Abstract
Cutinase from Thermobifida fusca is thermally stable and has potential application in the bioscouring of cotton in the textile industry. In the present study, the carbohydrate-binding modules (CBMs) from T. fusca cellulase Cel6A (CBMCel6A) and Cellulomonas fimi cellulase CenA (CBMCenA) were fused, separately, to the carboxyl terminus of T. fusca cutinase. Both fusion enzymes, cutinase-CBMCel6A and cutinase-CBMCenA, were expressed in Escherichia coli and purified to homogeneity. Enzyme characterization showed that both displayed similar catalytic properties and pH stabilities in response to T. fusca cutinase. In addition, both fusion proteins displayed an activity half-life of 53 h at their optimal temperature of 50°C. Compared to T. fusca cutinase, in the absence of pectinase, the binding activity on cotton fiber was enhanced by 2% for cutinase-CBMCel6A and by 28% for cutinase-CBMCenA, whereas in the presence of pectinase, the binding activity was enhanced by 40% for the former and 45% for the latter. Notably, a dramatic increase of up to 3-fold was observed in the amount of released fatty acids from cotton fiber by both cutinase-CBM fusion proteins when acting in concert with pectinase. This is the first report of improving the scouring efficiency of cutinase by fusing it with CBM. The improvement in activity and the strong synergistic effect between the fusion proteins and pectinase suggest that they may have better applications in textile bioscouring than the native cutinase.
Cotton fiber has a multilayered structure, with its outermost surface being the cuticle that is cross-linked to the primary cell wall of cotton fiber by esterified pectin substances. The major component of the cuticle is cutin, an insoluble polyester composed mainly of saturated C16 and C18 hydroxy and epoxy fatty acids (14, 16, 27, 38). During the process of scouring in the textile industry, the cuticle layer has to be removed in order to improve the wettability of cotton fiber, which then facilitates uniform dyeing and finishing. Traditionally, this process is performed by hot hydrolysis in alkaline medium, which not only consumes large quantities of water and energy but also causes severe pollution and fiber damage (20, 21, 33). Therefore, environment-friendly scouring methods based on biocatalysts have been actively sought (2, 30, 36).
Cutinase is a multifunctional esterase capable of degrading the cutin component of the cuticle. Earlier reports showed that the fungal cutinase from Fusarium solani pisi has potential use for cotton cuticle degradation and exhibits a good synergistic effect with pectinase, an enzyme utilized to degrade pectin, in the scouring of cotton fiber (1, 7, 8, 14). Moreover, site-directed mutagenesis has been performed to replace the specific amino acid residues near the active site of cutinase (3) to improve its hydrolytic activity toward polyesters. More recently, a cutinase from the thermophilic bacterium Thermobifida fusca has been identified and overexpressed in Escherichia coli in our laboratory (10). The good thermal stability and alkali resistance of this recombinant T. fusca cutinase make it potentially more amenable to textile bioscouring (10).
To further improve the applicability and/or catalytic efficiency of T. fusca cutinase, the present study attempts to engineer a novel cutin-degrading enzyme, based on analysis of the surface structure of cotton fiber. It has been observed that, in addition to cutin, pectin, proteins and other components, there is also a large amount of cellulose on the surface layer of cotton fiber (23). Thus, it is tempting to hypothesize that if the enzyme can be engineered to specifically bind to cellulose through a “gain of function” modification, its concentration on the surface of cotton fiber could increase significantly. Subsequently, its catalytic efficiency for cutin breakdown could be improved due to a proximity effect. In order to design such an enzyme, a fusion protein strategy in which a cellulose-binding protein/module will be attached to cutinase is considered.
It is well known that cellulase is capable of binding specifically to cellulose (25, 31). This enzyme has two separate modules: a catalytic module and a carbohydrate-binding module (CBM) (11). The two modules are discrete structural and functional units usually connected by a flexible linker (5, 17, 28). CBM has high specific capacities for cellulose binding. Previously, it has been reported that CBM is able to be fused to a chosen target protein by genetic manipulation (36), resulting in enhanced binding of this fusion protein to cellulose (6, 29). For example, fusion proteins were constructed by fusing CBM to β-glucose nucleotide enzyme (GUS) (13) or β-glycosidase (BglA) (19), which facilitates biochemical analysis of scouring efficiency for cotton fabrics.
In the present study, the CBM from T. fusca cellulase Cel6A (CBMCel6A) and the CBM from Cellulomonas fimi cellulase CenA (CBMCenA) were fused, separately, to the carboxyl terminus of T. fusca cutinase. The resulting fusion enzymes were compared to the native cutinase in terms of their biochemical properties, as well as the catalytic efficiency in cutin breakdown on cotton fiber. This is the first report of improving the scouring efficiency of cutinase by fusing it with CBM.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The T. fusca strain and plasmids pET20b(+)-Tfu_0883 and pBSK-CBMCenA were lab stocks (4, 10). Plasmid pET20b(+)-Tfu_0883 was used as the gene source of T. fusca cutinase. The T. fusca strain and plasmid pBSK-CBMCenA were used as the gene sources of CBMCel6A and CBMCenA, respectively. E. coli strain BL21(DE3) was used as the expression host, and pET20b(+) was used as the cloning and expression vector. Cells were grown in Luria-Bertani medium at 37°C and, if necessary, ampicillin in a final concentration of 100 μg/ml was added to the medium.
Construction of cutinase-CBM fusion protein expression vectors.
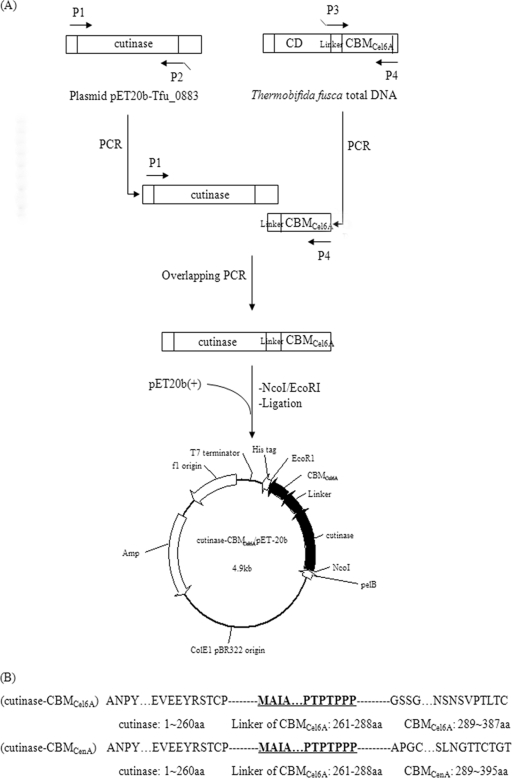
The genes encoding cutinase (NCBI accession number YP_288944) and CBMCenA (NCBI accession number AAA23084.1) were amplified by using plasmids pET20b-Tfu_0883 and pBSK-CBMCenA as the templates, respectively. The gene encoding CBMCel6A and its linker (NCBI accession number YP_289135) was amplified from T. fusca genomic DNA. Overlapping PCR was used to fuse the T. fusca CBMCel6A or C. fimi CBMCenA to the C terminus of T. fusca cutinase. The sequences of the primers are given in Table 1. The fusion procedure is shown in Fig. 1. An NcoI restriction site (in boldface) was introduced at the 5′ end of P1, while an EcoRI restriction site (in boldface) was introduced at the 3′ end of P4/P4′. The overlapping area is underlined. The primers were synthesized by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd.
TABLE 1.
Primers used in the construction of the fusion genes
| Primer | Sequence (5′-3′)a | Restriction enzyme |
|---|---|---|
| P1 | GGAATACCATATGTCCATGGCCAACCCCTACGAGCGCGG | NcoI |
| P2 | CGCGGCGATCGCCATGAACGGGCAGGTGGA | |
| P3 | TCCACCTGCCCGTTCATGGCGATCGCCGCG | |
| P4 | CATCTCGAGAGAATTCGGGCAGGTAAGGGTCGGAACAG | EcoRI |
| P2′ | GCGGCAGCCGGGAGCGGGAGGCGGCGTGGG | |
| P3′ | CCCACGCCGCCTCCCGCTCCCGGCTGCCGC | |
| P4′ | CATCTCGAGAGAATTCGGGGTGCCCGTGCAGGTGGTGC | EcoRI |
Restriction enzyme sites are indicated in boldface. Portions (15 bp) of the overlapping regions are underlined.
FIG. 1.
Construction of cutinase-CBM fusion proteins. (A) T. fusca cutinase was joined with the CBM from T. fusca Cel6A using overlapping extension PCR. The fusion of T. fusca cutinase with CBM from C. fimi CenA followed a similar strategy. (B) Amino acid composition of the fusion proteins.
The PCR was performed using 35 successive cycles as follows: denaturation at 94°C for 0.5 min, annealing at 56°C for 0.5 min, and primer extension at 72°C for 1.5 min. PrimeSTAR HS DNA polymerase was utilized during the process. The amplification product was isolated and ligated into the vector pMD18T-simple. The ligation mixture was used to transform chemically competent E. coli JM109. The plasmid isolated from these transformants was verified by restriction analysis, and the gene sequence was confirmed by DNA sequencing. The plasmids with the correct sequences for cutinase-CBMCel6A and cutinase-CBMCenA were named pMD18T/cutinase-CBMCel6A and pMD18T/cutinase-CBMCenA, respectively. The resulting plasmids were digested with NcoI and EcoRI and ligated into the similarly digested expression vector pET20b(+). The ligation mixture was used to transform chemically competent E. coli JM109 cells. The plasmid isolated from these transformants was verified by restriction analysis, and the gene sequence was confirmed again by DNA sequencing. The plasmids with the correct sequences for cutinase-CBMCel6A and cutinase-CBMCenA were named pET20b/cutinase-CBMCel6A and pET20b/cutinase-CBMCenA, respectively.
The enzymes used for DNA manipulations were purchased from TaKaRa Biotechnology Co., Ltd. DNA sequencing was performed by Shanghai Generay Biotechnology Co., Ltd. Genomic DNA extraction was performed according to the method of Sambrook et al. (32). Plasmid DNA was extracted by using the Sangon EZ-10 Spin Column Plasmid Mini-Preps kit. Plasmid and PCR products were recovered from agarose gel by using the Sangon purification kit.
Expression and purification of the cutinase-CBM fusion proteins.
E. coli BL21(DE3) cells harboring pET20b/cutinase-CBMCel6A or pET20b/cutinase-CBMCenA were grown in TB medium containing ampicillin (100 μg/ml) at 37°C. When the culture reached an A600 of 1.5 to 2.0, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.5 mM. The culture after 18 h of induction was centrifuged (10,000 × g, 30 min, 4°C), and the supernatant was collected.
The supernatant described above was treated with 70% (wt/vol) saturated ammonium sulfate solution, and the solution was kept at 4°C overnight. Precipitates were collected by centrifugation and dissolved in 50 ml of buffer A (20 mM sodium phosphate, 0.5 M NaCl, and 20 mM imidazole [pH 7.4]). The solutions were subsequently dialyzed against 2 liters of buffer A overnight and applied to a nickel affinity column preequilibrated with buffer A. The samples were allowed to bind with Ni-nitrilotriacetic acid agarose at a flow rate of 1 ml/min, followed by washing with buffer A until the UV baseline was reached. Elution was performed using a linear gradient from 0 to 500 mM imidazole in buffer A over 100 min. The fractions containing p-nitrophenyl butyrate (pNPB) hydrolase activity were pooled and dialyzed against 2 liters of buffer B (20 mM Tris-HCl [pH 8.0]) at 4°C overnight. The purified enzyme was concentrated by ultrafiltration and stored at −80°C.
Enzyme characterization of cutinase-CBMs.
For enzyme characterization, all of the values presented in graphs and tables are the means of three replications. Esterase activity, cutinase activity, and lipase activity were assayed as described previously (10), with pNPB, cutin, and triolein as the substrates, respectively.
Temperature optima of the fusion and native enzymes were measured at temperatures ranging between 20 and 70°C. The reaction was performed in a buffer containing 20 mM Tris-HCl, 10 mM NaCl, and 50 mM sodium taurodeoxycholate at pH 8 using pNPB as the substrate. Since the pH of the Tris buffer is temperature dependent, the buffers were adjusted to pH 8 at the desired temperature. The enzyme activity was measured by preincubating the buffer at the desired temperature for 2 min to allow it to reach the final pH of 8. The thermostability of the enzymes was determined by incubating the enzymes in 20 mM Tris-HCl (pH 8) at 50°C. At different intervals, samples were taken and assayed for residual activity using pNPB as the substrate.
pH optima of the fusion and native enzymes were investigated between pH 6 and 9 using either potassium phosphate buffer (pH 6 to 7) or Tris-HCl buffer (pH 7 to 9). To determine the pH stability, 20 mM concentrations of the following buffers were used: sodium acetate (pH 4 to 6), potassium phosphate (pH 6 to 7), Tris-HCl (pH 7 to 9), and glycine-NaOH (pH 9 to 11). The enzymes were preincubated in the various buffers at 37°C for 24 h, followed by the determination of residual activity using pNPB as the substrate.
The Michaelis-Menten parameters, Vmax and Km, were determined from Michaelis-Menten plots of specific activities at various substrate concentrations. Rates were measured in triplicate using pNPB (100 to 2,000 μM) as the substrate by continuous spectrophotometric analysis. Initial reaction velocities were calculated from the linear region (∼60 s) of the reaction progress curve and measured in triplicate by varying the concentration of the substrate. The apparent kinetic constant Km was calculated by using the GraphPad Prism program.
Protein quantification and SDS-PAGE analysis.
Protein concentrations were determined by using the Bio-Rad protein assay kit (Bio-Rad), with purified bovine serum albumin (Promega) as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in 10% acrylamide gels, and the proteins were visualized by staining with Coomassie brilliant blue R-250.
Binding and catalytic activity of cutinase-CBMs toward cotton fiber.
The adsorption property of the enzymes was tested with cotton fiber as described previously with minor modifications (4). Prior to treatment, raw cotton fiber was boiled for 1 min and then dried at 40°C overnight. Then, 1 g of pretreated cotton fiber was mixed with 0.1% penetrant and 0.5% bovine serum albumin in Tris-HCl buffer (20 mM, pH 8.0) at 25°C. The mixture was incubated for 30 min to avoid nonspecific binding. Equal units of native cutinase or fusion enzyme (10 U/ml toward pNPB) were then added to this solution, followed by incubation for another 1 h with shaking at 60 rpm. The reaction mixtures were centrifuged at 3,000 × g for 2 min, and the amount of unbound enzyme was estimated from the residual activity in the supernatant. The amount of cotton fiber-bound enzyme was calculated from the difference between the initial enzyme activity and unbound enzyme activity. Control assays were performed under the same conditions, except in the absence of raw cotton fiber. Pectinase was added when needed.
The desorption property of the enzymes was determined according to a method described previously (4). After 2 h of incubation, the reaction solution described above was diluted (1:20) with Tris-HCl buffer, and the mixture was incubated for another 60 min with shaking at 200 rpm at 25°C. The amount of desorbed enzyme was estimated from the enzymatic activity in the supernatant. All assays were performed in triplicate.
For the determination of released fatty acids catalyzed by cutinase or cutinase-CBM, a 10-ml reaction mixture containing 1 g of raw cotton fiber and 50 μM native or fusion cutinase was incubated in 20 mM Tris-HCl buffer with or without pectinase. The reaction mixture was shaken at 200 rpm for 18 h at 50°C. At various times, samples were removed and subjected to titration using 20 mM NaOH.
RESULTS
Construction and purification of cutinase-CBM fusion proteins.
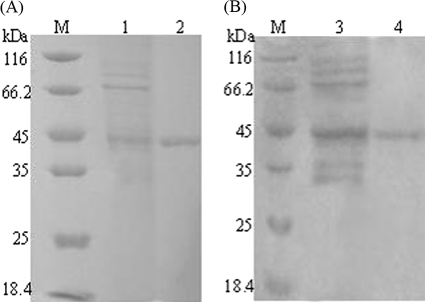
Cutinase-CBM fusion proteins were generated by fusing either the CBM of cellulase Cel6A from T. fusca (CBMCel6A) or the CBM of cellulase CenA from C. fimi (CBMCenA) to the C terminus of T. fusca cutinase through overlapping PCR amplification. The fused genes were subsequently inserted into the expression vector pET-20b(+), which encodes a C-terminal His6 tag and an N-terminal signal peptide PelB to allow the expressed proteins to be secreted. The resulting constructs pET20b/cutinase-CBMCel6A and pET20b/cutinase-CBMCenA were used for protein expression in E. coli BL21(DE3). The pNPB hydrolyzing activities in the culture supernatant were 94 U/ml for cutinase-CBMCel6A and 76 U/ml for cutinase- CBMCenA, values that were 230- and 190-fold greater, respectively, than that of the control culture [in which E. coli cells carried the vector pET-20b(+)]. Both fusion enzymes were purified by ammonium sulfate fraction and Ni-Sepharose affinity chromatography (see Tables S1 and S2 in the supplemental material). SDS-PAGE results demonstrated that they were purified to homogeneity with the same molecular mass of ∼45 kDa (Fig. 2). In addition, the purified enzymes were active with specific activities of 20,000 U/μmol of protein (see Table S1 in the supplemental material) for cutinase-CBMCel6A and 15,000 U/μmol of protein (see Table S2 in the supplemental material) for cutinase-CBMCenA.
FIG. 2.
SDS-PAGE analysis of the expression and purification of cutinase-CBMs. (A) Cutinase-CBMCel6A. Lanes: M, molecular mass markers; 1, culture supernatant of cutinase-CBMCel6A; 2, purified cutinase-CBMCel6A. (B) Cutinase-CBMCenA. Lanes: M, molecular mass markers; 1, culture supernatant of cutinase-CBMCenA; 2, purified cutinase-CBMCenA.
Enzymatic properties of cutinase-CBM fusion proteins.
The optimal temperature and pH of the fusion enzymes were determined at a temperature range of 20 to 70°C (see Fig. S1A in the supplemental material) and a pH range of 6 to 9 (see Fig. S1C in the supplemental material). For comparative purposes, T. fusca cutinase was also subjected to a similar analysis. The results showed that both cutinase-CBMCel6A and cutinase- CBMCenA exhibited an optimal temperature at 50°C, whereas the native cutinase displayed an optimal temperature at 60°C. Not surprisingly, all native and fusion enzymes exhibited the same optimal pH of 8. Subsequently, the thermostability was determined at the temperature of 50°C (see Fig. S1B in the supplemental material), whereas the pH stability was determined at pH values between 4 and 11 (see Fig. S1D in the supplemental material). The half-life of the native cutinase was 70 h at 50°C, while those of cutinase-CBM fusion enzymes were both 53 h at 50°C. Similar pH stabilities at a pH range of 6 to 9 were observed for all three enzymes.
Previously, it has been shown that T. fusca cutinase has broad substrate specificity against cutin, soluble esters, and insoluble triglycerides (10). The activity of cutinase-CBMs toward soluble ester had been confirmed in the purification and characterization analysis described above using pNPB as a substrate. For the insoluble triglycerides, both cutinase-CBMCel6A and cutinase-CBMCenA were found to be capable of hydrolyzing triolein with specific activities corresponding to 128 and 111% of the native cutinase, respectively. Furthermore, their cutin hydrolyzing activities were evaluated under their individual optimal temperature and pH. As shown in Table 2, the C16 and C18 family fatty acid monomers released after enzymatic reaction were 61% for cutinase-CBMCel6A, 64% for cutinase-CBMCenA, and 59% for the native cutinase. The hydroxy fatty acids, which are specific in cutin, were 3.6% for cutinase-CBMCel6A, 3.7% for cutinase-CBMCenA, and 3.1% for the native cutinase. These results demonstrated that both cutinase-CBMCel6A and cutinase-CBMCenA can hydrolyze cutin as efficiently as the native cutinase.
TABLE 2.
Monomeric products released from cutin hydrolysis by cutinase-CBMs and cutinasea
| Cutin hydrolysis product | Hydrolysis area (%) |
||
|---|---|---|---|
| Cutinase | Cutinase-CBMCel6A | Cutinase-CBMCenA | |
| Hexadecanoic acid | 27.6 | 30.6 | 31.4 |
| Octadecenoic acid | 26.7 | 25.2 | 27.5 |
| 9-Octadecenoic acid | 0.50 | 0.43 | 0.36 |
| 9,12-Octadecadienoic acid | 1.08 | 1.02 | 1.01 |
| 16-Hydroxyhexadecanoic acid | 0.54 | 0.54 | 0.58 |
| 18-Hydroxyoctadeca-9-enoic acid | 1.01 | 0.87 | 0.91 |
| 18-Hydroxyoctadeca-9,12-dienoic acid | 1.12 | 1.76 | 1.70 |
| 9,10,18-Trihydroxyoctadecanoic acid | 0.47 | 0.47 | 0.51 |
Cutin hydrolysis by cutinase-CBM and cutinase was carried out in 25 mM potassium phosphate buffer (pH 8.0) at 50°C, for 18 h. Each value represents the mean of three independent measurements, and the variation about the mean is below 5%.
The kinetics of the fusion enzymes was analyzed using pNPB as the substrate (Table 3). Their Km values were similar to that of the native cutinase, while their catalytic efficiencies (kcat/Km) were ca. 94% (cutinase-CBMCel6A) and 85% (cutinase-CBMCenA) of that of the native cutinase.
TABLE 3.
Kinetic parameters of cutinase-CBMs and cutinase
| Cutinase or cutinase-CBM | Mean ± SD |
kcat/Km (s−1) | |
|---|---|---|---|
| KmpNPB (μM) | kcat (s−1) | ||
| Cutinase | 640 ± 40 | 220 ± 10 | 0.34 |
| Cutinase-CBMCel6A | 620 ± 40 | 200 ± 10 | 0.32 |
| Cutinase-CBMCenA | 620 ± 30 | 180 ± 10 | 0.29 |
Binding and hydrolytic activity of cutinase-CBMs toward cotton fiber.
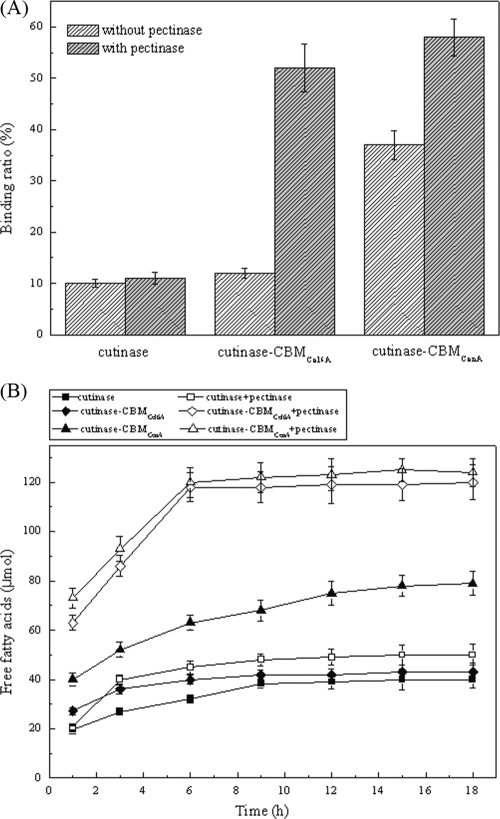
Adsorption of the enzyme on the surface of cotton fiber is the first step for cutinase to perform its hydrolysis toward cutin (37). The binding experiments (Fig. 3A) were performed under the conditions with or without the presence of pectinase, an enzyme utilized to remove pectin in bioscouring. The results showed that, compared to T. fusca cutinase, the binding of cutinase-CBMCel6A was enhanced by 2% in the absence of pectinase and 40% in the presence of pectinase, whereas the binding of cutinase-CBMCenA was enhanced by 28% in the absence of pectinase and 45% in the presence of pectinase. After dilution and reequilibration, almost all of the bound enzymes were desorbed. Thus, the binding of both cutinase and cutinase-CBMs with cotton fiber appeared to be reversible.
FIG. 3.
Scouring effects of cutinase-CBMs and cutinase on cotton fiber. (A) Adsorption on cotton fiber. Purified cutinase or cutinase-CBM fusion protein was incubated in 20 mM Tris-HCl buffer (pH 8.0, 25°C). (B) Analysis of products released from cotton fiber. A 50 μM concentration of purified cutinase or cutinase-CBM fusion protein was incubated in 20 mM Tris-HCl buffer (pH 8.0, 50°C) with or without pectinase, and the amount of released fatty acids was measured by titration with 0.02 N NaOH.
In addition to binding, their hydrolytic efficiencies toward cotton fiber were also compared (Fig. 3B). In the absence of pectinase, the amount of released fatty acids was similar to that of the native cutinase for cutinase-CBMCel6A but was 1.8-fold higher for cutinase-CBMCenA. In the presence of pectinase, however, both fusion enzymes released almost the same amount of fatty acids and exhibited a catalytic efficiency 3-fold higher than that of the native cutinase. This result is consistent with that of the above binding experiment. The increased binding capability resulted in enhanced cutin hydrolytic efficiency.
DISCUSSION
Previously, site-directed mutagenesis of the amino acid residues surrounding the active site has been performed in order to obtain higher catalytic efficiency for cutinase (3). In the present study, a fusion protein approach in which cutinase was fused with CBM for improved affinity to cotton fiber was developed. By taking advantage of the CBM's specific binding to cellulose on the surface of cotton fiber where cutin also exists, the concentration of fusion cutinase around cutin would be increased, which may then result in enhanced enzyme catalytic efficiency due to the proximity effect. Considering the conditions of the textile scouring process, the cutinase-CBM fusion protein has to meet the following requirements: (i) no significantly decreased cutin hydrolyzing activity compared with the native cutinase, (ii) good thermal stability and alkali resistance, (iii) good binding affinity to cotton fiber, and (iv) improved scouring effect for cotton fiber.
In order to meet the above requirements, the choice of a suitable CBM is critical. To date, characteristics of CBMs have been explored extensively (22, 35). They are found mainly in carbohydrate degrading enzymes from fungi (e.g., Trichoderma reesei) and bacteria (e.g., C. fimi), including cellulase, xylanase, mannanase, and a number of nonhydrolytic proteins (24). Considering the source of the cutinase used in the present study, naturally a CBM was first selected from the genome of T. fusca.
As identified by the CAZy ModO database (http://www.afmb.cnrs-mrs.fr/CAZY/), the genome of T. fusca encodes a total of 17 hydrolytic enzymes that possess a CBM (see Table S3 in the supplemental material), with 6 of them experimentally characterized (underlined in Table S3 in the supplemental material) (26). In addition, CBMs are divided into 59 different families (http://www.afmb.cnrs-mrs.fr/CAZY/), and those from the second family were found to be able to loosen crystalline cellulose without significant fiber damage (9). Among the six characterized CBMs from T. fusca, except for the CBM3 of Cel9A and the CBM4 of Cel9B, all of the others were identified to belong to the second family and thus were candidates for fusing with cutinase.
As for the directionality of the fusion, the homology structural model of cutinase showed that both the N and the C termini of the enzyme were exposed to the solvent side (10). Considering the presence of the N-terminal PelB signal sequence for transmembrane localization, it appears that the CBM is better to be fused to the C terminus of cutinase. Another consideration is a possible linker sequence between the cutinase and the CBM. For example, it has been reported that the linker between the CBM and catalytic domain of a bacterial cellulase is composed entirely of the Pro-Thr repetitive sequence (18). Such a linker in cellulases would possess certain flexibility and avoid possible structural hindrance, which ensures the uniform movement of the two domains on the fiber surface (18, 28, 34). Therefore, an appropriate linker between the cutinase and the CBM is desired.
Putting together the above considerations, the CBM from T. fusca cellulase Cel6A, which belongs to the second family of CBMs and has a 28-residue linker region, was chosen to be fused to the C terminus of cutinase.
Cutinase-CBMCel6A was well expressed and purified and was able to hydrolyze not only cutin but also insoluble triglycerides (triolein) and soluble esters (pNPB). In addition, it shares similar pH stability as cutinase and displayed an optimal temperature at 50°C and half-life of 53 h at 50°C. When their binding and catalytic efficiency toward cotton fiber were compared, cutinase-CBMCel6A did not appear to perform significantly better than the native cutinase. However, in the presence of pectinase, cutinase-CBMCel6A exhibited significant improvement in binding and a dramatic 3-fold increase in catalytic efficiency. This sharp contrast is likely because most of the cellulose on the surface of cotton fiber is not well exposed to the solvent and is embedded in the epidermis full of pectins, proteins, and other components, thus limiting the binding of CBM to cotton fiber. When pectinase was added in the reaction mixture, removal of pectin by this enzyme may have led to the exposure of cellulose, resulting in increased adsorption of cutinase-CBMCel6A, which eventually led to higher scouring efficiency of cotton fiber.
In addition to CBMCel6A, we also examined the possibility of using other CBMs that have been experimentally characterized. The CBM of endoglucanase A (CenA) from C. fimi, which also belongs to the second family of CBMs, was shown to have high affinity to cellulose (12, 15, 39) and appears to be a suitable candidate. Subsequently, CBMCenA was fused to the C terminus of cutinase using the same linker from T. fusca Cel6A. As expected, cutinase-CBMCenA displayed a substrate specificity and catalytic properties similar to those of the native cutinase. Interestingly, although the CBMCenA is from a mesophilic bacterium, this fusion enzyme still retained decent thermostability, which may be due to the presence of a disulfide bond in CBMCenA (15). Notably, significant improvement in the binding and catalytic efficiency toward cotton fiber was observed for cutinase-CBMCenA compared to the native cutinase and cutinase-CBMCel6A. In addition, similar to cutinase-CBMCel6A, a strong synergistic effect with pectinase was also observed with cutinase-CBMCenA. Thus, it appears that the scouring effect of cutinase-CBMCenA is better than that of cutinase-CBMCel6A.
In conclusion, cutinase-CBM fusion proteins were successfully created by fusing a CBM to the C terminus of T. fusca cutinase. Compared to the native cutinase, both fusion proteins, cutinase-CBMCel6A and cutinase-CBMCenA, share similar stabilities and catalytic properties but showed greatly enhanced binding and hydrolytic activity toward cotton fiber. These improvements, as well as the synergistic effect between the fusion proteins and the pectinase, suggest that the cutinase-CBM fusion proteins may have better application potential in textile bioscouring.
Supplementary Material
Acknowledgments
This study was supported financially by the National High-Tech Research and Development Program of China (2009AA02Z204), the National Natural Science Foundation of China (grant 30970057), the National Outstanding Youth Foundation of China (grant 20625619), the Research Program of State Key Laboratory of Food Science and Technology (SKLF-MB-200802), the Key Program of National Natural Science Foundation of China (grant 20836003), the Program of Innovation Team of Jiangnan University (2008CXTD01), and the Self-determined Research Program of Jiangnan University (Yao Zhang).
Footnotes
Published ahead of print on 20 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agrawal, P. B., V. A. Nierstrasz, G. H. Bouwhuis, and M. M. C. G. Warmoeskerken. 2008. Cutinase and pectinase in cotton bioscouring: an innovative and fast bioscouring process. Biocatal. Biotransformation 26:412-421. [Google Scholar]
- 2.Araujo, R., M. Casal, and A. Cavaco-Paulo. 2008. Application of enzymes for textile fibres processing. Biocatal. Biotransformation 26:332-349. [Google Scholar]
- 3.Araujo, R., C. Silva, A. O'Neill, N. Micaelo, G. Guebitz, C. M. Soares, M. Casal, and A. Cavaco-Paulo. 2007. Tailoring cutinase activity toward polyethylene terephthalate and polyamide 6,6 fibers. J. Biotechnol. 128:849-857. [DOI] [PubMed] [Google Scholar]
- 4.Azevedo, H., D. Bishop, and A. Cavaco-Paulo. 2000. Effects of agitation level on the adsorption, desorption, and activities on cotton fabrics of full-length and core domains of EGV (Humicola insolens) and CenA (Cellulomonas fimi). Enzyme Microb. Technol. 27:325-329. [DOI] [PubMed] [Google Scholar]
- 5.Black, G. W., J. E. Rixon, J. H. Clarke, G. P. Hazlewood, L. M. Ferreira, D. N. Bolam, and H. J. Gilbert. 1997. Cellulose-binding domains and linker sequences potentiate the activity of hemicellulases against complex substrates. J. Biotechnol. 57:59-69. [DOI] [PubMed] [Google Scholar]
- 6.Bolam, D. N., A. Ciruela, S. McQueen-Mason, P. Simpson, M. P. Williamson, J. E. Rixon, A. Boraston, G. P. Hazlewood, and H. J. Gilbert. 1998. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 331(Pt. 3):775-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho, C. M., M. R. Aires-Barros, and J. M. Cabral. 1999. Cutinase: from molecular level to bioprocess development. Biotechnol. Bioeng. 66:17-34. [DOI] [PubMed] [Google Scholar]
- 8.Cavaco-Paulo, A. 1998. Processing textile fibers with enzymes: an overview. ACS Symp. Ser. 1998:180-189. [Google Scholar]
- 9.Cavaco-Paulo, A., J. Morgado, J. Andreaus, and D. Kilburn. 1999. Interactions of cotton with CBD peptides. Enzyme Microb. Technol. 25:639-643. [Google Scholar]
- 10.Chen, S., X. Tong, R. W. Woodard, G. Du, J. Wu, and J. Chen. 2008. Identification and characterization of bacterial cutinase. J. Biol. Chem. 283:25854-25862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciolacu, D., J. Kovac, and V. Kokol. 2010. The effect of the cellulose-binding domain from Clostridium cellulovorans on the supramolecular structure of cellulose fibers. Carbohydr. Res. 345:621-630. [DOI] [PubMed] [Google Scholar]
- 12.Damude, H. G., S. G. Withers, D. G. Kilburn, R. C. Miller, Jr., and R. A. Warren. 1995. Site-directed mutation of the putative catalytic residues of endoglucanase CenA from Cellulomonas fimi. Biochemistry 34:2220-2224. [DOI] [PubMed] [Google Scholar]
- 13.Degani, O., S. Gepstein, and C. G. Dosoretz. 2004. A new method for measuring scouring efficiency of natural fibers based on the cellulose-binding domain-β-glucuronidase fused protein. J. Biotechnol. 107:265-273. [DOI] [PubMed] [Google Scholar]
- 14.Degani, O., S. Gepstein, and C. G. Dosoretz. 2002. Potential use of cutinase in enzymatic scouring of cotton fiber cuticle. Appl. Biochem. Biotechnol. 102-103:277-289. [DOI] [PubMed] [Google Scholar]
- 15.Din, N., I. J. Forsythe, L. D. Burtnick, N. R. Gilkes, R. C. Miller, Jr., R. A. Warren, and D. G. Kilburn. 1994. The cellulose-binding domain of endoglucanase A (CenA) from Cellulomonas fimi: evidence for the involvement of tryptophan residues in binding. Mol. Microbiol. 11:747-755. [DOI] [PubMed] [Google Scholar]
- 16.Fett, W. F., H. C. Gerard, R. A. Moreau, S. F. Osman, and L. E. Jones. 1992. Screening of nonfilamentous bacteria for production of cutin-degrading enzymes. Appl. Environ. Microbiol. 58:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilkes, N. R., B. Henrissat, D. G. Kilburn, R. C. Miller, Jr., and R. A. Warren. 1991. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol. Rev. 55:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilkes, N. R., D. G. Kilburn, R. C. Miller, Jr., and R. A. Warren. 1989. Structural and functional analysis of a bacterial cellulase by proteolysis. J. Biol. Chem. 264:17802-17808. [PubMed] [Google Scholar]
- 19.Ha, J. S., Y. M. Lee, S. L. Choi, J. J. Song, C. S. Shin, J. H. Kim, and S. G. Lee. 2008. Thermostable β-glycosidase-CBD fusion protein for biochemical analysis of cotton scouring efficiency. J. Microbiol. Biotechnol. 18:443-448. [PubMed] [Google Scholar]
- 20.Hardin, I. R., Y. Li, and D. Akin. 1998. Cotton wall structure and enzymatic treatments. ACS Symp. Ser. 687:190-203. [Google Scholar]
- 21.Hartzell, M. M., and Y. L. Hsieh. 1998. Enzymatic scouring to improve cotton fabric wettability. J. Textile Res. 68:233-241. [Google Scholar]
- 22.Hashimoto, H. 2006. Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. 63:2954-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krakhmalev, V. A., and A. A. Paiziev. 2006. Spiral structures of cotton fiber. Cellulose 13:45-52. [Google Scholar]
- 24.Levy, I., and O. Shoseyov. 2002. Cellulose-binding domains: biotechnological applications. Biotechnol. Adv. 20:191-213. [DOI] [PubMed] [Google Scholar]
- 25.Linder, M., J. Winiecka-Krusnell, and E. Linder. 2002. Use of recombinant cellulose-binding domains of Trichoderma reesei cellulase as a selective immunocytochemical marker for cellulose in protozoa. Appl. Environ. Microbiol. 68:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lykidis, A., K. Mavromatis, N. Ivanova, I. Anderson, M. Land, G. DiBartolo, M. Martinez, A. Lapidus, S. Lucas, A. Copeland, P. Richardson, D. B. Wilson, and N. Kyrpides. 2007. Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol. 189:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nini, L., L. Sarda, L. C. Comeau, E. Boitard, J. P. Dubes, and H. Chahinian. 2001. Lipase-catalyzed hydrolysis of short-chain substrates in solution and in emulsion: a kinetic study. Biochim. Biophys. Acta 1534:34-44. [DOI] [PubMed] [Google Scholar]
- 28.Quentin, M., M. Ebbelaar, J. Derksen, C. Mariani, and H. van Der Valk. 2002. Description of a cellulose-binding domain and a linker sequence from Aspergillus fungi. Appl. Microbiol. Biotechnol. 58:658-662. [DOI] [PubMed] [Google Scholar]
- 29.Richins, R. D., A. Mulchandani, and W. Chen. 2000. Expression, immobilization, and enzymatic characterization of cellulose-binding domain-organophosphorus hydrolase fusion enzymes. Biotechnol. Bioeng. 69:591-596. [DOI] [PubMed] [Google Scholar]
- 30.Sae-be, P., U. Sangwatanaroj, and H. Punnapayak. 2007. Analysis of the products from enzymatic scouring of cotton. Biotechnol. J. 2:316-325. [DOI] [PubMed] [Google Scholar]
- 31.Sakka, K., G. Takada, S. Karita, and K. Ohmiya. 1996. Identification and characterization of cellulose-binding domains in xylanase A of Clostridium stercorarium. Ann. N. Y. Acad. Sci. 782:241-251. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Sawada, K., and M. Ueda. 2001. Enzyme processing of textiles in reverse micellar solution. J. Biotechnol. 89:263-269. [DOI] [PubMed] [Google Scholar]
- 34.Shen, H., M. Schmuck, I. Pilz, N. R. Gilkes, D. G. Kilburn, R. C. Miller, Jr., and R. A. Warren. 1991. Deletion of the linker connecting the catalytic and cellulose-binding domains of endoglucanase A (CenA) of Cellulomonas fimi alters its conformation and catalytic activity. J. Biol. Chem. 266:11335-11340. [PubMed] [Google Scholar]
- 35.Shoseyov, O., Z. Shani, and I. Levy. 2006. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 70:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomme, P., A. Boraston, B. McLean, J. Kormos, A. L. Creagh, K. Sturch, N. R. Gilkes, C. A. Haynes, R. A. Warren, and D. G. Kilburn. 1998. Characterization and affinity applications of cellulose-binding domains. J. Chromatogr. B Biomed. Sci. Appl. 715:283-296. [DOI] [PubMed] [Google Scholar]
- 37.Tomme, P., D. P. Driver, E. A. Amandoron, R. C. Miller, Jr., R. Antony, J. Warren, and D. G. Kilburn. 1995. Comparison of a fungal (family I) and bacterial (family II) cellulose-binding domain. J. Bacteriol. 177:4356-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walton, T. J., and P. E. Kolattukudy. 1972. Determination of the structures of cutin monomers by a novel depolymerization procedure and combined gas chromatography and mass spectrometry. Biochemistry 11:1885-1896. [DOI] [PubMed] [Google Scholar]
- 39.Wong, W. K., B. Gerhard, Z. M. Guo, D. G. Kilburn, A. J. Warren, and R. C. Miller, Jr. 1986. Characterization and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene 44:315-324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.