Abstract
Predator-prey relationships among prokaryotes have received little attention but are likely to be important determinants of the composition, structure, and dynamics of microbial communities. Many species of the soil-dwelling myxobacteria are predators of other microbes, but their predation range is poorly characterized. To better understand the predatory capabilities of myxobacteria in nature, we analyzed the predation performance of numerous Myxococcus isolates across 12 diverse species of bacteria. All predator isolates could utilize most potential prey species to effectively fuel colony expansion, although one species hindered predator swarming relative to a control treatment with no growth substrate. Predator strains varied significantly in their relative performance across prey types, but most variation in predatory performance was determined by prey type, with Gram-negative prey species supporting more Myxococcus growth than Gram-positive species. There was evidence for specialized predator performance in some predator-prey combinations. Such specialization may reduce resource competition among sympatric strains in natural habitats. The broad prey range of the Myxococcus genus coupled with its ubiquity in the soil suggests that myxobacteria are likely to have very important ecological and evolutionary effects on many species of soil prokaryotes.
Predation plays a major role in shaping both the ecology and evolution of biological communities. The population and evolutionary dynamics of predators and their prey are often tightly coupled and can greatly influence the dynamics of other organisms as well (1). Predation has been invoked as a major cause of diversity in ecosystems (11, 12). For example, predators may mediate coexistence between superior and inferior competitors (2, 13), and differential trajectories of predator-prey coevolution can lead to divergence between separate populations (70).
Predation has been investigated extensively in higher organisms but relatively little among prokaryotes. Predation between prokaryotes is one of the most ancient forms of predation (27), and it has been proposed that this process may have been the origin of eukaryotic cells (16). Prokaryotes are key players in primary biomass production (44) and global nutrient cycling (22), and predation of some prokaryotes by others is likely to significantly affect these processes. Most studies of predatory prokaryotes have focused on Bdellovibrionaceae species (e.g., see references 51, 55, and 67). These small deltaproteobacteria prey on other Gram-negative cells, using flagella to swim rapidly until they collide with a prey cell. After collision, the predator cells then enter the periplasmic space of the prey cell, consume the host cell from within, elongate, and divide into new cells that are released upon host cell lysis (41). Although often described as predatory, the Bdellovibrionaceae may also be considered to be parasitic, as they typically depend (apart from host-independent strains that have been observed [60]) on the infection and death of their host for their reproduction (47).
In this study, we examined predation among the myxobacteria, which are also deltaproteobacteria but constitute a monophyletic clade divergent from the Bdellovibrionaceae (17). Myxobacteria are found in most terrestrial soils and in many aquatic environments as well (17, 53, 74). Many myxobacteria, including the model species Myxococcus xanthus, exhibit several complex social traits, including fruiting body formation and spore formation (14, 18, 34, 62, 71), cooperative swarming with two motility systems (64, 87), and group (or “wolf pack”) predation on both bacteria and fungi (4, 5, 8, 9, 15, 50). Using representatives of the genus Myxococcus, we tested for both intra- and interspecific variation in myxobacterial predatory performance across a broad range of prey types. Moreover, we examined whether prey vary substantially in the degree to which they support predatory growth by the myxobacteria and whether patterns of variation in predator performance are constant or variable across prey environments. The latter outcome may reflect adaptive specialization and help to maintain diversity in natural populations (57, 59).
Although closely related to the Bdellovibrionaceae (both are deltaproteobacteria), myxobacteria employ a highly divergent mode of predation. Myxobacteria use gliding motility (64) to search the soil matrix for prey and produce a wide range of antibiotics and lytic compounds that kill and decompose prey cells and break down complex polymers, thereby releasing substrates for growth (66). Myxobacterial predation is cooperative both in its “searching” component (6, 31, 82; for details on cooperative swarming, see reference 64) and in its “handling” component (10, 29, 31, 32), in which secreted enzymes turn prey cells into consumable growth substrates (56, 83). There is evidence that M. xanthus employs chemotaxis-like genes in its attack on prey cells (5) and that predation is stimulated by close contact with prey cells (48).
Recent studies have revealed great genetic and phenotypic diversity within natural populations of M. xanthus, on both global (79) and local (down to centimeter) scales (78). Phenotypic diversity includes variation in social compatibility (24, 81), the density and nutrient thresholds triggering development (33, 38), developmental timing (38), motility rates and patterns (80), and secondary metabolite production (40). Although natural populations are spatially structured and both genetic diversity and population differentiation decrease with spatial scale (79), substantial genetic diversity is present even among centimeter-scale isolates (78). No study has yet systematically investigated quantitative natural variation in myxobacterial predation phenotypes across a large number of predator genotypes.
Given the previous discovery of large variation in all examined phenotypes, even among genetically extremely similar strains, we anticipated extensive predatory variation as well. Using a phylogenetically broad range of prey, we compared and contrasted the predatory performance of 16 natural M. xanthus isolates, sampled from global to local scales, as well as the commonly studied laboratory reference strain DK1622 and representatives of three additional Myxococcus species: M. flavescens (86), M. macrosporus (42), and M. virescens (63) (Table 1). In particular, we measured myxobacterial swarm expansion rates on prey lawns spread on buffered agar (31, 50) and on control plates with no nutrients or with prehydrolyzed growth substrate.
TABLE 1.
List of myxobacteria used, with geographical origin
| Organism abbreviation used in text | Species | Strain | Geographic origin | Reference(s) |
|---|---|---|---|---|
| A9 | Myxococcus xanthus | A9 | Tübingen, Germany | 78 |
| A23 | Myxococcus xanthus | A23 | Tübingen, Germany | 78 |
| A30 | Myxococcus xanthus | A30 | Tübingen, Germany | 78 |
| A41 | Myxococcus xanthus | A41 | Tübingen, Germany | 78 |
| A46 | Myxococcus xanthus | A46 | Tübingen, Germany | 78 |
| A47 | Myxococcus xanthus | A47 | Tübingen, Germany | 78 |
| A75 | Myxococcus xanthus | A75 | Tübingen, Germany | 78 |
| A85 | Myxococcus xanthus | A85 | Tübingen, Germany | 78 |
| TV | Myxococcus xanthus | Tvärminne | Tvärminne, Finland | 79 |
| PAK | Myxococcus xanthus | Paklenica | Paklenica, Croatia | 79 |
| MAD | Myxococcus xanthus | Madeira 1 | Madeira, Portugal | 79 |
| WAR | Myxococcus xanthus | Warwick 1 | Warwick, UK | 79 |
| TOR | Myxococcus xanthus | Toronto 1 | Toronto, Ontario, Canada | 79 |
| SUL2 | Myxococcus xanthus | Sulawesi 2 | Sulawesi, Indonesia | 79 |
| KAL | Myxococcus xanthus | Kalalau | Kalalau, HI | 79 |
| DAV | Myxococcus xanthus | Davis 1A | Davis, CA | 79 |
| GJV1 | Myxococcus xanthus | GJV 1 | Unknown | 35, 72 |
| MXFL1 | Myxococcus flavescens | Mx fl1 | Unknown | 65 |
| MXV2 | Myxococcus virescens | Mx v2 | Unknown | 65 |
| CCM8 | Myxococcus macrosporus | Cc m8 | Unknown | 65 |
MATERIALS AND METHODS
Strains.
Myxobacterial strain information is provided in Table 1. The M. xanthus clones A9, A23, A30, A41, A46, A47, A75, and A85 examined here were among 78 isolates previously sampled from a 16- by 16-cm patch of soil in Tübingen, Germany (78). These particular isolates were selected because they had been previously screened for phenotypic variation in multiple traits (80, 81) and because they represented a wide range of the genotypic variation present among the original 78 isolates. A23, A46, and A47 were found to be genetically identical at nine multilocus sequencing typing (MLST) loci, including the highly variable pilA locus (78), yet A23 and A47 were found to be socially incompatible during group swarming, and A47 strongly antagonizes A23 in chimeric developmental cultures (81). M. xanthus isolates from greater spatial scales (four from Europe and four from widespread global locales) were isolated by the same method as the centimeter-scale isolates (79). The well-characterized M. xanthus laboratory strain DK1622 was included for comparison with more recently isolated strains. More specifically, we used a descendant of the original DK1622 strain, GJV1, which differs from DK1622 by five mutations of unknown effect that arose during laboratory cultivation (72). Myxococcus flavescens strain Mx fl1, Myxococcus virescens strain Mx v2, and Myxococcus macrosporus strain Cc m8 (65) were kindly provided by Hans Reichenbach.
Genotyping of myxobacteria.
Fragments of three highly conserved housekeeping genes commonly used in MLST studies (20, 23, 45) were sequenced for all myxobacterial strains. The genes examined were clpX (which encodes an ATP-dependent protease), icd (isocitrate dehydrogenase), and the nonessential dnaK homologue (heat shock protein 70 [HSP70] chaperone) sglK. Strands were sequenced in both directions, and sequences were aligned using the ClustalW algorithm in MEGA 4.0.2 (68). After alignment, the sequences were joined to make a concatemer of the three gene fragments and a neighbor-joining tree was constructed using MEGA 4.0.2 (1,000 bootstrap replicates).
Prey bacteria.
Ten species of soil bacteria representing a broad phylogenetic spectrum (84) were chosen as prey, including four Gram-positive species (two with high GC-content genomes and two with low GC content) and six Gram-negative species (two alpha-, two beta- and two gammaproteobacteria) (Table 2). These strains were from the strain collections of the Long-Term Ecological Research (LTER) soil plots at Kellogg Biological Station, Hickory Corners, MI, (37) and DSMZ. We also included the Gram-positive (low-GC) bacterium Micrococcus luteus and the Gram-negative gammaproteobacterium Escherichia coli (REL606) (43). The soil is not considered to be the usual niche of these latter two species, but myxobacteria frequently grow on animal dung, where they may feed on E. coli, and both species have been used in previous studies of myxobacterial predation (29, 30, 48, 73).
TABLE 2.
List of prey species used, including their phylogenetic classification and strain reference number
| Prey species | Phylum and subdivision | Source (reference)a | Strain reference no. |
|---|---|---|---|
| Arthrobacter globiformus | Gram positive, high-G+C subdivision | LTER (37) | LTER 27 |
| Bacillus bataviensis | Gram positive, low-G+C subdivision | DSMZ | 15601 |
| Curtobacterium citreum | Gram positive, high-G+C subdivision | LTER (37) | LTER 17 |
| Cytophaga johnsonae | Gram positive, low-G+C subdivision | ATCC | 17061 |
| Comamonas testosteroni | Gram negative, beta subdivision | DSMZ | 50244 |
| Escherichia coli REL606 | Gram negative, gamma subdivision | Richard Lenski (43) | N/A |
| Micrococcus luteus | Gram positive, low-G+C subdivision | ATCC | 4698 |
| Pseudomonas fluorescens | Gram negative, gamma subdivision | LTER (37) | LTER 56 |
| Rhizobium vitis | Gram negative, alpha subdivision | DSMZ | 6583 |
| Sinorhizobium fredii | Gram negative, alpha subdivision | LTER (37) | LTER 38 |
| Sphingobium yanoikuyae | Gram negative, beta subdivision | LTER (37) | LTER 19 |
| Xanthomonas fragariae | Gram negative, gamma subdivision | DSMZ | 3587 |
LTER, Kellogg LTER strain collection; ATCC, American Type Culture Collection; DSMZ, German Collection of Microorganisms and Cell Cultures.
Predation assays.
The predation assays were based on the lawn predation assay used by Pham et al. (50). The myxobacteria were spotted onto the center of a nutrient-free agar plate that had been inoculated with a lawn of prey, and the expansion of the myxobacteria swarm was measured after 5 days. We explain this methodology in further detail below.
(i) Preparation of prey lawns.
Prey species were inoculated into R2 broth (52) and grown for approximately 24 h at 32°C at 300 rpm. A 1-ml sample of each culture was removed and centrifuged at 5,000 rpm for 15 min. The supernatant was discarded, the cells were resuspended in TPM buffer (10 ml of 0.8 M MgSO4, 10 ml of 1 M Tris-HCl [pH 7.6], and distilled H2O to 1 liter) (7, 39), and optical densities were measured to provide biovolume estimates. Optical density had previously been calibrated to cell biovolume with a Beckman-Coulter particle counter. The remaining portion of each culture was centrifuged at 5,000 rpm and resuspended in TPM buffer to a biovolume density of 109 μm3/ml. Five hundred microliters of the cell suspension was spread onto a 10-cm-diameter TPM hard agar plate (TPM buffer with 1.5% agar) (7) and allowed to dry. Because TPM contains no carbon source, prey cells were the only substrate available for Myxococcus growth.
(ii) Control plates.
Three types of control plates were used in this experiment: one negative and two positive. The negative control plates did not have prey added, but were otherwise identical to the prey plates. For these plates, 500 μl of TPM buffer was spread across the surface of TPM hard agar and allowed to dry. Both types of positive control plates provided Casitone as a myxobacterial growth substrate. Casitone is composed of prehydrolyzed peptides (56), and myxobacteria feeding on this substrate are not required to kill prey cells or degrade prey cells and complex polymers to grow. The first type of positive control was a TPM hard agar plate on which 500 μl of Casitone-Tris (CTT) broth (10 g Casitone, 10 ml of 0.8 M MgSO4, 10 ml of 1 M Tris-HCl [pH 7.6], distilled H2O to 1 liter) (35) was spread and allowed to dry. The second type of positive control was composed of CTT hard agar plates (CTT broth with 1.5% agar). CTT agar plates are commonly used for M. xanthus swarming assays (61, 75). The degree of predator swarm expansion on plates with prey or Casitone that exceeded basal swarm expansion on the TPM plates containing no growth substrate was assumed to primarily reflect the degree of population growth on the relevant substrate.
(iii) Inoculation of prey with myxobacteria.
Myxobacteria were inoculated by placing a small sample of frozen culture on CTT agar plates, which were incubated at 32°C at 90% rH. After 2 days of growth, a small sample from the colony edge was transferred to 8 ml of CTT liquid broth, which was incubated at 32°C at 300 rpm for 2 to 3 days, depending on the growth rate of each strain. Exponentially growing cells were centrifuged at 5,000 rpm for 15 min and resuspended in TPM liquid to a density of 5 × 109 cells/ml after the supernatant had been discarded. A 10-μl spot of resuspended culture was placed on the center of each type of prey and control plates that had been prepared on the same day. After the culture aliquots had dried, the perimeter of the culture spot was marked on the plate bottom to indicate the starting point of swarming. Plates were incubated at 32°C with 90% rH for 5 days. After incubation, the distance swarmed from the marked starting point was measured in four equally spaced transects that passed through the center of the original spot. Measurements of the eight resulting radial “spokes” were then averaged to give a single “distance swarmed” value in millimeters. The experiment was replicated in five separate temporal blocks.
Statistical analyses. (i) General patterns.
To test for general effects, prey species, myxobacteria, and temporal block were fitted as factors in a general linear model (GLM) (26), using MINITAB software. An interaction term between prey species and myxobacterium was also included. This enabled us to decompose the phenotypic variance as follows: V(p) = V(predator) + V(prey) + V(predator·prey).
(ii) Responsiveness and inconsistency.
The interaction term in the GLM could be further decomposed into responsiveness and inconsistency, such that V(predator·prey) = R + I
The responsiveness component, R, is defined as
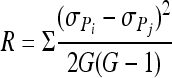 |
(1) |
The inconsistency component, I, is defined as
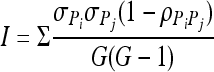 |
(2) |
G is the number of myxobacterial genotypes tested, P is the prey, i and j are two different myxobacterial genotypes, and σPi and σPj are the standard deviations of the responses to the prey types. ρPiPj is the correlation between performance on prey between two myxobacteria strains tested (76). The responsiveness component of the interaction term measures differences in the variances among prey species. The most straightforward biological interpretation of responsiveness is that it measures variability in prey utilization breadth among the myxobacteria. A low responsiveness value would indicate that the myxobacteria examined here tend to have very similar shapes of reaction norms across prey (i.e., most are specialists or most are generalists), while a high responsiveness value would indicate similar numbers of specialists and generalists. Inconsistency is a measure of noncorrelations (crossing-over of reaction norms or performance rank reversals) between different myxobacteria on different prey. For example, if predator A performed better on one prey type X than another predator B, but the predator B performed better than A on a different prey type Y, then there would be crossing reaction norms (a noncorrelation, or reversal of performance ranks). The prevalence of such performance rank reversals in the data set is captured by the inconsistency term. For a graphical representation of responsiveness and inconsistency, see Fig. S1 in the supplemental material.
Nucleotide sequence accession numbers.
All of the nucleotide sequences analyzed in this study have been deposited in GenBank. The accession numbers are as follows: clpX, DQ401890 to DQ401909 and GU733429 to GU733431; icd, DQ401910 to DQ401929 and GU733432 to GU733434; and sglK, DQ401990 to DQ402009 and GU733435 to GU733437.
RESULTS
General patterns.
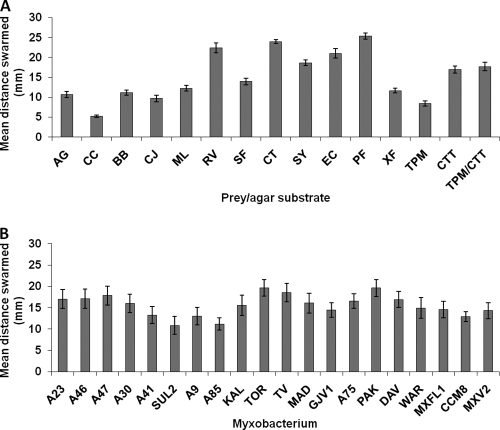
Myxococcus predatory performance, as reflected by swarming on prey-covered plates, was significantly affected by prey species type (Fig. 1A) (GLM, F11, 956 = 862.14, P < 0.001), and the main effect of prey type accounts for 71.6% of the total variance in swarming ability. Most prey species clearly fueled Myxococcus swarm expansion, as the average of all predator strain swarming rates on each prey type was significantly greater than on prey-free buffered TPM agar (paired t tests; see Table S3 in the supplemental material), except in two cases. Average predatory swarming on Cytophaga johnsonae was not significantly different than swarming on the prey-free TPM plates, and swarming on Curtobacterium citreum was actually significantly lower than on TPM plates, suggesting that C. citreum inhibits Myxococcus growth.
FIG. 1.
(A) Mean predator swarm expansion on each prey type and on TPM, CTT, and TPM/CTT controls. Prey abbreviations: AG, Arthrobacter globiformis; CC, Curtobacterium citreum; BB, Bacillus bataviensis; CJ, Cytophaga johnsonae; ML, Micrococcus luteus; RV, Rhizobium vitis; SF, Sinorhizobium fredii; CT, Comamonas testosteroni; SY, Sphingobium yanoikuyae; EC, Escherichia coli; PF, Pseudomonas fluorescens; XF, Xanthomonas fragariae. Error bars represent ±1 standard error (SE) of the mean (n = 20 Myxococcus strains). (B) Mean swarm expansion of each Myxococcus strain across all prey species. Error bars represent ±1 SE of the mean (n = 12 prey species).
There were significant differences in predatory swarming rates among myxobacteria when the performance of each was averaged across all prey (Fig. 1B) (GLM, F19, 956 = 76.49, P < 0.001), and the main effect of myxobacterium type accounts for 9.7% of the total variance. Average swarming rate across all prey was found to correlate significantly with swarming rate on both prey-free controls containing growth substrates (CTT spread onto TPM plate, r = 0.78, P < 0.001; CTT plates, r = 0.81, P < 0.001). Thus, most of this variance in average performance among myxobacteria can be attributed to intrinsic differences in population expansion rates manifest on any solid surface with nutrients rather than to variation in average performance of the “handling”-specific component of predation (i.e., the extraction and utilization of nutrients from prey cells). No measure of swarming rate in any environment with added growth substrate (prey or Casitone) was found to significantly correlate with swarming rate in the absence of nutrients on TPM agar (TPM versus average swarming on prey, r = 0.398, P = 0.082; TPM versus CTT, r = 0.294, P = 0.209; TPM versus CTT spread on TPM, r = 0.295, P = 0.207).
Despite the inference that much of the variation in average predator swarm expansion rates across all prey is not specific to the presence of prey cells, an important result is that there was a significant interaction between myxobacterium type and prey species (GLM, F209, 956 = 6.93, P < 0.001), accounting for 9.8% of total variance. This significant interaction is indicative of some degree of predatory specialization, which we analyze further below in the section “Responsiveness and inconsistency.” There was also a significant effect of temporal block (GLM, F4, 956 = 14.57, P < 0.001).
To determine whether there was a difference between predation on Gram-positive versus Gram-negative prey, we replaced the prey-species term in the GLM with a Gram-positive or Gram-negative term and found that Gram-negative prey support significantly more predatory growth by Myxococcus (average swarming distance, 19.6 mm) than do Gram-positives (average swarming distance 9.8 mm) (GLM, F1, 1,156 = 1,120.82, P < 0.001). There was a significant interaction between Gram-positive/negative and myxobacterium type (GLM, F19, 1,175 = 4.05, P < 0.001), indicating that there is some variation among predator strains in their relative performance on Gram positives versus Gram negatives. The effects of myxobacterium-type (GLM, F19, 1,156 = 13.92, P < 0.001) and temporal block (GLM, F4, 1,175 = 2.75, P < 0.022) were also significant in this model.
Specialization. (i) Variances.
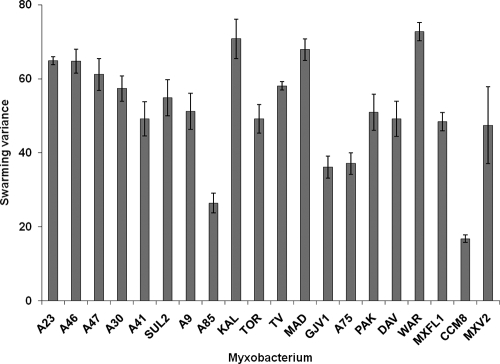
For a preliminary examination of possible specialization occurring among Myxococcus strains, we compared the variances in predation rates across all prey types for each predator strain. A low variance would indicate that the Myxoccocus genotype shows similar predatory performance across all prey types. High variance indicates substantial differences in predation rates of a Myxococcus genotype on different prey, which may be indicative of specialization. There were significant differences among the variances of predator performance across prey types (Kruskall-Wallace test [19], H19 = 69.85 P < 0.001) (Fig. 2), but variance did not correlate significantly with swarm expansion rate averaged across all prey (r = 0.39, P = 0.089). Thus, the differential variance in predatory growth across Myxococcus strains may be indicative of specialization and does not appear to be merely an artifact of a positive relationship between variance and swarm expansion rate.
FIG. 2.
Variance in predatory swarm expansion of each Myxococcus strain across all prey species. Error bars represent ±1 SE of the mean (n = 5 replicates).
(ii) Predator and prey ranks.
We ranked each prey species by the relative degree to which it supported predation for each Myxococcus strain (see Table S4 in the supplemental material). For example, out of all prey types M. xanthus strain A9 swarmed furthest on Comamonas testosteroni, giving C. testoteroni a rank of 1 for that predator strain. A9 swarmed the least on Curtobacterium citreum, giving C. citreum a rank of 12. A prey rank for a particular prey-predator combination that is much higher than the average for the respective prey type across other predators is suggestive of predator specialization on that prey. We also ranked each Myxococcus strain for its swarming performance on each prey type relative to the other predator strains (see Fig. S5 in the supplemental material). For example, the Toronto1 (TOR) M. xanthus strain swarmed the farthest (rank 1) on Arthrobactor globiformus, whereas the Sulawesi (SUL2) M. xanthus strain performed the worst on this prey (rank 12).
(iii) Responsiveness and inconsistency.
The significant interaction between prey type and predator type in the GLM analysis (∼10% of total phenotypic variance) means that distinct Myxococcus strains show different patterns of performance variation across prey species. The GLM interaction term can be further broken down into effects of “responsiveness,” which measures variation in prey utilization breadths among predator strains, and “inconsistency,” which measures the extent to which individual predator strains show specialized performance on particular prey species (see Fig. S1 in the supplemental material) (3, 76). Inconsistency accounts for the vast majority (82%) of the interaction term (and thus 8% of the total phenotypic variance), whereas the responsiveness component of the interaction is very small (18%, or 1.8% of the total variance).
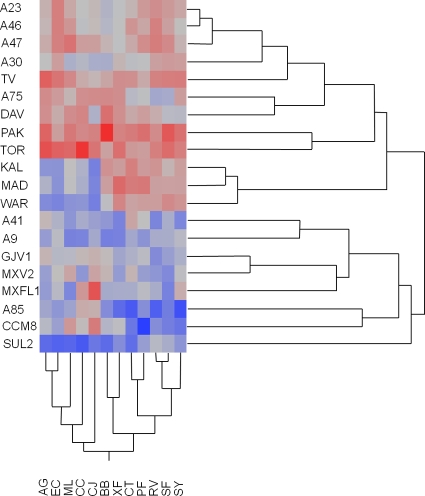
The interpretation of these results is straightforward. The low degree of responsiveness indicates that Myxococcus strains exhibit very little variation in prey utilization breadth (the number of prey types they can grow on), as most can predate each prey type to some degree. The relatively large effect of inconsistency shows that at least some Myxococcus strains exhibit a degree of specialization on some prey types, when specialization is defined as the crossing of reaction norms. Such specialization is evident in Fig. 3, which uses a color intensity scale to show the relative performance of all predators on each prey type, standardized to the average performance on each respective prey. Red indicates faster than average swarming for that prey type, whereas blue indicates slower than average swarming, with color intensity reflecting the degree of difference from the mean. Thus, the darkest red and blue squares within each column indicate maximum and minimum predatory performance on that prey type, respectively. Specialized performance by a given Myxococcus strain is reflected by large differences in color intensity within a row, which indicate the degree to which predators show different patterns of relative performance on different prey types. Differences in colors across rows (prey) indicate specialized performance. For example, strain TOR shows unusually high performance on C. citreum, PAK does so on Bacillus bataviensis, and KAL, MAD, and WAR all show relatively enhanced performance on the seven prey strains composing the right basal prey clade (Fig. 3) and relatively poor performance on the five prey strains composing the left clade.
FIG. 3.
Grid of standardized predatory performance and neighbor-joining trees of predators and prey based on predation phenotypes. Red indicates faster swarming than average by a Myxococcus strain on the respective prey type, whereas blue indicates slower swarming than average, with color intensity reflecting the degree of difference from the mean. See the main text for further explanation of the figure.
Genetic and phenotypic comparisons.
We examined whether patterns of predatory phenotype similarities and differences can be predicted by patterns of sequence similarity at MLST loci. To measure the phenotypic similarity between myxobacteria, we calculated a Euclidean distance for each possible pair of Myxococcus strains as Dx,y = [Σi(xi − yi)2]1/2, where xi and yi are the swarming distances of myxobacteria x and y, respectively, on prey species i and the summation is over all prey species. Using this distance matrix, we constructed a UPGMA (unweighted-pair group method with arithmetic mean) tree based on patterns of predatory phenotypes (Fig. 3).
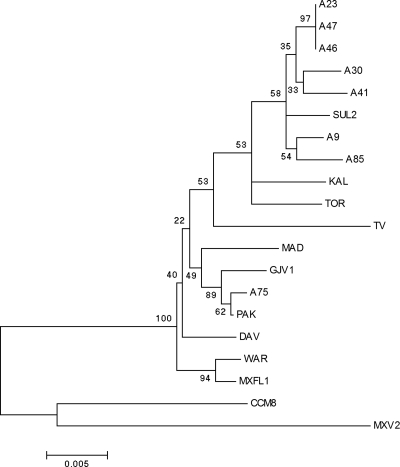
A phylogenetic tree based on the MLST loci clpX, icd, and sglK (Fig. 4) was found to be highly incongruent with the phenotype-based tree (Fig. 3). One exception to this incongruity was that the three most genetically similar strains (A23, A47, and A96), which come from the same centimeter-scale Tübingen population and are genetically identical at several MLST loci (78), clustered together in the phenotypic tree along with strain A30. However, other genetically related centimeter-scale isolates were placed within divergent clades in the predation phenotype tree (A9, A41, A75, and A85) (Fig. 3 and 4). We found no overall correlation between phenotypic distance and genetic distance based on the conserved MLST concatemer loci (Mantel test, r = 0.01, P = 0.24), which are expected to evolve neutrally and give a deeply rooted phylogenetic signal (20).
FIG. 4.
Neighbor-joining tree of relationships among Myxococcus strains based on the clpX-icd-sglK MLST concatemer. Bootstrap support (1,000 replicates) is shown for each node. The scale bar reflects genetic distance as amino acid substitutions per site.
DISCUSSION
We have found that both prey type and predator strain contribute significantly to variation in predatory performance across many Myxococcus isolates and prey species. Prey varied greatly in the degree to which they supported predatory population growth, with the Gram-positive species examined here being poorer prey for Myxococcus predators than the Gram-negative species (Fig. 1A). This result suggests a general difference in the susceptibility of Gram-positive and Gram-negative species to predation by Myxococcus, perhaps due to differences in cell wall degradation by predatory enzymes. The parasitic Bdellovibrionaceae are closely related to the myxobacteria and can only prey upon Gram-negative cells, as none have yet been found to consume a Gram-positive species (41, 46, 69). Both Bdellovibrionaceae (54) and Myxococcus species (85) carry genes for synthesis of type IV pili (TFP). In Myxococcus, TFP mediate social motility (S-motility) (35, 85), which, along with “adventurous motility” (A-motility) (49), allows predator cells to actively migrate in search of prey. Elimination of TFP production in M. xanthus reduces predation efficiency (4). It has been proposed that TFP play an important role in the entry of Bdellovibrio cells into the periplasm of their prey (21, 54). It is possible that TFP might play a similar role in the myxobacteria by adhering to prey cells and thereby facilitating predatory lysis. Thus, TFP may be involved in the apparent specialization of both Bdellovibrionaceae and myxobacterial predators on Gram-negative prey.
When the swarming rates of each Myxococcus strain on all 12 prey species were averaged, these mean swarming rates were found to vary significantly among the different Myxococcus strains examined (Fig. 1B). However, this meta-parameter of overall swarming rate on prey was found to correlate with swarming rate on the positive control plates containing no prey but rather prehydrolyzed nutrients (Casitone). Thus, a large proportion of the variation in overall growth ability in prey environments is not specific to consumption of bacterial prey cells, but rather reflects variation in the intrinsic population growth rates of strains on a solid surface irrespective of food source (prey cells versus prehydrolyzed amino acids). Nonetheless, some Myxococcus strains do vary in their prey-specific performance on individual prey species (Fig. 3), thus reflecting some degree of specialization. This specialization is not binary (ability versus complete inability to eat prey), and all of the Myxoccocus strains could grow on most of the prey bacteria in this study. Thus, specialization can be defined in terms of interactions between genotype and environment in which performance ranks among predator strains reverse across prey environments (28). The inconsistency term quantified here is a measure of such noncorrelations and reflects the degree of specialization occurring among the predator strains.
We note that some patterns of variation and specialization in predatory performance by Myxococcus isolates or variation in susceptibility to predation by prey may be specific to the experimental conditions of this study. For example, prey that are actively growing on a solid surface may differ in their susceptibility to predators compared to the nongrowing prey cultures examined here. Thus, we do not infer that patterns observed here are general across all experimental and soil environments in which predator-prey interactions might occur. Rather, our results suggest that specialized predatory performance (i.e., crossing reaction norms) might occur across a range of ecological environments. Further research may reveal some patterns observed here (e.g., the greater susceptibility of Gram-negative prey to predation) to hold generally across a variety of background environments.
Understanding the evolutionary mechanisms resulting in various forms of specialization is a key problem in evolutionary ecology, and several explanations might apply to Myxococcus (25). First, we have used the term “specialization” to refer to particular patterns of predatory performance observed under our experimental conditions. We note that particular instances of specialization in our assay do not necessarily reflect prior evolutionary adaptation by the relevant predator strain to the particular prey used here or closely related prey. First, we have no data regarding predator-prey interaction histories prior to isolation of the predator strains, but rather only the knowledge that most of the prey used here can be isolated from soil environments in which Myxococcus spp. also live. Second, the patterns observed here might reflect pleiotropic effects of selection on traits other than predation per se, such as production of anticompetitor toxins, that may indirectly affect the predation ability of a Myxococcus strain. Alternatively, instances of specialized performance on one particular prey might be due to relaxed selection for performance on other prey that are rarely encountered (36) or trade-offs on performance across prey (77).
To the degree that the specialization patterns found here do in fact represent prior specific adaptation to the relevant prey type(s) (or close relatives), such adaptation may have been facilitated by a higher encounter rate between the predator and prey involved in the specialized interaction or may reflect differences in the mutational accessibilities of adaptive peaks on different prey. Specialization may also reflect divergent coevolutionary histories between different predator-prey combinations (70). As for several other traits (33, 78, 79, 81), divergence in predatory phenotypes—including some instances of specialization—appears to occur within local populations as well as across isolated populations (Fig. 3 and 4). Given the spatial proximity of sympatric genotypes, they are likely to frequently compete for the same resources. Such competition may be reflected by antagonistic behaviors that have been documented between the Tübingen isolates (81). Strong resource competition might be reduced by ecological character displacement, the process whereby closely related organisms with similar phenotypes in a habitat diversify to exploit new resources (58). In the myxobacteria, character displacement might be brought about by the evolution of specialized predation abilities, such as we have observed in this study. Such predatory specialization may ultimately contribute to myxobacterial speciation.
The molecular basis of the predatory variation examined here remains to be investigated. However, M. xanthus produces a large number of secondary metabolites that may serve predatory functions (40). M. xanthus isolates are known to vary substantially in the range of secondary metabolites they produce (40), and such variation may contribute to variation in predatory phenotypes.
Myxobacteria are widespread in soils around the globe (53). This ubiquity, coupled with the ability to use a broad range of other microbial species (including fungi as well as bacteria) (8) as prey, suggests that myxobacteria are likely to strongly affect the population and evolutionary dynamics of many microbes that play important roles in both natural and agricultural processes. For example, myxobacteria may control populations involved in the carbon and nitrogen cycles (22), primary production by cyanobacteria (9, 44), plant host-parasite interactions (8), and nitrogen fixation in Rhizobium-legume mutualism (e.g., the predation of Myxococcus on Rhizobium vitis documented here).
Our understanding of myxobacterial predation, its ecological significance, its evolutionary origins and dynamics, and its potential applications remains very limited. This comparative study provides new insights into the range of variation in predatory ability that has evolved in the wild at the genus and species level within the myxobacteria. Due to their short generation times and large population sizes, evolution experiments with myxobacteria can also be conducted to address numerous questions regarding the evolution of predation, including the relative adaptation of searching versus handling components of predation in environments that differ in prey density (31) and patterns of specialization when predators evolve in environments with diverse prey that differ in quality.
Supplementary Material
Acknowledgments
We thank Susanne Kramer for advice on sequencing and Michiel Vos and Vicki Fleming for advice on sequence analysis.
This work was funded by NIH grant R01 GM079690.
Footnotes
Published ahead of print on 27 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abrams, P. A. 2000. The evolution of predator-prey interactions: theory and evidence. Annu. Rev. Ecol. Syst. 31:79-105. [Google Scholar]
- 2.Abrams, P. A. 1999. Is predator-mediated coexistence possible in unstable systems? Ecology 80:608-621. [Google Scholar]
- 3.Bell, G. 1990. The ecology and genetics of fitness in Chlamydomonas. 1. Genotype-by-environment interaction among pure strains. Proc. Biol. Sci. 240:295-321. [Google Scholar]
- 4.Berleman, J. E., T. Chumley, P. Cheung, and J. R. Kirby. 2006. Rippling is a predatory behavior in Myxococcus xanthus. J. Bacteriol. 188:5888-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berleman, J. E., J. Scott, T. Chumley, and J. R. Kirby. 2008. Predataxis behavior in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 105:17127-17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biesinger, Z., and J. W. Haefner. 2005. Proximate cues for predator searching: a quantitative analysis of hunger and encounter rate in the ladybird beetle, Coccinella septempunctata. Anim. Behav. 69:235-244. [Google Scholar]
- 7.Bretscher, A. P., and D. Kaiser. 1978. Nutrition of Myxococcus xanthus, a fruting myxobacterium J. Bacteriol. 133:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull, C. T., K. G. Shetty, and K. V. Subbarao. 2002. Interactions between myxobacteria, plant pathogenic fungi, and biocontrol agents. Plant Dis. 86:889-896. [DOI] [PubMed] [Google Scholar]
- 9.Burnham, J. C., S. A. Collart, and M. J. Daft. 1984. Myxococcal predation of the cyanobacterium Phormidium luridum in aqueous environments. Arch. Microbiol. 137:220-225. [Google Scholar]
- 10.Catania, K. C., and F. E. Remple. 2005. Asymptotic prey profitability drives star-nosed moles to the foraging speed limit. Nature 433:519-522. [DOI] [PubMed] [Google Scholar]
- 11.Chase, J. M., P. A. Abrams, J. P. Grover, S. Diehl, P. Chesson, R. D. Holt, S. A. Richards, R. M. Nisbet, and T. J. Case. 2002. The interaction between predation and competition: a review and synthesis. Ecol. Lett. 5:302-315. [Google Scholar]
- 12.Chesson, P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31:343-366. [Google Scholar]
- 13.Chesson, P., and J. J. Kuang. 2008. The interaction between predation and competition. Nature 456:235-238. [DOI] [PubMed] [Google Scholar]
- 14.Curtis, P. D., R. G. Taylor, R. D. Welch, and L. J. Shimkets. 2007. Spatial organization of Myxococcus xanthus during fruiting body formation. J. Bacteriol. 189:9126-9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daft, M. J., J. C. Burnham, and Y. Yamamoto. 1985. Lysis of Phormidium luridum by Myxococcus fulvus in continuous flow cultures. J. Appl. Bacteriol. 59:73-80. [Google Scholar]
- 16.Davidov, Y., D. Huchon, S. F. Koval, and E. Jurkevitch. 2006. A new alpha-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ. Microbiol. 8:2179-2188. [DOI] [PubMed] [Google Scholar]
- 17.Dawid, W. 2000. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 24:403-427. [DOI] [PubMed] [Google Scholar]
- 18.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dytham, C. 2004. Choosing and using statistics: a biologist's guide. Blackwell Publishing, Malden, MA.
- 20.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 21.Evans, K. J., C. Lambert, and R. E. Sockett. 2007. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J. Bacteriol. 189:4850-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falkowski, P. G., T. Fenchel, and E. F. Delong. 2008. The microbial engines that drive Earth's biogeochemical cycles. Science 320:1034-1039. [DOI] [PubMed] [Google Scholar]
- 23.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 24.Fiegna, F., and G. J. Velicer. 2005. Exploitative and hierarchical antagonism in a cooperative bacterium. PLoS Biol. 3:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Futuyma, D. J., and G. Moreno. 1988. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19:207-233. [Google Scholar]
- 26.Grafen, A., and R. Hails. 2007. Modern statistics for the life sciences. Oxford University Press, Oxford, United Kingdom.
- 27.Guerrero, R., I. Esteve, C. Pedrosalio, and N. Gaju. 1987. Predatory bacteria in prokayotic communities—the earliest trophic relationships. Ann. N. Y. Acad. Sci. 503:238-250. [Google Scholar]
- 28.Guntrip, J., and R. M. Sibly. 1998. Phenotypic plasticity, genotype-by-environment interaction and the analysis of generalism and specialization in Callosobruchus maculatus. Heredity 81:198-204. [Google Scholar]
- 29.Hillesland, K. L., R. E. Lenski, and G. J. Velicer. 2007. Ecological variables affecting predatory success in Myxococcus xanthus. Microb. Ecol. 53:571-578. [DOI] [PubMed] [Google Scholar]
- 30.Hillesland, K. L., and G. J. Velicer. 2005. Resource level affects relative performance of the two motility systems of Myxococcus xanthus. Microb. Ecol. 49:558-566. [DOI] [PubMed] [Google Scholar]
- 31.Hillesland, K. L., G. J. Velicer, and R. E. Lenski. 2009. Experimental evolution of a microbial predator's ability to find prey. Proc. Biol. Sci. 276:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeschke, J. M., M. Kopp, and R. Tollrian. 2002. Predator functional responses: discriminating between handling and digesting prey. Ecol. Monogr. 72:95-112. [Google Scholar]
- 33.Kadam, S. V., and G. J. Velicer. 2006. Variable patterns of density-dependent survival in social bacteria. Behav. Ecol. 17:833-838. [Google Scholar]
- 34.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 58:75-98. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawecki, T. J., N. H. Barton, and J. D. Fry. 1997. Mutational collapse of fitness in marginal habitats and the evolution of ecological specialisation. J. Evol. Biol. 10:407-429. [Google Scholar]
- 37.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraemer, S. A., M. A. Toups, and G. J. Velicer. 2010. Natural variation in developmental life-history traits of the bacterium Myxococcus xanthus. FEMS Microbiol. Ecol. 73:226-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 40.Krug, D., G. Zurek, O. Revermann, M. Vos, G. J. Velicer, and R. Muller. 2008. Discovering the hidden secondary metabolome of Myxococcus xanthus: a study of intraspecific diversity. Appl. Environ. Microbiol. 74:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambert, C., K. A. Morehouse, C. Y. Chang, and R. E. Sockett. 2006. Bdellovibrio: growth and development during the predatory cycle. Curr. Opin. Microbiol. 9:639-644. [DOI] [PubMed] [Google Scholar]
- 42.Lang, E., and E. Stackebrandt. 2009. Emended descriptions of the genera Myxococcus and Corallococcus, typification of the species Myxococcus stipitatus and Myxococcus macrosporus and a proposal that they be represented by neotype strains. Request for an opinion. Int. J. Syst. Evol. Microbiol. 59:2122-2128. [DOI] [PubMed] [Google Scholar]
- 43.Lenski, R. E., M. R. Rose, S. C. Simpson, and S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315-1341. [Google Scholar]
- 44.Liu, H., H. A. Nolla, and L. Campbell. 1997. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat. Microb. Ecol. 12:39-47. [Google Scholar]
- 45.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marbach, A., M. Varon, and M. Shilo. 1976. Properties of marine Bdellovibrios. Microb. Ecol. 2:284-295. [DOI] [PubMed] [Google Scholar]
- 47.Martin, M. O. 2002. Predatory prokaryotes: an emerging research opportunity. J. Mol. Microbiol. Biotechnol. 4:467-477. [PubMed] [Google Scholar]
- 48.McBride, M. J., and D. R. Zusman. 1996. Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli. FEMS Microbiol. Lett. 137:227-231. [DOI] [PubMed] [Google Scholar]
- 49.Mignot, T., J. W. Shaevitz, P. L. Hartzell, and D. R. Zusman. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315:853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pham, V. D., C. W. Shebelut, M. E. Diodati, C. T. Bull, and M. Singer. 2005. Mutations affecting predation ability of the soil bacterium Myxococcus xanthus. Microbiology 151:1865-1874. [DOI] [PubMed] [Google Scholar]
- 51.Pineiro, S. A., G. E. Sahaniuk, E. Romberg, and H. N. Williams. 2004. Predation pattern and phylogenetic analysis of Bdellovibrionaceae from the Great Salt Lake, Utah. Curr. Microbiol. 48:113-117. [DOI] [PubMed] [Google Scholar]
- 52.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reichenbach, H. 1999. The ecology of the myxobacteria. Environ. Microbiol. 1:15-21. [DOI] [PubMed] [Google Scholar]
- 54.Rendulic, S., P. Jagtap, A. Rosinus, M. Eppinger, C. Baar, C. Lanz, H. Keller, C. Lambert, K. J. Evans, A. Goesmann, F. Meyer, R. E. Sockett, and S. C. Schuster. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689-692. [DOI] [PubMed] [Google Scholar]
- 55.Rogosky, A. M., P. L. Moak, and E. A. B. Emmert. 2006. Differential predation by Bdellovibrio bacteriovorus 109J. Curr. Microbiol. 52:81-85. [DOI] [PubMed] [Google Scholar]
- 56.Rosenberg, E., K. H. Keller, and M. Dworkin. 1977. Cell density-dependent growth of Myxococcus xanthus on casein. J. Bacteriol. 129:770-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schluter, D. 1996. Ecological causes of adaptive radiation. Am. Nat. 148:S40-S64. [Google Scholar]
- 58.Schluter, D. 2000. Ecological character displacement in adaptive radiation. Am. Nat. 156:S4-S16. [Google Scholar]
- 59.Schluter, D. 2000. The ecology of adaptive radiation. Oxford University Press, Oxford, United Kingdom.
- 60.Seidler, R. J., and M. P. Starr. 1969. Isolation and characterization of host-independent bdellovibrios. J. Bacteriol. 100:769-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi, W. Y., and D. R. Zusman. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. U. S. A. 90:3378-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 63.Smith, D. R., and M. Dworkin. 1994. Territorial interactions between 2 Myxococcus species. J. Bacteriol. 176:1201-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spormann, A. M. 1999. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 63:621-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sproer, C., H. Reichenbach, and E. Stackebrandt. 1999. The correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Bacteriol. 49:1255-1262. [DOI] [PubMed] [Google Scholar]
- 66.Sudo, S., and M. Dworkin. 1972. Bacteriolytic enzymes produced by Myxococcus xanthus. J. Bacteriol. 110:236-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sutton, D. C., and P. J. Besant. 1994. Ecology and characteristics of bdellovibrios from three tropical marine habitats. Mar. Biol. 119:313-320. [Google Scholar]
- 68.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 69.Taylor, V. I., P. Baumann, J. L. Reichelt, and R. D. Allen. 1974. Isolation, enumeration, and host range of marine Bdellovibrios. Arch. Microbiol. 98:101-114. [DOI] [PubMed] [Google Scholar]
- 70.Thompson, J. N. 1999. Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 153:S1-S14. [Google Scholar]
- 71.Velicer, G. J., L. Kroos, and R. E. Lenski. 2000. Developmental cheating in the social bacterium Myxococcus xanthus. Nature 404:598-601. [DOI] [PubMed] [Google Scholar]
- 72.Velicer, G. J., G. Raddatz, H. Keller, S. Deiss, C. Lanz, I. Dinkelacker, and S. C. Schuster. 2006. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc. Natl. Acad. Sci. U. S. A. 103:8107-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Velicer, G. J., and K. L. Stredwick. 2002. Experimental social evolution with Myxococcus xanthus. Antonie Van Leeuwenhoek 81:155-164. [DOI] [PubMed] [Google Scholar]
- 74.Velicer, G. J., and M. Vos. 2009. Sociobiology of the Myxobacteria. Annu. Rev. Microbiol. 63:599-623. [DOI] [PubMed] [Google Scholar]
- 75.Velicer, G. J., and Y. T. N. Yu. 2003. Evolution of novel cooperative swarming in the bacterium Myxococcus xanthus. Nature 425:75-78. [DOI] [PubMed] [Google Scholar]
- 76.Venail, P. A., R. C. MacLean, T. Bouvier, M. A. Brockhurst, M. E. Hochberg, and N. Mouquet. 2008. Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities. Nature 452:210-214. [DOI] [PubMed] [Google Scholar]
- 77.Via, S. 1990. Ecological genetics and host adaptation in herbivorus insects—the experimental study of evolution in natural and agricultural systems. Annu. Rev. Entomol. 35:421-446. [DOI] [PubMed] [Google Scholar]
- 78.Vos, M., and G. J. Velicer. 2006. Genetic population structure of the soil bacterium Myxococcus xanthus at the centimeter scale. Appl. Environ. Microbiol. 72:3615-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vos, M., and G. J. Velicer. 2008. Isolation by distance in the spore-forming soil bacterium Myxococcus xanthus. Curr. Biol. 18:386-391. [DOI] [PubMed] [Google Scholar]
- 80.Vos, M., and G. J. Velicer. 2008. Natural variation of gliding motility in a centimetre-scale population of Myxococcus xanthus. FEMS Microbiol. Ecol. 64:343-350. [DOI] [PubMed] [Google Scholar]
- 81.Vos, M., and G. J. Velicer. 2009. Social conflict in centimeter and global-scale populations of the bacterium Myxococcus xanthus. Curr. Biol. 19:1763-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Werner, E. E., and D. J. Hall. 1974. Optimal foraging and size selection of prey by Bluegill Sunfish (Lepomis macrochirus). Ecology 55:1042-1052. [Google Scholar]
- 83.West, S. A., S. P. Diggle, A. Buckling, A. Gardner, and A. S. Griffins. 2007. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38:53-77. [Google Scholar]
- 84.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu, S. S., and D. Kaiser. 1995. Gentic and functional evidence that type-IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 86.Yamanaka, S., A. Kawaguchi, and K. Komagata. 1987. Isolation and identification of myxobacteria from soils and plant materials, with special reference to DNA-base composition, quinone system, and cellular fatty-acid composition, and with a description of a new species, Myxococcus flavescens. J. Gen. Appl. Microbiol. 33:247-265. [Google Scholar]
- 87.Zusman, D. R., A. E. Scott, Z. Yang, and J. R. Kirby. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 5:862-872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.