Abstract
Resistance to lysostaphin, a staphylolytic glycylglycine endopeptidase, is due to a FemABX-like immunity protein that inserts serines in place of some glycines in peptidoglycan cross bridges. These modifications inhibit both binding of the recombinant cell wall targeting domain and catalysis by the recombinant catalytic domain of lysostaphin.
Lysostaphin is a glycylglycine endopeptidase produced by Staphylococcus simulans biovar staphylolyticus (18) that lyses susceptible staphylococci by hydrolyzing the polyglycine cross bridges in their cell wall peptidoglycans (3). The lysostaphin gene sequence was independently determined in 1987 by two groups (8, 13). BLAST analysis (1) of mature lysostaphin revealed two domains: an N-terminal catalytic domain (CAT), which is a member of the M23 family of zinc metalloendopeptidases, and a C-terminal cell wall targeting domain (CWT), which is a member of the SH3b domain family (Fig. 1 A).
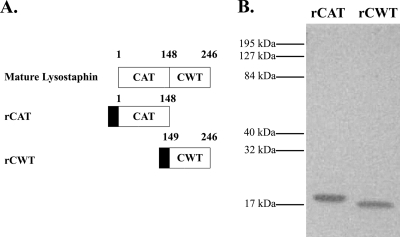
FIG. 1.
(A) Schematic diagram of mature lysostaphin, the recombinant catalytic domain (rCAT) (lysostaphin residues 1 to 148), and the recombinant cell wall targeting domain (rCWT) (lysostaphin residues 149 to 246). The numbers represent the beginning and end of the domains, and the solid boxes indicate the N-terminal His6 tag of the recombinant proteins. (B) SDS-PAGE analysis of rCAT and rCWT purified by a nickel affinity column. Mobilities of molecular mass standards are given on the left side of the gel.
The lysostaphin endopeptidase resistance gene (epr or lif) encodes a FemABX-like immunity protein that is located adjacent to the lysostaphin gene on the plasmid pACK1 in S. simulans bv. staphylolyticus (4, 7, 20). Members of the FemABX family of proteins are nonribosomal peptidyl transferases that are involved in the addition of cross bridge amino acids during peptidoglycan subunit synthesis in the cytoplasm (15). In S. simulans bv. staphylolyticus, the lysostaphin immunity protein inserts serines in place of some glycines during peptidoglycan synthesis, which provides resistance to lysostaphin (4, 20).
Originally it was suggested that the incorporation of serines in these peptidoglycan cross bridges gave increased resistance to lysostaphin because of the inability of the enzyme to hydrolyze glycyl-serine or seryl-glycine bonds (4, 14, 16). Others later reported that the CWT specifically binds to the polyglycine cross bridges in staphylococci (6) and the binding of CWT to producer-strain cells was less than that to susceptible cells (2). However, the ability of the enzyme or its targeting domain to bind to purified peptidoglycans from staphylococci containing the lysostaphin resistance gene has not been determined. Therefore, we determined if the modification to staphylococcal peptidoglycan cross bridges made by the lysostaphin immunity protein affected the activity of the binding domain, the catalytic domain, or both.
Generation of rCAT and rCWT of lysostaphin.
Primers for CAT (5′ ACA GCT GGA TCC GCT GCA ACA CAT GAA CAT TCA GC 3′ and 5′ TTC GGA AGC TTA GTT ACT GTA CCA CCT GCT TTT CCA TAT C 3′) and for CWT (5′ TAC AGG ATC CCC AAC GCC GAA TAC AGG TTG GAA AA 3′ and 5′ TAA AAA AAG CTT TCA CTT TAT AGT TCC CCA AAG AAC ACC 3′) were used to amplify the regions encoding the domains. The PCR products and pQE80L, which provides an N-terminal His6 tag, were digested with BamHI and HindIII (Roche Diagnostics GmbH, Mannheim, Germany), and the reaction products were ligated using T4 DNA ligase (Roche) to create pQELSSCAT and pQELSSCWT. Electrocompetent Escherichia coli M15/pREP4 was used for transformation. The cells were made competent by use of the protocol described by Sheng et al. (17). Plasmid DNA was extracted and purified from transformants using the QIAprep spin miniprep kit (Qiagen, Valencia, CA) and sequenced using the primers PR and RS (Qiagen) to ensure sequence fidelity before protein expression. Each recombinant protein (rCAT and rCWT) was purified by the procedure described by Lai et al. (11). Recombinant proteins were analyzed by SDS-PAGE using a 12.5% gel (10) and stained with Biosafe Coomassie brilliant blue (Bio-Rad, Hercules, CA) (Fig. 1B).
Binding of rCWT to peptidoglycans from strains with and without the lysostaphin immunity protein.
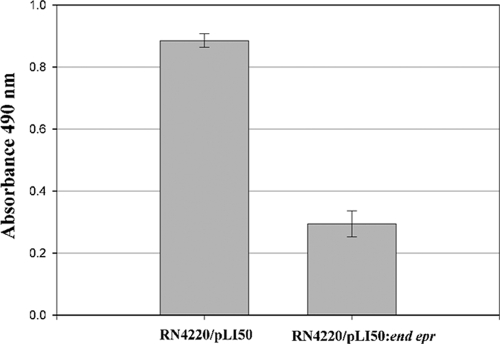
Previously it was reported by Baba and Schneewind (2) that lysostaphin is unable to bind to the producer cell, S. simulans bv. staphylolyticus, and that if the binding domain is removed, the enzyme cannot attach to susceptible cells. These authors suggested that this was due to the lysostaphin immunity protein inserting serines in the place of some glycines in the peptidoglycan cross bridge (2). Other wall-associated polymers, though, such as wall teichoic acids, have been shown to inhibit the binding of lysostaphin (6). Therefore, the ability of rCWT to attach to purified peptidoglycans from an S. aureus strain containing an 8.4-kb fragment from pACK1 that has the gene for the lysostaphin immunity protein (RN4220/pLI50::end epr) and an S. aureus strain without that gene (RN4220/pLI50) (4) was determined using a modification of our previously described binding assay (5). In the assay, rCWT was at a final concentration of 10 μg/ml, which was determined in preliminary experiments to be in the middle of the linear range of the assay (1 to 30 μg/ml; data not shown), and color was allowed to develop for 5 min instead of 1 h. Peptidoglycans were purified as previously described; the cross bridge composition for strain RN4220/pLI50 is Gly4.5Ser0.2, and that for RN4220/pLI50::end epr is Gly2.7Ser1.6 (4). As seen in Fig. 2, rCWT did not bind as well to peptidoglycan from strain RN4220/pLI50::end epr as it did to peptidoglycan from strain RN4220/pLI50. In contrast to the findings of Baba and Schneewind (2), we do see some binding of the rCWT to Epr-modified peptidoglycan. Although our binding assay is not directly comparable to theirs (different strains and whole cells versus purified peptidoglycans), both their results and our results are consistent with Epr modification of peptidoglycans inhibiting binding of the lysostaphin CWT.
FIG. 2.
Binding of rCWT to peptidoglycans from S. aureus RN4220/pLI50 and RN4220/pLI50::end epr. Binding of rCWT was detected using nickel-coupled peroxidase with o-phenylenediamine dihydrochloride as a substrate, and absorbance was monitored spectrophotometrically at 490 nm. The bars represent the means ± standard deviations of results for triplicate samples.
Sensitivity of peptidoglycans to rCAT.
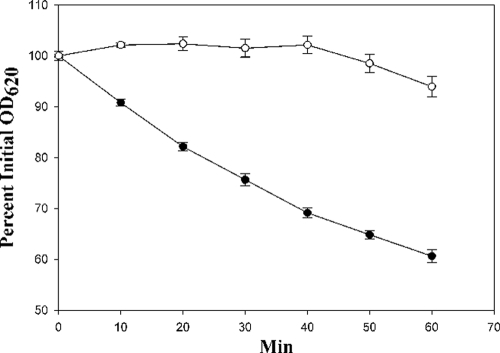
Lysostaphin has been shown to be unable to cleave artificial peptide substrates containing both serines and glycines, whereas it can hydrolyze glycyl peptides (16, 19). This suggested that the catalytic domain of lysostaphin may be unable to hydrolyze the cross bridge of the producer cell due to the insertion of serines (4, 16, 19); however, the catalytic domain has never been separated from the binding domain to see if this is the case. Therefore, peptidoglycans from strains RN4420/pLI50 and RN4220/pLI50::end epr were incubated with rCAT (0.1 mg/ml) for 1 h, and the decrease in turbidity was monitored spectrophotometrically. As seen in Fig. 3, the peptidoglycan from strain RN4220/pLI50 was hydrolyzed more readily by rCAT than the peptidoglycan from strain RN4220/pLI50::end epr, indicating that the lysostaphin immunity protein-specified modification also inhibited the activity of the catalytic domain in addition to inhibiting the binding of the rCWT to peptidoglycans. For comparison, a molar equivalent of N-terminal His6-tagged recombinant lysostaphin caused in 10 min or less reductions in turbidities similar to those seen in 60 min for rCAT.
FIG. 3.
Relative sensitivities of peptidoglycans from S. aureus RN4220/pLI50 (•) and RN4220/pLI50::end epr (○) to rCAT were analyzed by monitoring the reduction in the optical density at 620 nm (OD620) over 1 h. Results were normalized to the percent initial OD, and the error bars represent ± standard deviations of results for triplicate samples.
These results are similar to the inhibition of the cell wall binding and catalytic domains of the streptococcolytic enzyme zoocin A by streptococcal peptidoglycans from cells with the zoocin A immunity factor, Zif (5). Zif is a FemABX-like immunity protein that inserts an additional l-alanine into the majority of the peptidoglycan cross bridges in the producer cell (5). Thus, the peptidoglycan modifications due to FemABX immunity proteins associated with lysostaphin and zoocin A affect both their binding and catalytic domains. Binding of the CWT domain from another staphylolytic enzyme, ALE-1 (which contains the M23 [89% identical, 96% similar to lysostaphin] and SH3b [84% identical, 89% similar to lysostaphin] domains), to purified peptidoglycans also was inhibited when serines were substituted for some glycines in the cross bridges by the ALE-1-associated FemABX-like immunity protein (12). The effect of these substitutions on the catalytic activity of the CAT domain of ALE-1 has not been tested. We expect that the peptidoglycan modifications caused by other FemABX-like immunity proteins will affect the activities of both domains of such peptidoglycan hydrolases, as we have shown for lysostaphin and zoocin A.
Lysostaphin has potential for use as an antistaphylococcal agent for treatment of infections caused by antibiotic-resistant strains of S. aureus, as recently reviewed by Kumar (9). Therefore, understanding resistance to this potent enzyme due to peptidoglycan modifications should be of interest to those who are evaluating lysostaphin for possible clinical use.
Footnotes
Published ahead of print on 20 August 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., and O. Schneewind. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 15:4789-4797. [PMC free article] [PubMed] [Google Scholar]
- 3.Browder, H. P., W. A. Zygmunt, J. R. Young, and P. A. Tavormina. 1965. Lysostaphin: enzymatic mode of action. Biochem. Biophys. Res. Commun. 19:383-389. [DOI] [PubMed] [Google Scholar]
- 4.DeHart, H. P., H. E. Heath, L. S. Heath, P. A. LeBlanc, and G. L. Sloan. 1995. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl. Environ. Microbiol. 61:1475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gargis, S. R., A. S. Gargis, H. E. Heath, L. S. Heath, P. A. LeBlanc, M. M. Senn, B. Berger-Bächi, R. S. Simmonds, and G. L. Sloan. 2009. Zif, the zoocin A immunity factor, is a FemABX-like immunity protein with a novel mode of action. Appl. Environ. Microbiol. 75:6205-6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gründling, A., and O. Schneewind. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 188:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heath, H. E., L. S. Heath, J. D. Nitterauer, K. E. Rose, and G. L. Sloan. 1989. Plasmid-encoded lysostaphin endopeptidase resistance of Staphylococcus simulans biovar staphylolyticus. Biochem. Biophys. Res. Commun. 160:1106-1109. [DOI] [PubMed] [Google Scholar]
- 8.Heinrich, P., R. Rosenstein, M. Böhmer, P. Sonner, and F. Götz. 1987. The molecular organization of the lysostaphin gene and its sequences repeated in tandem. Mol. Gen. Genet. 209:563-569. [DOI] [PubMed] [Google Scholar]
- 9.Kumar, J. K. 2008. Lysostaphin: an antistaphylococcal agent. Appl. Microbiol. Biotechnol. 80:555-561. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lai, A. C.-Y., S. Tran, and R. S. Simmonds. 2002. Functional characterization of domains found within a lytic enzyme produced by Streptococcus equi subsp. zooepidemicus. FEMS Microbiol. Lett. 215:133-138. [DOI] [PubMed] [Google Scholar]
- 12.Lu, J. Z., T. Fujiwara, H. Komatsuzawa, M. Sugai, and J. Sakon. 2006. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J. Biol. Chem. 281:549-558. [DOI] [PubMed] [Google Scholar]
- 13.Recsei, P. A., A. D. Gruss, and R. P. Novick. 1987. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc. Natl. Acad. Sci. U. S. A. 84:1127-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson, J. M., J. K. Hardman, and G. L. Sloan. 1979. Relationship between lysostaphin endopeptidase production and cell wall composition in Staphylococcus staphylolyticus. J. Bacteriol. 137:1158-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohrer, S., and B. Berger-Bächi. 2003. FemABX peptidyl transferases: a link between branched-chain wall peptide formation and β-lactam resistance in gram-positive cocci. Antimicrob. Agents Chemother. 47:837-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose, K. E., J. M. Robinson, J. W. Ross, J. K. Hardman, H. E. Smith, and G. L. Sloan. 1983. Chemical and ultrastructural studies on the cell wall of Staphylococcus simulans biovar staphylolyticus. Curr. Microbiol. 8:37-43. [Google Scholar]
- 17.Sheng, Y., V. Mancino, and B. Birren. 1995. Transformation of Escherichia coli with large DNA molecules by electroporation. Nucleic Acids Res. 23:1990-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sloan, G. L., J. M. Robinson, and W. E. Kloos. 1982. Identification of “Staphylococcus staphylolyticus” NRRL B-2628 as a biovar of Staphylococcus simulans. Int. J. Syst. Bacteriol. 32:170-174. [Google Scholar]
- 19.Sloan, G. L., E. C. Smith, and J. H. Lancaster. 1977. Lysostaphin endopeptidase-catalysed transpeptidation reactions of the imino-transfer type. Biochem. J. 167:293-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thumm, G., and F. Götz. 1997. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol. Microbiol. 23:1251-1265. [DOI] [PubMed] [Google Scholar]