Abstract
As a comparison to a similar study on Photorhabdus strains, 15 Xenorhabdus bacterial strains and secondary phenotypic variants of two strains were screened for proteolytic activity by five detection methods. Although the number and intensity of proteolytic activities were different, every strain was positive for proteolytic activity by several tests. Zymography following native PAGE detected two groups of activities with different substrate affinities and a higher and lower electrophoretic mobility that were distinguished as activity 1 and 2, respectively. Zymography following SDS-PAGE resolved three activities, which were provisionally named proteases A, B, and C. Only protease B, an ∼55-kDa enzyme, was produced by every strain. This enzyme exhibited higher affinity to the gelatin substrate than to the casein substrate. Of the chromogenic substrates used, three were hydrolyzed: furylacryloyl-Ala-Leu-Val-Tyr (Fua-ALVY), Fua-LGPA (LGPA is Leu-Gly-Pro-Ala) (a substrate for collagen peptidases), and succinyl-Ala-Ala-Pro-Phe-thiobenzyl (Succ-AAPF-SBzl). All but the Fua-LGPA-ase activity seemed to be from secreted enzymes. According to their substrate preference profiles and inhibitor sensitivities, at least six such proteolytic enzymes could be distinguished in the culture medium of Xenorhabdus strains. The proteolytic enzyme that was secreted the earliest, protease B and the Succ-AAPF-SBzl-hydrolyzing enzyme, appeared from the early logarithmic phase of growth. Protease B could also be detected in the hemolymph of Xenorhabdus-infected Galleria mellonella larvae from 15 h postinfection. The purified protease B hydrolyzed in vitro seven proteins in the hemolymph of Manduca sexta that were also cleaved by PrtA peptidase from Photorhabdus. The N-terminal sequence of protease B showed similarity to a 55-kDa serralysin type metalloprotease in Xenorhabdus nematophila, which had been identified as an orthologue of Photorhabdus PrtA peptidase.
Xenorhabdus and Photorhabdus bacteria are highly virulent, fatal pathogens for insects. Phylogenetically, they are sister genera in the family Enterobacteriaceae (3, 4). There are some differences between Xenorhabdus and Photorhabdus in their biology (e.g., light production), and they also differ in their interaction with their symbiotic nematode partners, which are in the Steinernematidae and Heterorhabditidae genera, respectively (8, 9). At the same time, they also have several properties in common. For example, due to their similar strategy of infection, their entrance into the hemocoel is absolutely dependent on the invasion of insects by their symbiotic nematode partners. An interesting feature of both genera is that they have two phenotypic (form) variants, primary and secondary (9). The primary form is natural, while the secondary form can be observed (generated) mostly in the laboratory. They differ in, for example, antibiotic production, outer membrane proteins, and cell surface structures (fimbriae and flagellae [23], symbiotic capabilities with nematode partners, and exoenzyme production [9]). The secondary form variants were found, with nonbiochemical detection methods, to produce less or no proteolytic activity compared to the primary phenotypic variants (see references 9 and 23 and references therein). The high pathogenicity makes Xenorhabdus and Photorhabdus good model organisms of infection, which can be exploited—by studying the function of their virulence factors—for the investigation of the immune system of insects and the mechanisms the pathogens use to cope with the immune defense of hosts. The comparative analysis of these bacterial partners provides an opportunity to study the question of how similar the infection mechanisms can be at the molecular level of two evolutionarily different insect pathogen bacterium-nematode complexes that, at the same time, have similar infection strategies.
Of the virulence factors, we have been interested in secreted proteases that may be used by the pathogens during the first stage of infection in the penetration of the tissues of host or in the suppression of its immune response. The secretion and biochemistry of these enzymes are better studied in Photorhabdus, where four secreted proteases could be detected in a screen of 20 strains by a combination of five methods (15). The earliest secreted Photorhabdus protease is PrtA peptidase, a metzincin in the M10B family of serralysins. The others are PhpC (Photorhabdus protease C), which belongs to the M4 metallopeptidase family of thermolysin-like proteases, OpdA, a collagen peptidase in the family of thimet oligopeptidases and PhpD, a furylacryloyl-Ala-Leu-Val-Tyr (Fua-ALVY)-cleaving enzyme, the identity of which is still unknown. In contrast, although a number of Xenorhabdus strains were tested for proteolytic activity with simple bacteriological plate assays (2, 25), only one (Xenorhabdus nematophila) was investigated by a biochemical detection method of protease activities, zymography. Two activities have been found by this method, and one of these activities has been partially characterized (5).
As an approach to establish the similarity between Xenorhabdus and Photorhabdus in the mechanism of infection regarding the type and role of proteolytic enzymes, we investigated 15 Xenorhabdus strains for the secretion of proteases employing the same five detection methods that we had previously used for Photorhabdus strains. Two of the strains (Xenorhabdus nematophila AN6 and Xenorhabdus cabanillassii RIO-HU) were represented with their phenotypic variant pairs.
MATERIALS AND METHODS
Substrates.
The substrates were purchased from Sigma-Aldrich (St. Louis, MO) (succinyl-Ala-Ala-Pro-Phe-thiobenzyl [Succ-AAPF-SBzl]) and Bachem (Bubendorf, Switzerland) (Fua-Leu-Gly-Pro-Ala [Fua-LGPA]) or prepared as described previously (succinyl-Ala-Ala-Pro-Phe-Lys-aminomethyl-coumarine [Succ-AAPX-AMC] [X is Phe and Lys] [13] and Fua-ALVY [16]). For solutions, the substrates were dissolved in dimethylformamide.
Enzymes.
The purification of protease B, which was used as the standard in gel electrophoresis and zymography and in hemolymph protein digestion, will be published elsewhere (M. K. Massaoud, J. Marokházi, and I. Venekei, submitted for publication). The N-terminal sequence of the enzyme was determined by András Patthy at the ELTE-MTA Biotechnology Research Group. Recombinant Photorhabdus PrtA peptidase (a gift from G. Felföldi from our laboratory) was purified from the culture medium of Escherichia coli HB101 strain harboring pUC19 plasmid vector which contained a gene encoding Photorhabdus PrtA (kindly provided by R. ffrench-Constant, University of Bath, United Kingdom) with anion-exchange chromatography on PAE silica 300 column as described previously (6). Bovine pancreatic trypsin, chymotrypsin, and elastase, as well as Clostridium histolyticum collagenase were purchased from Sigma-Aldrich.
Bacterial strains and cell culturing conditions.
For taxonomic details, see reference 4. The strains were obtained from the collection of entomopathogenic nematode-bacterium strains established and maintained at the Department of Genetics, Eötvös University, Budapest, Hungary, by one of us (A. Fodor). Information on the strains used in this study are summarized in Table 1. Several additional important pieces of information about the strains follow. X. nematophila is the first characterized (1) and the most studied species. It has recently been sequenced (GenBank accession number FN667742). The natural nematode symbiotic partner of X. nematophila is Steinernema carpocapsae. The X. nematophila type strain DSM 3370 (19, 20) was isolated in the Czech Republic in Europe. The American AN6/1 strain was originally isolated by R. Akhurst and forwarded to R. E. Hurlbert (United States). The secondary variant (AN6/2) spontaneously segregated out in S. Forst's laboratory (see details in references 11, 23, and 24). Xenorhabdus cabanillassii is the natural symbiont of Steinernema riobrave (21). The strain (RIO-HU) was isolated and characterized by us (19) from the S. riobrave that Byron Adams kindly provided. Two strains of Xenorhabdus kozodoii were isolated by us from an unknown Steinernema species from Morocco and from Steinernema arenarium from Russia, kindly provided by Ralf-Udo Ehlers (Germany) and Marek Tomalak (Poland) and were named Morocco and AZ, respectively. The type strains of Xenorhabdus budapestensis (DSM 16342), Xenorhabdus szentirmaii (DSM 16338), Xenorhabdus ehlersii (DSM 16337), and Xenorhabdus innexi (DSM 16336) were isolated by E. Szállás from Steinernema bicornutum (kindly provided by Béla Tallósi), Steinernema rarum (Uruguay), Steinernema longicaudatum (China), and Steinernema scapterisci (sent by B. Adams to us). Their taxonomic identification was made by one of us (14).
TABLE 1.
Bacterial Xenorhabdus strains and Steinernema species (nematode symbiont) used in this studya
| Xenorhabdus species (bacterium) | Xenorhabdus strain or isolateb | Steinernema species (nematode symbiont) | Source of: |
Geographic origin of the nematode | 16S rRNA gene accession no. | |
|---|---|---|---|---|---|---|
| Nematode | Bacterium | |||||
| X. nematophila | DSM 3370T | S. carpocapsae | Z. Mracek | E. Stackebrandt | Czech Republic | X82251 |
| AN6/1 | R. Akhurst | S. A. Forst | US | AY278674 | ||
| AN6/2 | No host | S. A. Forst | In the lab | |||
| X. cabanillassii | RIO-HU/1 | S. riobrave | B. Adams | A. Fodor | Texas | Z7638 |
| RIO/2 | No host | A. Fodor | In the lab | |||
| X. bovienii | DSM 4766T | S. feltiae | I. N. Filipjev 1934 | E. Stackebrandt | Russia | X82252 |
| “Krausseii” | S. kraussei | J. Gunter | J. Gunter | Switzerland | ||
| X. kozodoii | “Anomali AZ” | S. arenarium | N. Simoes | A. Fodor | Russia | |
| “Intermedium BIO” | S. intermedium | N. Simoes | A. Fodor | South Carolina | ||
| “Morocco” | Steinernema sp. | R.-U. Ehlers | A. Fodor | Morocco | ||
| X. poinarii | DSM 4768T | S. glaserii | G. Steiner 1929 | E. Stackebrandt | North Carolina | |
| “Cubanum” | S. cubanum | Z. Mracek | A. Lucskai | Cuba | ||
| X. beddingii | DSM 4764T | S. longicaudatum | C. P. Sheng and G. H. Wang | E. Stackebrandt | China | |
| X. budapestensis | DSM 16342T | S. bicornutum | B. Tallosi | A. Fodor | Vojvodina, Serbia | AJB10293 |
| X. szentirmaii | DSM 16338T | S. rarum | B. Adams | A. Fodor | Cordoba, Spain | AJB10295 |
| X. ehlersii | DSM 16337T | S. longicaudatum | B. Adams | A. Fodor | China | |
| X. innexi | DSM 16336T | S. scapterisci | B. Adams | A. Fodor | Uruguay | |
The bacterial Xenorhabdus species and strain or isolate, the nematode symbiont Steinernema species, source of nematode and bacterium (the individuals who provided the nematode and bacterium), geographic origin of the nematode, and EMBL accession number of 16S rRNA gene are shown (18a).
For further information about the bacterial strains, see “Bacterial strains and cell culturing conditions” in Materials and Methods. The numbers after the slashes indicate the phenotypic variants (2, 7-9). Isolates are shown within double quotation marks. Type strains are indicated by a superscript T. DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen.
Liquid cultures were grown in Luria-Bertani (LB) medium without antibiotics at 30°C in a rotary shaker. They were inoculated with single colonies from LB plates which had been incubated for 48 h at 28°C and replica tested on NBTA plates (nutrient agar supplemented with 25 mg of bromothymol blue and 40 mg of triphenyl-2,3,5-tetrazolium chloride per liter) to confirm the phenotypic variation status of strains.
Insects and Manduca sexta hemolymph.
Galleria mellonella (greater wax moth) (Lepidoptera) larvae were bred in our laboratory. They were reared at 25°C on beeswax sheets supplemented with granulated pollen. Fifth-instar larvae were used in all experiments. The hemolymph from nonimmunized Manduca sexta larvae were provided by G. Felföldi from our laboratory. The larvae were bled through their horn cut, and their hemolymph was immediately mixed with 10 volumes of phosphate-buffered saline (PBS) that contained 10 mM phenylthiourea. The cellular fraction was sedimented with centrifugation at 2,800 × g for 20 min. The supernatants were stored at −80°C.
Plate assay for gelatin hydrolysis.
Frazier's method (10) for detecting hydrolysis of gelatin caused by bacteria and the evaluation of activities were performed as described previously (15).
Polyacrylamide gel electrophoresis and zymography.
The composition of gels, the conditions of sample preparation and electrophoresis, and the incubation of gels for the development of zymographic activity were as described previously for analysis of Photorhabdus strains (15). To test the effect on the zymographic activity of phenylmethylsulfonyl fluoride (PMSF) and the complex-forming compounds EDTA and 1,10-phenanthroline, they were applied in 5.0 mM concentration during both electrophoresis sample preparation and incubation of gels after the gels were run. The samples were loaded on the gel at a distance of three wells from each other to prevent cross contamination during electrophoresis due to the diffusion of inhibitors.
Enzyme activity measurements.
Enzyme activities were measured in a final volume of 1.0 ml at 30°C in an enzyme assay buffer (50 mM Tris-HCl [pH 8.0], 10 mM CaCl2, 0.1 M NaCl, and 50 μg/ml bovine serum albumin) at a final substrate concentration of 50 μM using 50 μl of culture supernatant or cell lysate. The activities on fluorometric substrate with AMC chromophore were measured at a wavelength of 380 nm for excitation light and 460 nm for emission light. The activities on 2-furylacryloyl group blocked substrates and on the substrate with the thiobenzyl leaving group (Succ-AAPF-SBzl) were measured in a photometer at 324 nm. In the latter case, the enzyme assay buffer was supplemented with 25 μM SH reagent, 4,4-dithiodipyridine. The reactions were started with the addition of the appropriate substrate. The precise concentration of the furylacryloyl substrate was determined spectrophotometrically using a ɛ of 24,700 M−1 cm−1 (22). The hydrolysis rates of Succ-AAPF-SBzl and the furylacroyl substrates were calculated from the first, linear part of the time dependence curves, using the values ɛ = 19,000 M−1cm−1 and Δɛ = 2,510 M−1cm−1, respectively. (Δɛ is the difference between the absorption coefficient of the furylacryloyl substrate and the furylacryloyl-amino acid product.)
Infection experiments and hemolymph sample preparation.
An overnight LB culture of X. kozodoii Morocco strain, which was started with several colonies from a fresh LB plate, was 20-fold diluted into LB medium and grown to an optical density at 600 nm (OD600) of 0.3 (2- to 3-h incubation at 30°C). Then the cells in 7.0 ml of such a culture were sedimented, washed twice in 1.0 ml of PBS (137.9 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4 [pH 6.5]), and finally resuspended in 1.0 ml of PBS. To determine cell counts, serial dilutions (101 to 106) were made in PBS, and 20-μl amounts of the dilutions were plated onto LB plates. Three groups of fifth-instar G. mellonella (greater wax moth) (Lepidoptera) larvae (5 larvae in each group) were injected with 5 μl of Xenorhabdus cell suspension (∼50 cells). Hemolymph samples (5 to 8 μl) were taken from the larvae in the three groups 12, 18, and 24 h postinfection via cutting a proleg and diluted 10-fold immediately with PBS that contained 10 mM phenylthiourea. The cellular fraction was sedimented as described above. For zymographic analysis and enzyme assays, 4.0-μl and 7.0-μl portions of these samples were used, respectively.
Digestion of M. sexta hemolymph proteins.
Prior to digestion, the hemolymph proteins were fractioned by DEAE anion-exchange chromatography by the method of Felföldi et al. (6) to separate proteins into fractions of distinctive protein composition. Six such fractions (A to F) were obtained (see Fig. 4A). For digestion, 1 to 16 μg hemolymph protein (depending on the protein content of the fraction) was incubated at 25°C for 90 min in the presence of 1.0 pmol (∼0.06 μg) of the following proteases: bovine pancreatic trypsin, chymotrypsin, elastase, Clostridium histolyticum collagenase, and purified protease B.
RESULTS
Screening for protease activity with zymography.
As an initial step in the investigation of secreted proteolytic activities, we screened the cultures of Xenorhabdus strains using five detection methods that we had employed earlier on Photorhabdus strains (15). The results of screening protease activity by zymography and SDS-PAGE and native PAGE are summarized in Table 2.
TABLE 2.
Summary of results of proteases secreted by 17 Xenorhabdus strains and phase variants detected by five methods
| Xenorhabdus species and strain or isolatea | Gelatin liquefaction assay resultb | Zymographic result for band following: |
Activity (×10−7 M min−1) on: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Substrate | SDS-PAGEc |
Native PAGEc |
||||||||
| Band A | Band B | Band C | Band 1 | Band 2 | Succ-AAPF-Sbzld | Fua-LGPAe | Fua-ALVYe | |||
| X. nematophila strains | ||||||||||
| DSM 3370 | + | Gelatin | (+) (s) | — | — | (+) (ll) | — | 2.5 (el) | 10.5 (l) | 6.0 (el) |
| Casein | — | (x) (l)f | — | — | (x) (ll) | |||||
| AN6/1 | + | Gelatin | — | — | — | — | — | 2.3 (el) | 0.0 (l) | 7.1 (el) |
| Casein | — | (x) (ll)f | — | — | — | |||||
| AN6/2 | + | Gelatin | (+) (s) | (+) (l) | 3.5 (el) | 6.5 (l) | 8.2 (el) | |||
| Casein | — | xx (l)f | — | — | (x) (ll) | |||||
| X. cabanillassii strains | ||||||||||
| Riobrave/1 | + | Gelatin | (+) (ll) | — | — | — | — | 23.0 (el) | 23.5 (l) | 11.0 (el) |
| Casein | — | (x) (l)f | — | — | x (ll) | |||||
| Riobrave/2 | + | Gelatin | (+) (ll) | — | — | — | — | 2.1 (el) | 10.6 (l) | 13.4 (el) |
| Casein | — | (x) (l)f | — | — | — | |||||
| X. bovienii strains | ||||||||||
| DSM 4766 | + | Gelatin | (+) (ll) | + (s) | — | (+) (s) | — | 22.0 (el) | 10.1 (l) | 10.4 (el) |
| Casein | — | x (ll) | (x) (s) | — | (x) (s) | |||||
| Kraussei | + | Gelatin | — | (+) (ll) | — | + (ll) | — | 67.0 (el) | 15.6 (l) | 31.9 (el) |
| Casein | — | xx (ll) | (x) (ll) | x (ll) | xx (l) | |||||
| X. kozodoii strains | ||||||||||
| Intermedium | + | Gelatin | (+) (s) | (+) (ll) | — | (+) (ll) | — | 6.1 (el) | 8.5 (l) | 9.3 (el) |
| Casein | — | xx (el) | (x) (ll) | — | x (ll) | |||||
| Anomali AZ | + | Gelatin | — | (+) (l)f | — | + (ll) | — | 3.1 (el) | 26.7 (l) | 39.0 (el) |
| Casein | — | (x) (el)f | (x) (l) | xx (ll) | xx (ll) | |||||
| Morocco | + | Gelatin | (+) (ll) | + (l) | — | + (l) | — | 18.0 (el) | 9.6 (l) | 8.7 (el) |
| Casein | — | xx (el)f | (x) (l) | xx (el) | xx (ll) | |||||
| X. poinarii strains | ||||||||||
| DSM 4768 | + | Gelatin | (+) (s) | ++ (l)f | — | (+) (ll) | — | 1.8 (el) | 11.3 (l) | 15.4 (el) |
| Casein | — | xx (l)f | — | — | (x) (ll) | |||||
| Cubanum | + | Gelatin | (+) (s) | + (l) | — | ++ (ll) | — | 7.9 (el) | 10.0 (l) | 11.9 (el) |
| Casein | — | xx (el) | (x) (ll) | — | xx | |||||
| X. beddingii DSM 4764 | + | Gelatin | — | ++ (el)f | — | (+) (ll) | — | 2.0 (el) | 8.3 (l) | 12.9 (el) |
| Casein | — | xx (el)f | — | — | (x) (ll) | |||||
| X. budapestensis DSM 16342 | + | Gelatin | — | + (s) | — | ++ (ll) | — | 2.8 (el) | 9.2 (l) | 13.7 (el) |
| Casein | — | xx (ll) | — | — | xx (ll) | |||||
| X. szentirmaii DSM 16338 | + | Gelatin | — | (+) (ll) | — | + (ll) | — | 4.7 (el) | 31.2 (l) | 11.9 (el) |
| Casein | — | xx (el)f | — | — | x (ll) | |||||
| X. ehlersii DSM 16337 | NT | Gelatin | — | + (el) | — | ++ (l) | — | 2.8 (el) | 14.4 (l) | 27.0 (el) |
| Casein | — | x (el)f | — | — | (x) (l) | |||||
| X. innexi DSM 16336 | NT | Gelatin | — | + (ll) | — | ++ (ll) | — | 4.5 (el) | 4.0 (l) | 10.6 (el) |
| Casein | — | xx (el)f | — | — | xx (ll) | |||||
The identities of strains are given in Table 1.
Symbols: +, clearing zone is visible; −, clearing zone is not visible. NT, not tested.
The semiquantitative ratings of the activity bands by zymography on the gelatin substrate were as follows: —, no activity; (+), weak; +, medium (easily detected); ++, strong. The semiquantitative ratings of the strongest activities for activity band by zymography on the casein substrate were as follows: —, no activity; (x), weak; x, medium; xx, strong. The time of first detection of an activity is shown in parentheses behind the semiquantitative ratings as follows: el, early logarithmic phase (0- to 4-h culture); l, logarithmic phase (4- to 8-h culture); ll, late logarithmic or early stationary phase (8- to 20-h culture); s, stationary phase (longer than 20 h growth). (For the optical densities in these phases, see the growth curves of two strains in Fig. 2. The zymographic activities are distinguished according to their electrophoretic mobility such that band A [SDS-PAGE] and band 1 [native PAGE] have the lowest mobility.)
The activities were measured using 50-μl supernatant of 4-h cultures.
The activities were measured using 50-μl supernatant samples of 24-h cultures. For the dynamics of production of Fua-ALVY-ase and Fua-LGPA-ase activities, see Fig. 2. The calculation of enzyme activities is given in Materials and Methods.
When the cultures were incubated longer than 20 to 24 h, the production of these activities declined.
In a nonspecific, semiquantitative plate assay for gelatinase activity, Xenorhabdus strains performed similarly to Photorhabdus strains: the majority of activities differed by only severalfold. However, unlike Photorhabdus, each of the Xenorhabdus strains studied gave positive results, including the two secondary form variants. The strongest activity (by X. nematophila DSM 4768) was 20-fold higher than the weakest one (X. nematophila AN6/1).
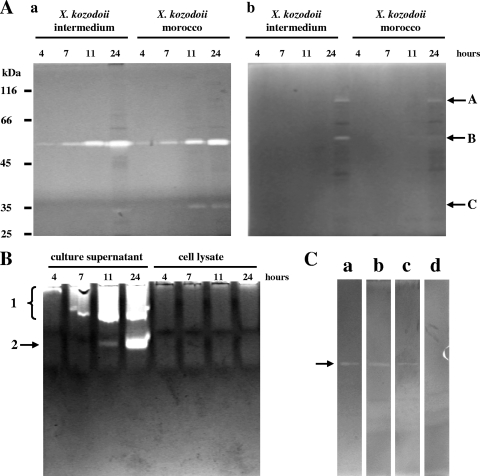
Another nonspecific and semiquantitative method is zymography after gel electrophoresis which, however, is able to resolve activities according to their molecular properties. We applied this method in four variations: after both SDS-PAGE and native PAGE and by using gelatin and casein as the substrate. Gelatin allowed a more sensitive detection of Xenorhabdus proteases especially after SDS-PAGE. We could separate three different activity bands with SDS-PAGE that we distinguished by letters (bands A to C in Fig. 1A) and two activities with native PAGE that we labeled with numbers (bands 1 and 2 in Fig. 1B). When the lysed cell fractions were analyzed, the activities remained below the level of detection, indicating that they are not from intracellular proteases (see Fig. 1B for activity bands 1 and 2). Sometimes, activity bands 2 and B were split, which we attributed either to molecular variants or degradation products of the enzymes that are still active. At the same time, activity 1 was usually a wide band which with the casein substrate could often be resolved in four to six component bands. Molar mass estimation gave 90, 55, and 35 kDa for activities A, B, and C, respectively. We could observe different substrate preferences: activities A and C were mainly detectable with gelatin and casein substrates, respectively, while activity 1 had an apparently stronger affinity to substrates than activity 2, as indicated by substrate cleavage during electrophoresis (generating a long stripe of activity) and the concomitant dependence of migration on protein amount. To investigate which of the activities detected after native PAGE and SDS-PAGE were from the same enzyme, we ran gels with the purified protease generating activity B and employed inhibitors. (The X. kozodoii Morocco and Intermedium strains were used in these experiments because they expressed the most stable activities A and C.) We found that activity B as well as activity band 1 on gelatin substrate and activity band 2 on casein substrate were from the same enzyme (data not shown). We called this enzyme Xenorhabdus protease B. The inhibitor sensitivities of all the zymographic activities were similar: they were inhibited by chelators (EDTA and 1,10-phenanthroline), but not with serine protease inhibitor (PMSF [data not shown]). Therefore, the question of whether the protease generating band 1 (on casein substrate) and any of those generating band A or C are the same cannot be answered with certainty. However, because no correlation can be observed between them in their occurrence (Table 2), we suppose that they are different enzymes and that there are only technical reasons why activities A and C cannot be detected after native PAGE and activity 1 cannot be detected (on casein) after SDS-PAGE. It is important to note that, interestingly, EDTA did not inhibit protease B activity completely. Therefore, we also performed zymography with purified protease B using a reduced amount of the enzyme (∼5.0 pmol), but we obtained the same result (Fig. 1C). Thus, altogether at least four proteolytic activities could be distinguished with zymographies in the culture supernatant of Xenorhabdus strains, a number which is higher than that for Photorhabdus strains (15).
FIG. 1.
Zymographic detection of Xenorhabdus proteases in culture. (A and B) Enzyme activities produced by X. kozodoii Morocco and Intermedium strains were monitored with zymography coupled to SDS-PAGE (A) and zymography coupled to native PAGE (B). (A) The activities were tested with both casein (a) and gelatin (b) as the substrate. The positions of enzyme activity bands A, B, and C are shown to the right of the gels. (B) Protease production of X. kozodoii Morocco strain is shown using casein as the substrate. The positions of enzyme activity bands 1 and 2 are shown to the left of the gel. (C) Effects of inhibitors of catalytic serine (PMSF) and metal ion (EDTA and 1,10-phenanthroline) are shown on the activity of ∼5.0 pmol of protease B (arrow), purified from X. kozodoii Morocco strain. The gel slices are as follows: a, enzyme not treated (control); b, PMSF-treated enzyme; c, EDTA-treated enzyme; d, 1,10-phenanthroline-treated enzyme. The concentration of inhibitors during both sample treatment and gel slice incubation was 5.0 mM.
The strains tested exhibited large differences in both the number and intensity of protease production (Table 2). Strains AN6/1, RIO-HU/1, and DSM 16337 were the weakest, while strains Morocco, Anomali AZ, Kraussei, and Intermedium were the strongest in this regard, indicating that this property was strain and not species specific. The comparisons of strains also showed that the production of protease B (activity B) was the most intensive and the most common (being produced by all the strains) and that the molecular properties of the enzymes, which produced activity bands A, B, and C as well as 1 and 2, were different (reflected by their relative mobility). The appearance of zymographic activities allowed an estimation of the dynamics in the secretion of the corresponding enzymes, except for the heterogeneous activity band 1 (on casein [see above]). According to this, protease B was produced the earliest (detected even in the early logarithmic phase, i.e., in 4-h cultures), while all the others were produced from the late logarithmic or stationary phase. Protease B was also different from the other enzymes in that its secretion declined in the stationary phase.
Screening for proteolytic activities with chromogenic substrates.
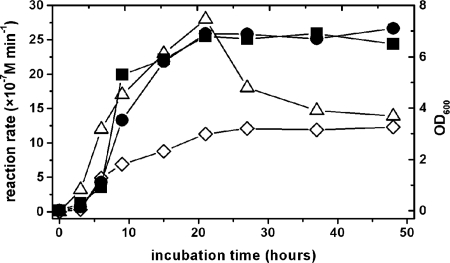
For a quantitative detection of proteolytic activities, we used several oligopeptide substrates, the cleavage of which can also be informative to the type of the enzyme. For example, Succ-AAPX-AMC (where X is Phe and Lys) and Succ-AAPF-SBzl are used to measure the activity of serine proteinases, while Fua-LGPA is readily hydrolyzed by collagen peptidases. Every strain showed activity on Succ-AAPF-SBzl (Table 2), but we could not find activity with the Succ-AAPX-AMC substrate. This indicates that Succ-AAPF-SBzl is a suboptimal substrate; therefore, when the easily hydrolyzable (thiol)ester bond is replaced with the more stable amide and/or the P1′ position is occupied by aminomethyl-coumarin and not thiobenzyl group in the substrate, the enzyme is unable to cleave it. We also employed two other substrates, Fua-LGPA and Fua-ALVY. We had previously found the former to be hydrolyzed by oligopeptidase A of Photorhabdus (16), while the latter was cleaved by an as yet uncharacterized enzyme in Photorhabdus luminescens Brecon strain (15). Essentially every Xenorhabdus strain showed activity on these substrates (only Fua-LGPA was not cleaved by strain AN6/1 [Table 2]). The Fua-LGPA-ase activity was supposedly from the Xenorhabdus orthologue of oligopeptidase A. The dynamics of production were different for the three activities observed with the chromogenic substrates. While the Fua-LGPA-ase activity exhibited a steady increase from its first appearance in the logarithmic growth phase (similarly to that in Photorhabdus strains), the other two showed a peak, the Fua-ALVY-ase activity in the late logarithmic or early stationary phase (Fig. 2) and the Succ-AAPF-SBzl-ase activity at the beginning of the logarithmic phase (data not shown). When cell lysates were compared to the culture supernatants, the Fua-LGPA-ase and Succ-AAPF-SBzl-ase activities were an order of magnitude higher in the lysates (not shown). These results indicated intracellular enzymes that leaked out to the medium on cell death. This conclusion was supported for the Fua-LGPA-ase activity by the dynamics of its appearance in the medium, which was also the same for the Fua-LGPA-hydrolyzing intracellular enzyme, oligopeptidase A, of Photorhabdus. However, leaking cannot explain either the early appearance or the peak in the activity of the Succ-AAPF-SBzl-hydrolyzing enzyme. Instead, the higher activity in the cellular faction might be explained by a periplasmic localization of the Succ-AAPF-SBzl-ase enzyme or by an enzyme(s) in the cytosol that can hydrolyze this substrate. (Succ-AAPF-Bzl is an easy-to-hydrolyze, ester-type substrate.)
FIG. 2.
Culture growth and detection of proteolytic activities with chromogenic substrates. Growth of the culture growth was monitored by measuring the optical density at 600 nm (OD600) (filled symbols • and ▪ for X. kozodoii Morocco and Intermedium strains, respectively). The data shown are representative of four measurements for both cultures. Enzyme activities were measured with Fua-LGPA and Fua-ALVY substrates (open symbols ⋄ and ▵, respectively). The data are representative of two measurements on both substrates in the culture of X. kozodoii Morocco strain. The activities were measured using 50 μl of culture supernatant at a final concentration of substrate of 50 μM.
Since the distribution among strains of the activities on chromogenic substrates was different from both each other and the zymographic activities, we suppose that they were produced by different enzymes. To investigate this question further, we tested the inhibitor sensitivity of these activities from X. kozodoii Morocco and Intermedium BIO strains (Table 3). The activity on Succ-AAPF-SBzl was dependent on active serine, since it was inhibited by PMSF, while because they were inhibited by EDTA and 1,10-phenanthroline, the Fua-LGPA-ase and Fua-ALVY-ase activities proved metal ion dependent. These, at the same time, differed from each other in their sensitivity to thiol reagents (Cys, thimerosal, and dithiothreitol [DTT]). The results of these experiments could not completely exclude the possibility that either the Fua-LGPA-ase or Fua-ALVY-ase activity is from protease B, but the purified enzyme did not cleave either of these substrates (Massaoud et al., submitted). Thus, by using chromogenic substrates, the number of secreted proteases detected in Xenorhabdus increased to six (protease B, as well as activities A, C, 1 [on casein substrate], and those on Fua-ALVY and Succ-AAPF-SBzl substrates). This number is three times more than what has been found for Photorhabdus strains using the same detection methods.
TABLE 3.
Effects of inhibitors on the observed protease activities
| Inhibitor | Remaining activitya on: |
||
|---|---|---|---|
| Succ-AAPF-Sbzl | Fua-LGPA | Fua-ALVY | |
| EDTA | 99.1 ± 11.2 | 1.3 ± 2.2 | 0.1 ± 0.1 |
| 1,10-Phenanthroline | 105.3 ± 15.1 | 0.1 ± 1.5 | 0.9 ± 0.5 |
| PMSF | 14.8 ± 7.5 | 86.3 ± 5.4 | 16.6 ± 6.2 |
| Cys | NT | 29.7 ± 11.0 | 0.2 ± 0.3 |
| Thimerosal | NT | 97.0 ± 5.2 | 56.3 ± 8.8 |
| DTT | NT | 68.5 ± 7.8 | 0.5 ± 0.2 |
Activity expressed as a percentage of the activity of the control. The results are from measurements on two different culture supernatants, and the data shown are the means ± standard errors of the means of at least three measurements. The control (100%) values for the Succ-AAPF-SBzl-ase, Fua-LGPA-ase, and Fua-ALVY-ase activities are (2.90 ± 0.21) × 10−7 M/min, (7.6 ± 0.5) × 10−7 M/min, and (3.4 ± 0.42) × 10−6 M/min, respectively. NT, not tested.
Protease B in the hemolymph.
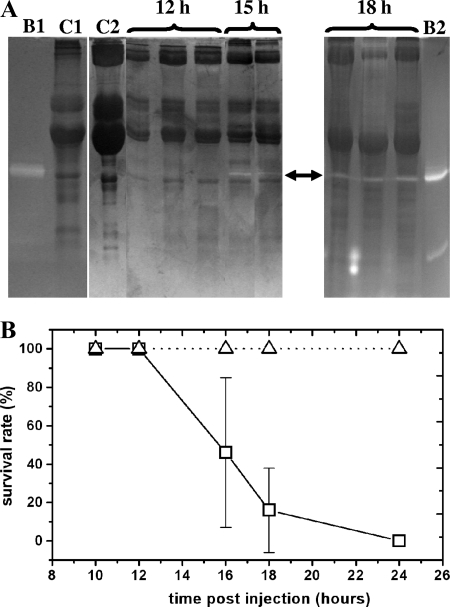
On the basis of our previous observations on Photorhabdus PrtA peptidase, we hypothesized that some of the secreted proteases of Xenorhabdus can also take part in the establishment of infection. The best candidate enzymes for such a role are those that are secreted early and by every strain. Of the proteases we have found in Xenorhabdus, protease B, the Fua-ALVY-ase enzyme, and the Succ-AAPF-SBzl-cleaving enzyme met these criteria. Consequently, we investigated the dynamics of appearance of these activities in the hemolymph of Xenorhabdus-infected G. mellonella larvae. We could detect protease B in the hemolymph from the 15th hour of infection when the larvae were still alive but moved only when they were prodded (Fig. 3A). According to the mortality assay (Fig. 3B), this is relatively late during the molecular interactions between the pathogen and the host, i.e., in the late pathogenicity phase of infection. (We note that samples from dead insects were not suitable for analysis with gel electrophoresis because of the high content of insoluble material in the dead insects that could not be sedimented even with longer and faster centrifugation than what was used for pelleting cellular fraction.) At the same time, we did not observe an increase in the activity on the Succ-AAPF-SBzl and Fua-ALVY substrates, which was also rather high in the hemolymph of PBS-injected insects (data not shown).
FIG. 3.
Detection of protease B in insects and insect mortality on Xenorhabdus infection. (A) SDS-PAGE-coupled zymographic detection with casein substrate of protease B production during G. mellonella infection (the detected activity is indicated by the arrow). The numbers above the lanes indicate the time the sample was taken in hours postinfection. Purified protease B was also run in the gels as a standard (lanes B1 and B2, ∼10.0 and ∼20.0 pmol of enzyme loaded, respectively). Lane C1, control hemolymph from PBS-injected insects; lane C2, control hemolymph from naïve insects. (B) Survival rate of G. mellonella larvae injected with ∼100 cells of X. kozodoii Morocco strain (□) or with PBS (▵). Data from four experiments are averaged. For the conditions of infection and sample preparation, see Materials and Methods.
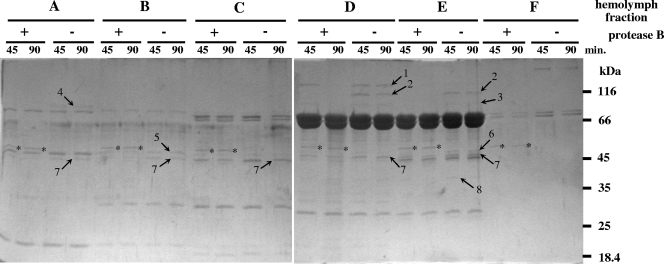
To obtain information about the possible role(s) of a secreted protease, identification of its target proteins is needed. In order to find such proteins, we exposed six separated fractions of Manduca sexta hemolymph (see Materials and Methods) to digestion with purified protease B at a ratio of 1 to 6 μg hemolymph protein to ∼0.06 μg (1.0 pmol) of protease B in vitro. As shown in Fig. 4, eight proteins were cleaved which, at the same time, were not digested by the nonspecific pancreatic serine proteinases, trypsin and chymotrypsin (data not shown). However, all eight proteins were cleaved when the hemolymph fractions were exposed to digestion by PrtA peptidase of Photorhabdus (data not shown). This made identification of many of protease B targets or PrtA peptidase possible, because in an earlier study we had already established the identities of substrate proteins of PrtA peptidase in M. sexta hemolymph with N-terminal sequencing and database search (6). These are band 8, serine protease homolog 3, band 7, Manduca serpin 1, band 5 (or 6), hemocyte aggregation inhibitor protein, which are all immune-related proteins of M. sexta. In contrast, when tested on several other native proteins (albumin, fibrinogen, collagen types I and IV), protease B (like PrtA peptidase [17]) did not show activity (not shown). Such a target protein profile is more suggestive of an enzyme that functions to enhance virulence, rather than an enzyme that is involved in the bioconversion of the insect cadaver. The fact that the production of protease B declines from about 24 h postinfection (shortly before the death of insects) also supports this notion.
FIG. 4.
Cleavage with protease B of proteins in the hemolymph of M. sexta. The in vitro digestion of hemolymph fractions with purified protease B is shown. The hemolymph fraction (fractions A to F) (see Materials and Methods) and whether the hemolymph fraction was incubated in the presence (+) or absence (−) of protease B are indicated above the gels. The incubation times (in minutes) are shown immediately above the gels. The bands from protease B are indicated by asterisks on the gels.
For its apparent functional and molecular similarity to PrtA peptidase, we purified protease B (Massaoud et al., submitted) and determined its N-terminal sequence to establish its relationship with PrtA peptidase. The interrogation of the MEROPS database of proteases with the N-terminal sequence resulted in two hits with Xenorhabdus metalloproteases (Fig. 5). Although the match of these two hits was not perfect, it was by far the best match if the comparison included all 15 amino acids of protease B. The sequence similarity is in accordance also with the metalloprotease character of protease B (not seen in the N-terminal sequence but in other sequence regions) and with the similarities between protease B and Photorhabdus PrtA peptidase (in molar mass and target proteins). Both PrtA peptidase and the X. nematophila enzymes are metzincins and belong to the M10 subfamily of Zn-metalloproteases in the M10B group. (These are the serralysins, the bacterial group of interstitial collagenases [18].)
FIG. 5.
Comparison of the N-terminal sequence of protease B to several N-terminal sequences in the serralysin (M10B) subfamily of metalloproteases. Amino acids that are identical to the amino acid in protease B are shown by capital letters. UniProt identifiers are given in parentheses.
DISCUSSION
With five detection methods, we have found six secreted proteolytic activities and one intracellular proteolytic activity in the screening of 17 Xenorhabdus strains that included two primary-secondary form variant pairs. Although exhibiting differences in the number and intensity, three of the secreted activities were present in every strain. Similar to the results of a study by Boemare and Akhurst (2) on phenotypic variant pairs of 18 Xenorhabdus strains with bacteriological plate assays employing various protein substrates and to our observations of Photorhabdus strains (15), we did not find characteristic differences between the primary and secondary phenotypic variant pairs of Xenorhabdus strains in their protease production such that the latter ones would be less active. The small number of primary-secondary pairs in our study does not allow us to draw further conclusions regarding a form variant pair-specific difference in either the intensity or number of activity types. A comparison with data from other groups (2, 23) regarding proteolytic activities is further limited by, e.g., differences in the substrates used, which determines the sensitivity of detection. For example, casein is less sensitive to digestion than gelatin (a denatured protein) is, while the amino acid sequence in gelatin provides a smaller variety of cleavage sites. A further complication might arise from inactivation of enzymes if during preparation the gel electrophoresis samples are precipitated or exposed to heat.
The identical methods of detection permit a comparison, which has not been possible before, of the secretion and usage of proteolytic enzymes between the two closely related bacterial genera, Xenorhabdus and Photorhabdus, which also have comparable pathogen strategies and symbiotic roles. As for similarities, the Fua-LGPA-ase activity of Xenorhabdus appears to be the same as oligopeptidase A of Photorhabdus. Also, protease B of Xenorhabdus appears to be similar to PrtA peptidase (PrtA) of Photorhabdus, despite their different dynamics of production. The latter two are metalloenzymes of the same molar mass (and probably of the same type [see below]), which are produced from a relatively early stage of infection. They have similar preference of native substrate proteins as well, suggesting a similar function, which might be a role in immune suppression (6). Furthermore, there is a lack in both pathogens of those secreted activities that could be detected with substrates which are sensitive for trypsin- and chymotrypsin-like serine proteinase activities (enzymes that can cleave the Succ-AAPX-AMC substrates). On the other hand, there are also numerous differences between the two genera, which is surprising, given their very close relationship. For instance, unlike Photorhabdus, Xenorhabdus strains did not secrete thermolysin-like enzyme, while Photorhabdus does not secrete such activities as activities A and C of several Xenorhabdus strains. Similarly, the activities on Fua-ALVY and on Succ-AAPF-SBzl substrates are common in Xenorhabdus but are missing in Photorhabdus, making protease secretion by the former bacterium more abundant in the number of secreted protease types (according to the detection methods we used). Even if we consider only those four activities that are secreted by almost all of the tested Xenorhabdus strains, a larger set of potential proteolytic virulence factors is available for Xenorhabdus than for Photorhabdus. The interesting question of whether this difference influences the mechanism of infection or toxicity (e.g., the speed of insect killing between the two bacterial genera) needs further investigation.
Although we did not compare the pathogenicity of our strains, other studies showed that pathogenicity does not correlate with phenotype variation of Xenorhabdus (23) and Photorhabdus (15) or with protease production of Photorhabdus (15). Therefore, we would not expect a correlation between the pathogenicity of Xenorhabdus strains and their protease production either. At the same time, this does not mean that a secreted protease cannot be a virulence factor. Pathogenicity is a complex function of the strategies followed by the host and the pathogen, both trying to maximize their survival in the interaction between them by numerous factors. On the part of the pathogen, this is best seen in “overkilling,” attempted by cocktails of compounds with toxic or immunosuppressive effects. (The significance of proteases in the latter, as in the case of protease B and PrtA, is conceivable.) However, in such a multicomponent “arsenal of arms,” it is less likely that the role of a participant is indispensable (a typical property of complex systems). Indeed, the pathogenicity of a PrtA knockout mutant of Photorhabdus luminescens subsp. laumondii TT01 strain remained the same as that of the wild type (J. Marokházi, unpublished result).
The dynamics of production of the detected activities were different if we compared them in culture (e.g., occurring from the early logarithmic to stationary phase) or if we compared the occurrence in culture and during infection. Unfortunately, for technical reasons (see Results), we could make the latter comparison for protease B because of the three earliest secreted activities, Fua-ALVY-ase, Succ-AAPF-SBzl-ase, and protease B (which might be the most interesting for potential role in the early, pathogenicity phase of infection), we could investigate the production of only protease B during infection. Our observations with zymography of hemolymph samples suggest that this activity is produced later than 12 but earlier than 15 h postinfection, which is significantly later than the occurrence in culture (4 to 7 h, or early to mid logarithmic phase). According to mortality curves, it appears that when we could detect protease B activity in the hemolymph with certainty, it might be about 3 h before insect death. If we compare this occurrence of protease B to that of Photorhabdus PrtA peptidase, the former is relatively later taking into account also the dynamics of infection: on injection of the same number of bacterial cells, G. mellonella larvae died 28 to 34 h after Photorhabdus infection with the first detection of PrtA peptidase occurring 18 to 21 h after infection (15). At the same time, it cannot be ruled out that, similarly to PrtA peptidase (17), some protease B activity, which might be detectable only with a sensitive and specific substrate, is present in the tissues or in the hemolymph even at 7 to 9 h after infection.
In an earlier study, Caldas et al. detected two proteases in X. nematophila culture that they distinguished as protease I and protease II (5). We suppose that protease I and protease A (activity A) are the same because of their similar molar masses (both are ∼90 kDa). For the same reason, protease II and protease B (molar masses of 60 and 55 kDa, respectively) might be the same enzyme. The identity of the latter two is also supported by the fact that the production of protease II, like that of protease B, declines on longer incubation. However, the relationship of protease B with protease II of Xenorhabdus and Photorhabdus PrtA peptidase requires further investigation, because protease II, but not protease B, was inhibited with serine proteinase inhibitors (5) and because protease B could not be completely inhibited with EDTA, an inhibitor of metalloenzymes, including PrtA peptidase. We try to answer this question with a detailed enzymological characterization of protease B and with a comparison of the available amino acid sequences and tertiary structures (Massaoud et al., submitted).
Acknowledgments
We acknowledge András Patthy for N-terminal sequencing.
Mustafa K. Massaoud was supported by a Ph.D. fellowship by the Libyan Ministry of Education.
Footnotes
Published ahead of print on 27 August 2010.
REFERENCES
- 1.Akhurst, R. J. 1980. Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect parasitic nematodes. J. Invertebr. Pathol. 62:68-72. [Google Scholar]
- 2.Boemare, N. E., and R. J. Akhurst. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus ssp. (Enterobacteriaceae). J. Gen. Microbiol. 134:751-761. [DOI] [PubMed] [Google Scholar]
- 3.Boemare, N. E., R. J. Akhurst, and R. G. Mourant. 1993. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 43:249-255. [Google Scholar]
- 4.Boemare, N. 2002. Biology, taxonomy and systematics of Photorhabdus and Xenorhabdus, p. 35-56. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, Oxon, United Kingdom.
- 5.Caldas, C., A. Cherqui, A. Pereira, and N. Simoes. 2002. Purification and characterization of an extracellular protease from Xenorhabdus nematophila involved in insect immunosuppression. Appl. Environ. Microbiol. 68:1297-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felföldi, G., J. Marokházi, M. Képiró, and I. Venekei. 2009. Identification of natural target proteins indicates functions of a serralysin-type protease, PrtA, in anti-immune mechanisms. Appl. Environ. Microbiol. 75:3120-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forst, S., and K. Nealson. 1996. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 60:21-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 9.Forst, S., and D. Clarke. 2002. Bacteria-nematode symbiosis, p. 57-77. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, Oxon, United Kingdom.
- 10.Frazier, W. C. 1926. A method for the detection of changes in gelatin due to bacteria. J. Infect. Dis. 39:302-306. [Google Scholar]
- 11.Furgani, G., E. Böszörményi, A. Fodor, A. M. Fodor, S. Forst, J. Hogan, Z. Katona, M. G. Klein, E. Stackebrandt, A. Szentirmai, F. Sztaricskai, and S. Wolf. 2008. Xenorhabdus antibiotics: a comparative analysis and potential utility for controlling mastitis caused by bacteria. J. Appl. Microbiol. 104:745-758. [DOI] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Gráf, L., A. Jancsó, L. Szilágyi, G. Hegyi, K. Pintér, G. Náray-Szabó, J. Hepp, K. Medzihradszky, and W. J. Rutter. 1988. Electrostatic complementarity within the substrate-binding pocket of trypsin. Proc. Natl. Acad. Sci. U. S. A. 85:4961-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lengyel, K., E. Lang, A. Fodor, E. Szállás, P. Schumann, and E. Stackebrandt. 2005. Description of four novel species of Xenorhabdus, family Enterobacteriaceae: Xenorhabdus budapestensis sp. nov., Xenorhabdus ehlersii sp. nov., Xenorhabdus innexi sp. nov., and Xenorhabdus szentirmaii sp. nov. Syst. Appl. Microbiol. 28:115-122. [DOI] [PubMed] [Google Scholar]
- 15.Marokházi, J., K. Lengyel, S. Pekár, G. Felföldi, A. Patthy, L. Gráf, A. Fodor, and I. Venekei. 2004. Comparison of proteolytic activities produced by entomopathogenic Photorhabdus bacteria: strain- and phase-dependent heterogeneity in composition and activity of four enzymes. Appl. Environ. Microbiol. 70:7311-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marokházi, J., G. Kóczán, F. Hudecz, L. Gráf, A. Fodor, and I. Venekei. 2004. Enzymatic characterization with progress curve analysis of a collagen peptidase from an entomopathogenic bacterium, Photorhabdus luminescens. Biochem. J. 379:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marokházi, J., N. Mihala, F. Hudecz, A. Fodor, L. Gráf, and I. Venekei. 2007. Cleavage site analysis of a serralysin-like protease, PrtA, from an insect pathogen Photorhabdus luminescens and development of a highly sensitive and specific substrate. FEBS J. 274:1946-1956. [DOI] [PubMed] [Google Scholar]
- 18.Rawlings, N. D., D. P. Tolle, and A. J. Barrett. 2000. MEROPS: the peptidase database. Nucleic Acids Res. 28:323-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Stackebrandt, E. 1999. The hidden prokaryotic diversity, p. 13-27. In N. Boemare, P. Richardson, and F. Coudert (ed.), Entomopathogenic nematodes—taxonomy, phylogeny and gnotobiological studies of entomopathogenic nematode complexes. EUR 18832 EN, COST 819. Office for Official Publications of the EC, Luxembourg, Luxembourg.
- 19.Szállás, E., C. Koch, A. Fodor, J. Burghardt, O. Buss, A. Szentirmai, K. H. Nealson, and E. Stackebrandt. 1997. Phylogenetic evidence for the taxonomic heterogeneity of Photorhabdus luminescens. Int. J. Syst. Bacteriol. 47:402-407. [DOI] [PubMed] [Google Scholar]
- 20.Szállás, E., R. Pukall, H. Pamjav, G. Kovács, Z. Buzás, A. Fodor, and E. Stackebrandt. 2001. Passengers who missed the train: comparative sequence analysis, PhastSystem PAGE PCR-RFLP and automated RiboPrint phenotypes of Photorhabdus strains, p. 36-53. In C. T. Griffin, A. M. Burnell, M. J. Downes, and R. Mulder (ed.), Developments in entomopathogenic nematode/bacterial research. European Commission Publications, Luxembourg, Luxembourg.
- 21.Tailliez, P., S. Pagès, N. Ginibre, and N. Boemare. 2006. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int. J. Syst. Evol. Microbiol. 56:2805-2818. [DOI] [PubMed] [Google Scholar]
- 22.Van Wart, H. E., and D. R. Steinbrink. 1981. A continuous spectrophotometric assay for Clostridium histolyticum collagenases. Anal. Biochem. 113:356-365. [DOI] [PubMed] [Google Scholar]
- 23.Völgyi, A., A. Fodor, A. Szentirmai, and S. Forst. 1998. Phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 64:1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volgyi, A., A. Fodor, and S. Forst. 2000. Inactivation of a novel gene produces a phenotypic variant cell and affects the symbiotic behavior of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 66:1622-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamanaka, S., A. Hagiwara, Y. Nishimura, H. Tanabe, and N. Ishibashi. 1992. Biochemical and physiological characteristics of Xenorhabdus species, symbiotically associated with entomopathogenic nematodes including Steinernema kushidai and their pathogenicity against Spodoptera litura (Lepidoptera: Noctuidae). Arch. Microbiol. 158:387-393. [Google Scholar]