (58). However, a sizeable proportion (46%) of ORFs showed best BLAST hits to genomes other than those in Acidobacteria. Previous studies have suggested an evolutionary link between the Acidobacteria and the Proteobacteria, with the suggestion that the Acidobacteria may be a sister group of the Deltaproteobacteria (9). This link is understandable given the results of the best BLAST hits from the current study, where the greatest number of sequences that did not match with available Acidobacteria genome sequences (Fig. 1) showed best matches with Deltaproteobacteria. However, it is still not clear if this is more due to the prevalence of available proteobacterial genome sequences in available databases as opposed to a true evolutionary link to the Acidobacteria. This may be resolved as additional Acidobacteria genome sequences become available.
FIG. 1.
Proportion of best matches to bacterial phyla other than the Acidobacteria (Proteobacteria shown at the class level) ORFs identified in a fosmid library constructed from soil-extracted DNA. Analysis was performed using tBLASTx.
Gene order and regions of synteny between fosmid clones.
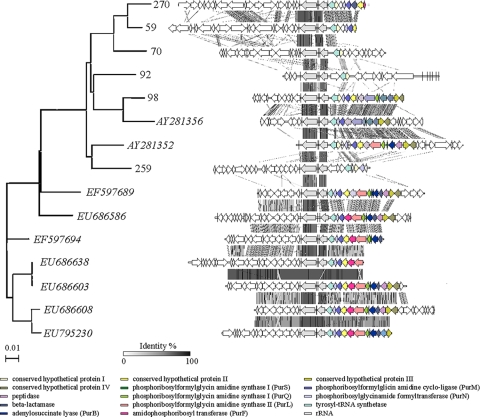
Among the 10 clones that were affiliated with subdivision 6, 6 clones contained a homologous region of variable length (clones 59, 70, 92, 98, 259, and 270). Remarkably, this region of colinearity is also present in two previously published soil metagenomic clones (33, 45), as well as several metagenomic clones from deep-sea samples (44). Thus, it appears that a large fraction of the total acidobacterial communities in diverse habitats share this feature. The region of synteny contains a tyrosyl-tRNA transferase, a hypothetical protein, and genes involved in de novo purine synthesis (Fig. 2). The degree of similarity of genome fragment organization generally followed the phylogenetic distances determined based on 16S rRNA gene sequence analysis. Further research will be required to determine whether there is some special feature of these genes that has led to their conservation adjacent to rRNA operons within this phylum and whether these Acidobacteria populations also share syntenous regions elsewhere on their chromosomes.
FIG. 2.
Comparative genomic organization of genomic fragments affiliated with acidobacterial subdivision 6 (fosmid clones 59, 70, 92, 98, 259, and 270 from this study; fosmid clones with GenBank accession numbers AY281356 and AY281352 from the study of Quaiser et al. [46]; and fosmid clones with GenBank accession numbers EF597689, EU686586, EF597694, EU686638, EU686603, EU686608, and EU795230 from another study of Quaiser et al. [45]). A neighbor-joining tree of 16S rRNA gene sequences is shown on the left as a reference. Sequences are aligned according to the ribosomal operon localization. 16S rRNA and 23S rRNA genes are indicated as gray arrows. Conserved regions between genomic fragments are indicated by gray-shaded areas, with gray intensity being a function of sequence similarity by tBLASTx over lengths greater than 150 bp. Particular ORFs mentioned in the text are highlighted in color.
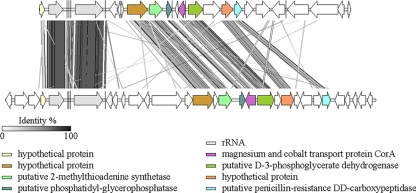
A region of colinearity was also identified for two fosmid inserts affiliated with subdivision 4, clone 253 from this study and clone 46h5 (GenBank accession numbers AY281357 and AY281358) from the study of Quaiser et al. (45) (Fig. 3). However, there are only five genomic fragments that have been recovered to date from members of this subdivision (three from this study and two from the study of Quaiser et al. [45]). Thus, it remains to be seen if this observation represents a lucky coincidence or a more widespread phenomenon, as seen for the syntenous regions within subdivision 6 clones. None of the recovered fosmid clones had any significant regions of colinearity with the genomes available for A. capsulatum, strain Ellin345, or strain Ellin607.
FIG. 3.
Comparative genomic organization of fosmid 253 (subdivision 4) (top) in relation to previously described fosmid 46h5 (46) (bottom). Conserved genomic regions between fosmid clones are indicated by gray-shaded areas, with gray intensity being a function of sequence similarity by tBLASTx over lengths greater than 150 bp. Particular ORFs mentioned in the text are highlighted in color.
Phylogenetic analysis of members of subdivision 6 based on 16S rRNA genes and genes shared between fosmid clones.
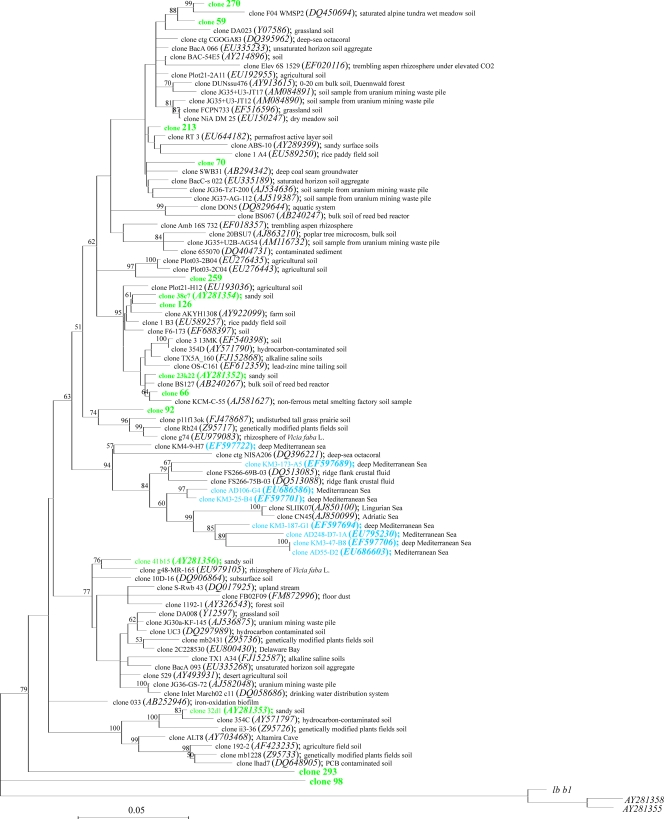
Given that the majority of metagenomic clones were affiliated with subdivision 6, we chose to focus special attention on the phylogenetic analysis of this subdivision. There is a large degree of sequence variation within this subdivision, and the diversity of sequences derived from soils appears to be much greater than that found in aquatic environments (Fig. 4). Phylogenetic analysis of the 16S rRNA gene sequences belonging to subdivision 6 revealed that sequences recovered from aquatic environments seemed to cluster rather tightly, with sequence similarities ranging from 0.911 to 0.999 (see Table S18 in the supplemental material). Soil clones, on the other hand, were spread across the breadth of the subdivision, with sequence similarities ranging from 0.822 to 0.999. Thus, soilborne Acidobacteria appeared to be more diverse, with a total 16S rRNA gene diversity that was comparable to those observed within many phyla (47).
FIG. 4.
Neighbor-joining tree of subdivision 6 of the Acidobacteria, highlighting the sequences recovered by this and two other metagenomic studies (45, 46). Names in italics indicate sequences retrieved from GenBank. Green, this study and soil metagenome library (46); blue, deep-sea metagenomic library (45). Numbers near nodes indicate bootstrap values. Sequences from subdivision 4 were used as an outgroup.
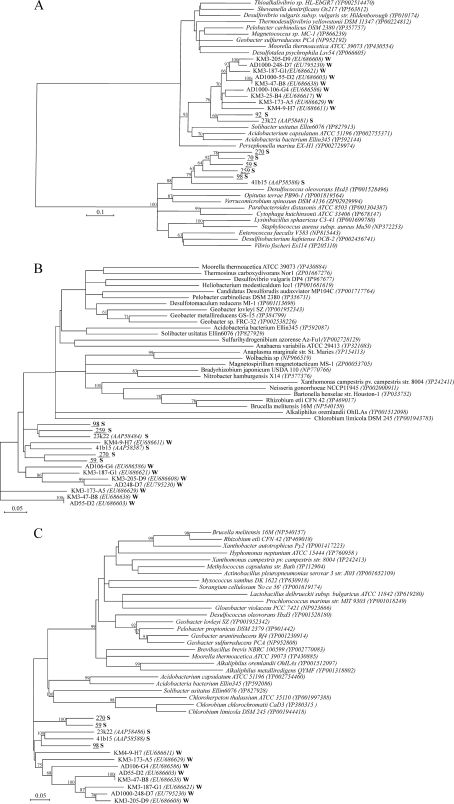
The recovery of regions of synteny (see below) between several clones within this and previous studies (45, 46) also allowed for a more in-depth analysis of several protein-encoding genes for comparison with 16S rRNA gene-based phylogeny. The clustering of soil and aquatic sequences within the SSU gene-based tree is generally mirrored in the phylogenies of the protein-encoding genes that we examined, as well as comparisons across the full lengths of the inserts that exhibit regions of synteny (Fig. 2). The phylogenetic reconstructions derived from PurM and PurN show the Acidobacteria as a monophyletic group, with a clear separation of environmental sequences based upon habitat (Fig. 5 B and C). Interestingly, TyrS exhibits a different pattern, with two distinct groups of sequences, affiliated with different bacterial groups (Fig. 5A). All the sequences recovered from deep-sea environments (Fig. 5A, designated with “W”) clustered tightly and were affiliated with the sequences derived from two soil clones as well as the three fully sequenced Acidobacteria genomes. The remaining soil clones belonged to the other TyrS subclass and clustered together, showing greatest similarity to the sequence available for Desulfococcus oleovorans (GenBank accession number YP_001528496). It remains to be investigated why these genes display such contrasting phylogenies.
FIG. 5.
Maximum likelihood phylogenetic analysis of tyrosyl-tRNA synthetase (TyrS) (A), phosphoribosylformylglycinamidine cyclo-ligase (PurM) (B), and trifunctional purine biosynthetic protein (PurN) (C). Amino acid sequences obtained in this study are underlined. A “W” after the name indicates sequences obtained from deep-sea clones, and an “S” after the name indicates sequences obtained from soil clones. The analysis was performed on 220 (A), 262 (B), and 125 (C) unambiguously aligned amino acid positions. Numbers near nodes indicate bootstrap values (only bootstrap values of ≥60 are shown).
To detect potential hot spots for recombination within the overlapping region of subdivision 6 clones, a composite alignment (12 clones, 8,750 bp, and using clone 270 as a reference) was subjected to several tests designed to detect sequence regions for which recombination events may explain sequence patterns (38). All predicted events with statistical significance of P of <0.05 detected by one or more of the five algorithms used (RDP, GENECONV, MaxChi, Chimera, and SiScan) are listed in the Table S19 in the supplemental material. Breakpoints were not evenly distributed across the examined region (see Fig. S1 in the supplemental material), with the highest number of breakpoints detected within PurM (22), followed by TyrS, PurN, and two conserved hypothetical proteins (16, 12, 10, and 3 breakpoints, respectively) (see Fig. S1 in the supplemental material). The various methods used yielded different results but generally revealed the same patterns of hot and cold spots for potential recombination across the analyzed region, and only a single site was identified to be significant by all algorithms used. Thus, recombination analysis suggested that the presence of hot spots of recombination within the regions of synteny were observed within subdivision 6 clones, but these regions did not appear to be unusual in their G+C contents, codon usage, or tetranucleotide usage patterns (not shown).
Conclusions.
The metagenomic screening method employed in this study proved to be successful for the recovery of genomic information from diverse Acidobacteria directly from the environment. However, although a large amount of genomic information could be obtained, insights into ecology are still limited due to the lack of niche-defining genes and physiological knowledge for members of this phylum. Despite this limitation, the information recovered did allow for the most extensive phylogenetic and evolutionary study of Acidobacteria subdivision 6 to date. Also, the information gathered via the rRNA gene-based PCR screening method required a large number of clones to be screened and provided only insight into the genomic regions directly adjacent to rRNA operons. Additional genome sequencing efforts for a broad range of strains of Acidobacteria will no doubt facilitate metagenomic efforts, as this should allow the identification of metagenomics clones of acidobacterial origin by end sequencing and binning. Also, novel high-throughput screening methods (24) and single-cell sequencing approaches (25, 32) hold great promise in expanding our understanding of the genomic diversity of this phylum.
It is amazing that the observed region of gene synteny within subdivision 6 fosmid clones was found so broadly and frequently. The significance of this finding and the potential for other regions of homology across the chromosomes of subdivision 6 members remain to be determined. Given the widespread nature of strains exhibiting this feature, and the large proportion of Acidobacteria subdivision 6 in diverse ecosystems, this represents a clear research priority.
Supplementary Material
Acknowledgments
This work was funded by the BSIK program Ecogenomics (http://www.ecogenomics.nl/).
Eiko E. Kuramae is acknowledged for her support and valuable discussions regarding the work presented in the manuscript.
Footnotes
Published ahead of print on 20 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in-situ detection of individual microbial-cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barns, S. M., E. C. Cain, L. Sommerville, and C. R. Kuske. 2007. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 73:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezemer, T. M., and W. H. van der Putten. 2007. Ecology-diversity and stability in plant communities. Nature 446:E6-E7. [DOI] [PubMed] [Google Scholar]
- 7.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 8.Chan, O. C., X. D. Yang, Y. Fu, Z. L. Feng, L. Q. Sha, P. Casper, and X. M. Zou. 2006. 16S rRNA gene analyses of bacterial community structures in the soils of evergreen broad-leaved forests in south-west China. FEMS Microbiol. Ecol. 58:247-259. [DOI] [PubMed] [Google Scholar]
- 9.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283-1287. [DOI] [PubMed] [Google Scholar]
- 10.Curtis, T. P., and W. T. Sloan. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221-226. [DOI] [PubMed] [Google Scholar]
- 11.Dedysh, S. N., T. A. Pankratov, S. E. Belova, I. S. Kulichevskaya, and W. Liesack. 2006. Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes—characterization of a gene coding for 16S-ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichorst, S. A., J. A. Breznak, and T. M. Schmidt. 2007. Isolation and characterization of soil bacteria that define Teniglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 73:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle, WA.
- 16.Foerstner, K. U., C. von Mering, S. D. Hooper, and P. Bork. 2005. Environments shape the nucleotide composition of genomes. EMBO Rep. 6:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, J. A. L., F. Bartumeus, D. Roche, J. Giraldo, H. E. Stanley, and E. O. Casamayor. 2008. Ecophysiological significance of scale-dependent patterns in prokaryotic genomes unveiled by a combination of statistic and genometric analyses. Genomics 91:538-543. [DOI] [PubMed] [Google Scholar]
- 18.Gomez-Alvarez, V., G. M. King, and K. Nusslein. 2007. Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiol. Ecol. 60:60-73. [DOI] [PubMed] [Google Scholar]
- 19.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansel, C. M., S. Fendorf, P. M. Jardine, and C. A. Francis. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74:1620-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, J. F., X. Q. Zhao, Z. Zhang, and J. Yu. 2007. Compositional dynamics of guanine and cytosine content in prokaryotic genomes. Res. Microbiol. 158:363-370. [DOI] [PubMed] [Google Scholar]
- 22.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurst, L. D., and A. R. Merchant. 2001. High guanine-cytosine content is not an adaptation to high temperature: a comparative analysis amongst prokaryotes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 268:493-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingham, C. J., A. Sprenkels, J. Bomer, D. Molenaar, A. van den Berg, J. Vlieg, and W. M. de Vos. 2007. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. U. S. A. 104:18217-18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishoey, T., T. Woyke, R. Stepanauskas, M. Novotny, and R. S. Lasken. 2008. Genomic sequencing of single microbial cells from environmental samples. Curr. Opin. Microbiol. 11:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kielak, A., A. S. Pijl, J. A. van Veen, and G. A. Kowalchuk. 2008. Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol. Ecol. 63:372-382. [DOI] [PubMed] [Google Scholar]
- 29.Kielak, A., A. S. Pijl, J. A. van Veen, and G. A. Kowalchuk. 2009. Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J. 3:378-382. [DOI] [PubMed] [Google Scholar]
- 30.Kowalchuk, G. A., A. Speksnijder, K. Zhang, R. M. Goodman, and J. A. van Veen. 2007. Finding the needles in the metagenome haystack. Microb. Ecol. 53:475-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasken, R. S. 2007. Single-cell genomic sequencing using multiple displacement amplification. Curr. Opin. Microbiol. 10:510-516. [DOI] [PubMed] [Google Scholar]
- 33.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Garcia, P., S. Duperron, P. Philippot, J. Foriel, J. Susini, and D. Moreira. 2003. Bacterial diversity in hydrothermal sediment and epsilon proteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 5:961-976. [DOI] [PubMed] [Google Scholar]
- 35.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K.-H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 39.Musto, H., H. Naya, A. Zavala, H. Romero, F. Alvarez-Valin, and G. Bernardi. 2004. Correlations between genomic GC levels and optimal growth temperatures in prokaryotes. FEBS Lett. 573:73-77. [DOI] [PubMed] [Google Scholar]
- 40.Musto, H., H. Naya, A. Zavala, H. Romero, F. Alvarez-Valin, and G. Bernardi. 2006. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem. Biophys. Res. Commun. 347:1-3. [DOI] [PubMed] [Google Scholar]
- 41.Nylander, J. A. A. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 42.Penn, K., D. Y. Wu, J. A. Eisen, and N. Ward. 2006. Characterization of bacterial communities associated with deep-sea corals on Gulf of Alaska seamounts. Appl. Environ. Microbiol. 72:1680-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polymenakou, P. N., S. Bertilsson, A. Tselepides, and E. G. Stephanou. 2005. Bacterial community composition in different sediments from the Eastern Mediterranean Sea: a comparison of four 16S ribosomal DNA clone libraries. Microb. Ecol. 50:447-462. [DOI] [PubMed] [Google Scholar]
- 44.Quaiser, A., P. Lopez-Garcia, Y. Zivanovic, M. R. Henn, F. Rodriguez-Valera, and D. Moreira. 2008. Comparative analysis of genome fragments of Acidobacteria from deep Mediterranean plankton. Environ. Microbiol. 10:2704-2717. [DOI] [PubMed] [Google Scholar]
- 45.Quaiser, A., T. Ochsenreiter, C. Lanz, S. C. Schuster, A. H. Treusch, J. Eck, and C. Schleper. 2003. Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics. Mol. Microbiol. 50:563-575. [DOI] [PubMed] [Google Scholar]
- 46.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 47.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamames, J., and A. Moya. 2008. Estimating the extent of horizontal gene transfer in metagenomic sequences. BMC Genomics 9:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 53.Tringe, S. G., C. von Mering, A. Kobayashi, A. A. Salamov, K. Chen, H. W. Chang, M. Podar, J. M. Short, E. J. Mathur, J. C. Detter, P. Bork, P. Hugenholtz, and E. M. Rubin. 2005. Comparative metagenomics of microbial communities. Science 308:554-557. [DOI] [PubMed] [Google Scholar]
- 54.Van der Putten, W. H., S. R. Mortimer, K. Hedlund, C. Van Dijk, V. K. Brown, J. Leps, C. Rodriguez-Barrueco, J. Roy, T. A. D. Len, D. Gormsen, G. W. Korthals, S. Lavorel, I. S. Regina, and P. Smilauer. 2000. Plant species diversity as a driver of early succession in abandoned fields: a multi-site approach. Oecologia 124:91-99. [DOI] [PubMed] [Google Scholar]
- 55.van Elsas, J. D., A. Speksnijder, and L. S. van Overbeek. 2008. A procedure for the metagenomics exploration of disease-suppressive soils. J. Microbiol. Methods 75:515-522. [DOI] [PubMed] [Google Scholar]
- 56.Venter, J. C., K. Remington, J. Hoffman, H. Baden-Tillson, C. Pfannkoch, H. O. Smith, A. L. Halpern, D. Rusch, S. Levy, J. F. Heidelberg, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, O. White, J. Peterson, Y.-H. Rogers, K. Nealson, A. H. Knap, M. W. Lomas, and R. Parsons. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 57.Wang, H. C., E. Susko, and A. J. Roger. 2006. On the correlation between genomic G+C content and optimal growth temperature in prokaryotes: data quality and confounding factors. Biochem. Biophys. Res. Commun. 342:681-684. [DOI] [PubMed] [Google Scholar]
- 58.Ward, N. L., J. F. Challacombe, P. H. Janssen, B. Henrissat, P. M. Coutinho, M. Wu, G. Xie, D. H. Haft, M. Sait, J. Badger, R. D. Barabote, B. Bradley, T. S. Brettin, L. M. Brinkac, D. Bruce, T. Creasy, S. C. Daugherty, T. M. Davidsen, R. T. DeBoy, J. C. Detter, R. J. Dodson, A. S. Durkin, A. Ganapathy, M. Gwinn-Giglio, C. S. Han, H. Khouri, H. Kiss, S. P. Kothari, R. Madupu, K. E. Nelson, W. C. Nelson, I. Paulsen, K. Penn, Q. Ren, M. J. Rosovitz, J. D. Selengut, S. Shrivastava, S. A. Sullivan, R. Tapia, L. S. Thompson, K. L. Watkins, Q. Yang, C. Yu, N. Zafar, L. Zhou, and C. R. Kuske. 2009. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75:2046-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmermann, J., J. M. Gonzalez, C. Saiz-Jimenez, and W. Ludwig. 2005. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in altamira cave using 23S rRNA sequence analyses. Geomicrobiol. J. 22:379-388. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.