Abstract
In this study, we analyzed the influence of phosphate (Pi) limitation on the metabolism of Corynebacterium glutamicum. Metabolite analysis by gas chromatography-time-of-flight (GC-TOF) mass spectrometry of cells cultivated in glucose minimal medium revealed a greatly increased maltose level under Pi limitation. As maltose formation could be linked to glycogen metabolism, the cellular glycogen content was determined. Unlike in cells grown under Pi excess, the glycogen level in Pi-limited cells remained high in the stationary phase. Surprisingly, even acetate-grown cells, which do not form glycogen under Pi excess, did so under Pi limitation and also retained it in stationary phase. Expression of pgm and glgC, encoding the first two enzymes of glycogen synthesis, phosphoglucomutase and ADP-glucose pyrophosphorylase, was found to be increased 6- and 3-fold under Pi limitation, respectively. Increased glycogen synthesis together with a decreased glycogen degradation might be responsible for the altered glycogen metabolism. Independent from these experimental results, flux balance analysis suggested that an increased carbon flux to glycogen is a solution for C. glutamicum to adapt carbon metabolism to limited Pi concentrations.
Phosphorus is an essential nutrient for all cells and is required for, e.g., the biosynthesis of nucleotides, NAD(P)H, DNA, and RNA but also for the regulation of protein activity by phosphorylation of histidine, aspartate, serine, threonine, or tyrosine residues. A common phosphorus source is inorganic phosphate (Pi), and cells have developed mechanisms for the acquisition, assimilation, and storage of Pi. When Pi becomes limiting, many bacteria induce the synthesis of proteins that enable them to capture the residual Pi resources more efficiently and to make alternative phosphorus sources accessible. The corresponding genes are collectively named Pi starvation-inducible genes, or psi genes. The Pi starvation response, and in particular its regulation, has been most carefully studied in Escherichia coli (45) and Bacillus subtilis (14).
We recently started to characterize the Pi starvation response in Corynebacterium glutamicum, a Gram-positive soil bacterium used industrially for the production of more than two millions tons of amino acids per year, mainly l-glutamate and l-lysine (12). An overview of the biology, genetics, physiology, and application of C. glutamicum can be found in two recent monographs (3, 6). Phosphorus constitutes 1.5% to 2.1% of the cell dry weight of C. glutamicum (24), part of which can be present as polyphosphate (22, 29). Several of the enzymes involved in polyphosphate metabolism have been characterized recently, such as a class II polyphosphate kinase (28), the exopolyphosphatases Ppx1 and Ppx2 (26), a polyphosphate/ATP-dependent glucokinase (25), and a polyphosphate/ATP-dependent NAD+ kinase (27). The Pi starvation stimulon of C. glutamicum was determined using whole-genome DNA microarrays (15). Comparison of the mRNA profiles before and at different times after a shift from Pi excess to Pi starvation led to the identification of a group of genes that are presumably required to cope with limited Pi supply. This group includes the following: the pstSCAB operon, encoding an ABC transporter for high-affinity Pi uptake; the ugpAEBC operon, encoding an ABC transporter for uptake of glycerol 3-phosphate; glpQ1, encoding a glycerophosphoryl diester phosphodiesterase; ushA, encoding a secreted enzyme with UDP-sugar hydrolase and 5′-nucleotidase activities (33); nucH, encoding a putative secreted nuclease which possibly plays a role in liberating Pi from extracellular nucleic acids; phoC (NCgl2959/cg3393), which may encode a cell wall-associated phosphatase (46); phoH1, encoding an ATPase of unknown function; and the pctABCD operon, encoding an ABC transport system which might be involved in the uptake of a yet-unknown phosphorus-containing compound (15). C. glutamicum lacks homologs of genes for phosphonate degradation, as well as the capability to utilize phosphonates as P sources (15).
In most bacteria analyzed in this respect, the Pi starvation response is controlled by two-component signal transduction systems, e.g., the PhoBR system in E. coli (13) and the PhoPR system in B. subtilis (14). Our previous studies revealed that in C. glutamicum, a two-component system composed of the sensor kinase PhoS and the response regulator PhoR is involved in the activation of phosphate starvation-inducible genes (21). Studies with purified proteins showed that phosphorylation by PhoS increased the DNA-binding affinity of PhoR, which bound to many of the Pi starvation-inducible genes, but with different affinities (34).
The study reported here was initiated by the question how the metabolism of C. glutamicum responds to Pi limitation. Our results reveal a link between Pi limitation and glycogen metabolism, which was also used for metabolic simulations based on a genome-wide metabolic model.
MATERIALS AND METHODS
Strains and cultivation.
The strains and plasmids used in this study are listed in Table 1. The wild-type C. glutamicum strain ATCC 13032, its glgC disruption mutant (36), and its ΔsugR mutant (9) were precultivated aerobically at 30°C in baffled 500-ml shake flasks on a rotary shaker at 120 rpm using CGIII complex medium (10 g peptone, 10 g yeast extract, and 25 g NaCl per liter) supplemented with 222 mM glucose. After cells had been washed with 0.9% (wt/vol) NaCl, they were transferred to defined CGXII minimal medium (18) supplemented with protocatechuic acid (30 mg/liter) as an iron chelator and either 222 mM glucose or 300 mM potassium acetate as a carbon source. For the analysis of the response of C. glutamicum to Pi limitation at the metabolite level, the cells were precultivated twice in CGXII glucose medium with 0.13 mM Pi, and after being washed, they were inoculated into CGXII glucose medium under different Pi conditions (13 mM, 0.65 mM, 0.26 mM, and 0.13 mM). Pi was added as KH2PO4 and K2HPO4. Samples for metabolite analysis were taken after 8, 12, and 24 h cultivation. For in vivo 13C labeling of the metabolites, cells were grown in CGXII medium containing 222 mM uniformly 13C-labeled glucose (Cambridge Isotope Laboratory, Andover, MA) as the sole carbon source under the conditions described above, including two precultivations with 13C-labeled glucose.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−thi-1 endA1 hsdR17(r− m−) supE44 ΔlacU169 (φ80lacZΔM15) recA1 gyrA96 relA1 | Invitrogen |
| C. glutamicum ATCC 13032 | Biotin-auxotrophic wild-type strain | 19 |
| C. glutamicum ΔsugR | ATCC 13032 derivative with an in-frame deletion of sugR | 9 |
| C. glutamicum IMC | ATCC 13032 derivative with a disruption of glgC | 36 |
| Plasmids | ||
| pET2 | Kanr; promoter-probe vector for C. glutamicum | 44 |
| pET2-pgm | Kanr; pET2 with a 423-bp fragment covering the pgm promoter (−413 to +10 with respect to the proposed translational start site) | This study |
| pET2-glgC | Kanr; pET2 with a 406-bp fragment covering the glgC promoter (−402 to +4 with respect to the proposed translational start site) | This study |
Extraction of metabolites and sample preparation for metabolite profiling.
One-milliliter samples of triplicate cultures with known optical densities at 600 nm (OD600) were added to 2-ml Eppendorf tubes containing 500 mg of silicon oil (δ = 1.05 g/cm3) and 300 μl 20% (vol/vol) perchloric acid (HClO4, δ = 1.18 g/cm3). The tubes were centrifuged immediately at 13,000 rpm for 30 s in order to separate the cells from the culture supernatant and to inactivate metabolism by the acid treatment. After careful removal of the supernatant, the samples were mixed and neutralized with 185 μl of 6 N potassium hydroxide. The pH was controlled by the indicator bromthymol blue, which is green at pH 7. Aliquots of the polar phase, corresponding to 5.0 mg cells (dry weight) [an OD600 of 1 corresponds to 0.25 mg cells (dry weight) per ml (16)] were lyophilized for at least 2 days. Subsequently, the dried cell extracts were treated for 90 min at 35°C with 50 μl methoxyamine hydrochloride in pyridine (20 mg/ml) and subsequently trimethylsilated with 80 μl N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) for 4 h at 35°C to derivatize the metabolites for gas chromatography-time-of-flight (GC-TOF) mass spectrometry (MS) analysis.
Metabolite pattern analysis.
Metabolite pattern analysis was used as an initial approach to determine whether the metabolite patterns of cells differ depending on Pi availability. In a first series of experiments, GC-TOF MS data sets were obtained from cells cultured for 24 h in CGXII glucose medium with different Pi concentrations (13 mM, 0.65 mM, 0.25 mM, and 0.13 mM Pi). In a second series, GC-TOF MS data sets were obtained from three time points (8 h, 12 h, and 24 h) of cultures grown in CGXII glucose medium under either Pi excess (13 mM) or Pi starvation (0.13 mM). To perform metabolite pattern analysis, for each sample a data set containing 2,517 mass fragments (whose chemical identities were unknown) differing in either mass or retention time and identified in all analyzed samples by MarkerLynx software was exported to SIMCA-P+ software (Umetrics AB, Umeå, Sweden). These datasets were analyzed by partial least-squares discriminant analysis (PLS-DA) (47).
GC-(EI/CI)-TOF MS.
The derivatized metabolite samples prepared as described above were analyzed by GC-TOF MS using both the electron ionization (EI+) mode and chemical ionization (CI+) mode. The GC-TOF MS system was composed of an Agilent gas chromatograph 6890N (Agilent, Santa Clara, CA) equipped with a Gerstel MPS2 multipurpose sampler (Gerstel, Mülheim, Germany) and a GCT Premier benchtop orthogonal acceleration time-of-flight mass spectrometer (Waters, Milford, MA). The system was operated using MassLynx software (version 4.1; Waters). GC was performed using a 30-m by 0.25-μm DB-5MS column (J&W Scientific, Folsom, CA) with a constant flow rate of 1 ml/min of helium as the carrier gas. After the needle of the injector syringe had been washed with hexane and methanol, 1 μl of sample was injected with a split ratio of 2:1 at 280°C to a glass liner (4-mm inner diameter) filled with glass wool. For GC separation, the oven temperature stayed initially for 2 min at 85°C and then increased to 320°C by 15°C/min, where it was held for 5 min. Transfer of the samples from the GC to the mass spectrometer was performed at 150°C.
Evaporated chemicals were ionized by either the EI or CI method. For EI+, the ionization energy was tuned at 70 eV according to the operation manual using heptacosafluorotributylamine [(C4F9)3N] as an internal reference. For positive chemical ionization (CI+), isobutene (0.7 bar, 10−5 mbar in the source) as a reagent gas generated ionized [MH]+, [M + C4H9]+, and [M + C3H3]+ quasi-molecules at an electron energy of 50 eV. Tuning for the CI+ mode was performed according to the operation manual using 2,4,6-tri(trifluoromethyl)-1,3,5-triazine as an internal reference. Mass ions were detected with the scan time set to 0.9 s and the interscan set to 0.1 s in the centroid mode using lock mass at 218.9856 m/z. Mass fragment patterns were analyzed to identify metabolites using NIST MS search with several public libraries and a homemade library.
Metabolite identification by GC-(EI/CI)-TOF MS based on the use of naturally and uniformly 13C-labeled metabolites.
GC-MS has been widely used to analyze volatile chemicals and derivatized chemicals because of high sensitivities and the availability of standard mass fragment libraries. In general, the identification process in GC-MS depends on the matching score of mass fragment patterns between analytes and standards with the same retention time or the index number only when mass fragment libraries of standards are available. However, a number of peaks with low matching scores (below 700) show ambiguous metabolite identification or remain unknown because of the incompleteness of biological metabolite databases and the possibility of undesired reactions by using highly reactive derivatization chemicals. Therefore, in this study an additional procedure was used to identify “real” metabolites among possible chemicals from derivatized cell extracts. The method is based on the comparison of uniformly 13C-labeled metabolites with naturally labeled metabolites, which were measured separately. By combining all information obtained from the extracts of the cultures grown with either unlabeled or 13C-labeled glucose using the EI/CI ionization methods, metabolites were identified by several criteria as outlined in Fig. S1 in the supplemental material. In the case of mass fragments that still remained unknown after this protocol, identification was attempted based on the exact mass measurement using a Matlab script, which searches possible chemical compositions online in publicly available chemical databases (Pubchem and KEGG compound DB). The details of this procedure are described in the supplemental material.
Analysis of phosphorus-containing metabolites using 31P in vivo NMR spectroscopy.
31P nuclear magnetic resonance (NMR) analysis was performed essentially as described previously (28). Cultures were harvested in the mid-exponential growth phase (8 h) and in the stationary phase (24 h). One gram (wet weight) of cell pellet was suspended in 4 ml absolute ethanol, mixed for 1 min, and centrifuged for 10 min at 4,400 × g and 4°C. The supernatant was discarded, and the pellet was resuspended in a mixture of 1.1 ml fresh bi-distilled water, 0.3 ml 1 M EDTA (pH 8.2), and 0.6 ml D2O. Seven hundred microliters of a 2-ml cell suspension was transferred into a 5-mm NMR tube and analyzed. During preparation, all samples were kept on ice or were kept frozen until further use.
The 31P-NMR spectra were measured at 5°C on a Varian Inova 400 MHz spectrometer. An amount of D2O sufficient to obtain a stable lock signal was added to each sample prior to measurement. The following parameters were used: frequency, 161,985 MHz; excitation pulse width, 9.25 μs; pulse repetition delay, 1 s; and spectral width, 18.35 kHz. Routine spectra were acquired with 4,096 scans. Chemical shifts were referenced to 85% orthophosphoric acid (0 ppm). Standards of Pi and polyphosphate (“P68” with polymerization from 10 to 40; BK Giulini Chemie, Ladenburg, Germany) were prepared with final concentrations of 10 mM, in terms of Pi (29). Signals were integrated with the MestRe Nova software (Mestrelab Research, Santiago, Spain) to quantify total intracellular Pi and phosphorus-containing metabolites, e.g., phosphomonoesters such as sugar phosphates, NDP-glucose, or polyphosphate.
Measurement of intracellular glycogen contents.
The glycogen contents of C. glutamicum were determined by the enzymatic method as described previously (31, 35). A culture volume corresponding to 12.5 mg cells (dry weight) [an OD600 of 1 corresponds to 0.25 mg/ml cells (dry weight) (16)] was centrifuged, and the cells were washed twice with TN buffer (50 mM Tris-HCl [pH 6.3], 50 mM NaCl). After centrifugation, the cell pellet was resuspended in 1 ml of 40 mM potassium acetate buffer (pH 4.2) and transferred to 2-ml safe-lock Eppendorf tubes filled with 250 mg zirconia/silica beads (0.1 mm diameter). After inactivation of cell-bound glycosidic activity by incubation at 99°C for 5 min, the cells were disrupted by two 30-s rounds of bead beating at 4,500 rpm using a Silamat S5 (Ivoclar Vivadent, Ellwangen, Germany). The cell debris and glass beads were separated from the supernatant by centrifugation (13,000 × g, 20 min), and the supernatant was collected and stored at −20°C until use. Each sample was divided into two 100-μl aliquots (labeled sample A and sample B). Two microliters of amyloglucosidase (10 mg/ml; Roche Diagnostics, Mannheim, Germany) was added to sample A to degrade glycogen to free glucose, whereas sample B served as a reference. Both samples were incubated for 2 h at 57°C with shaking at 850 rpm. Subsequently, the glucose concentration in the two samples was determined using a coupled enzymatic assay with hexokinase and glucose 6-phosphate dehydrogenase (Roche Diagnostics, Mannheim, Germany) by measuring the NADH formed at 340 nm. Finally, the glycogen content was calculated in mg per g of cells (dry weight) after subtraction of the glucose concentration of the reference sample B from that of the test sample A.
Construction of transcriptional fusions and chloramphenicol acetyltransferase (CAT) assays.
For CAT assays, DNA fragments covering the promoter regions of the pgm and glgC genes were amplified and cloned into the corynebacterial promoter-probe vector pET2 (44), resulting in the plasmids pET2-pgm and pET2-glgC, respectively (Table 1). The cloned fragments were controlled by DNA sequence analysis. The plasmids were introduced into wild-type C. glutamicum, and the transformed strains were cultivated as outlined above. The CAT assays were performed as described previously (9).
Constraint-based analysis using a genome-scale model of C. glutamicum.
Flux balance analysis (FBA) was performed using a slightly modified version of the genome-scale model of C. glutamicum ATCC 13032 (20) consisting of 446 metabolic reactions and 411 metabolites. The following additional reactions for glycogen formation and degradation were considered as described in reference 36: for GlgC, G1P + ATP → ADP-GLC + PPi; for GlgA, ADP-GLC → GLGN + ADP; and for GlgP and MalP, GLGN + Pi → G1P. For this network, steady-state flux distributions were calculated by FBA using linear programming-based optimization of growth as cellular objective function. In order to obtain feasible phenotypic spaces related to the experimentally observed Pi starvation response of C. glutamicum, the flux cone was constrained to different combinations of uptakes rates for Pi and glucose or acetate as carbon sources (32). In addition, only by-products that were observed during the growth experiments were allowed to be formed. For these fluxes, upper bounds were defined that were estimated from the quantitative experimental data generated by high-pressure liquid chromatography (HPLC) analysis. This refers to lactate, acetate, fumarate, and malate for glucose-grown cells, whose formation rates were estimated to be 0.10, 0.02, 0.01, and 0.01 mmol g of cells (dry weight)−1 h−1, respectively. All simulations were performed under MATLAB (R2008b; Mathworks) using the COBRA toolbox with the LP solver GLPK (1).
RESULTS
Influence of different Pi concentrations on the metabolite pattern of C. glutamicum cells.
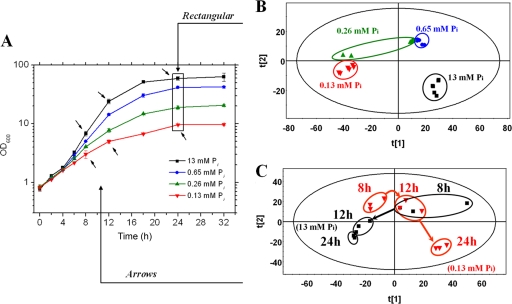
In order to determine the consequences of Pi limitation at the metabolite level, metabolome analysis was performed by GC-TOF mass spectrometry (GC-TOF MS) and the resulting data were analyzed by PLS-DA as a tool for multivariate data analysis (23). For this purpose, Corynebacterium glutamicum wild type was cultivated at four different initial Pi concentrations (13 mM, 0.65 mM, 0.26 mM, and 0.13 mM) in CGXII glucose minimal medium. The highest concentration is the one regularly used in this medium. Growth of the cells was comparable to that reported previously (15) and is shown in Fig. 1A. The cells were collected after 24 h of cultivation, intracellular metabolites were extracted and analyzed by GC-TOF MS, and PLS-DA was performed with the data from 2,517 mass fragments that were identified. The resulting score plot for metabolite pattern analysis is shown in Fig. 1B. It is obvious that the samples obtained from cells grown with 13 mM, 0.65 mM, and 0.13 mM Pi formed three distinct groups, whereas the samples obtained from cells grown with 0.26 mM Pi formed two subgroups, one being located close to the 0.65 mM group and the other next to the 0.13 mM group. This analysis was based on mass fragment patterns only rather than on a set of identified metabolites, and it clearly indicated that different Pi concentrations in the medium affect the metabolite composition within the cells. For the following experiments, Pi was used at 13 mM for Pi excess conditions and 0.13 mM for Pi-limiting conditions.
FIG. 1.
Influence of growth with different Pi concentrations on the metabolome of C. glutamicum. (A) Growth of C. glutamicum ATCC 13032 in CGXII minimal medium with 222 mM glucose and different concentration of Pi. Cells were precultured twice in CGXII glucose medium with 0.13 mM potassium Pi and then transferred to CGXII medium containing 0.13 mM, 0.26 mM, 0.65 mM, or 13 mM inorganic Pi. The experiment was performed in triplicate, and mean values and standard deviations are shown. After 24 h, samples of all cultures were taken and used for metabolite analysis by GC-TOF MS. The 2,517 mass fragments detected in all samples were used for PLS-DA, representing one symbol of the score plots. (B) PLS-DA score plot of the metabolome samples taken after 24 h of growth with different Pi concentrations. t[1] and t[2] represent vectors for the most significant components of the matrix x of mass ion abundances. The plot shows a directionality of the metabolite pattern from Pi excess to limitation. (C) PLS-DA score plot of the samples taken after 8, 12, and 24 h from cultures grown in CGXII glucose medium with either 13 mM Pi or 0.13 mM Pi.
In a second series of experiments, C. glutamicum was grown in glucose medium either with 13 mM Pi or with 0.13 mM Pi and samples for GC-TOF MS were taken after 8 h, 12 h, and 24 h of cultivation. The experiment was performed in triplicate starting with independent cultures and resulted in 18 metabolite data sets that again were analyzed by PLS-DA. As shown in Fig. 1C, except for two data sets (Pi excess culture after 8 h and Pi-limited culture after 12 h) which overlapped, all other data sets formed distinct groups on the score plot. For further analysis, cells cultivated for 24 h were used.
Semiquantitative comparison of metabolites in cells cultivated under Pi excess or Pi starvation.
In order to get more detailed insights into the changes at the metabolite level that occur under Pi limitation, a semiquantitative comparison was performed in which the metabolomes of cells grown in triplicate for 24 h under Pi excess (13 mM) or Pi starvation (0.13 mM) were analyzed by GC-TOF MS using both electron ionization and chemical ionization. For this purpose, one culture was grown with naturally labeled glucose, whereas the other one was grown with 13C6-glucose. The availability of uniformly 13C-labeled metabolites was useful for the identification of the mass fragments detected by GC MS (see the supplemental material).
Table 2 lists metabolites that could be unequivocally identified in the samples. The pool of cytoplasmic Pi was calculated to be 3-fold lower in the Pi-limited cells, which is in contrast to the 100-fold-lower Pi concentration in the medium at the start of the cultivation. It reflects the capability of the cells to maintain a comparably high cytoplasmic Pi concentration when the external Pi is limiting, due to the activation of the Pi starvation response. The lower level of lactic acid in Pi-limited cells corresponds to the fact that Pi-limited cultures did not produce l-lactate, whereas the Pi excess cultures did (Fig. 2A). Lactate excretion during growth on glucose is usually caused by oxygen limitation, and because of the much lower glucose consumption rate of the Pi-limited cells, no oxygen limitation occurred. The detection of glycolic acid in C. glutamicum extracts was unexpected, as this compound has not been described yet as a metabolite in this organism and no pathway leading to glycolate in C. glutamicum is known. l-Alanine was detected in two forms, as a 2-fold trimethylsilyl-modified form and a 3-fold trimethylsilyl-modified form. Both forms showed a reduced level in the Pi-limited cells. As l-alanine is derived from pyruvate, this result might be a consequence of a reduced glycolytic flux and a reduced pyruvate pool during Pi limitation. Also, three intermediates of the tricarboxylic acid (TCA) cycle showed reduced levels in Pi-limited cells, namely, succinate, malate, and fumarate. l-Proline, which is formed from l-glutamate, showed a greatly reduced pool under Pi limitation, which could be due to a reduced TCA cycle flux and reduced levels of NADPH or ATP.
TABLE 2.
Relative ratio of identified metabolites during growth of C. glutamicum under Pi limitation and Pi excess
| Name | Derivatization(s) | Monoisotopic mass (Da) | RT (min)a | RIb | Areac (Pi limitation/Pi excess) | Area ratio (Pi limit./Pi excess) | P valued |
|---|---|---|---|---|---|---|---|
| Lactic acid | 2TMSe | 234.1108 | 5.48 | 1,033 | 7,337 ± 295/384,667 ± 48,336 | 0.02 | 0.005 |
| Glycolic acid | 2TMS | 220.0951 | 5.67 | 1,041 | 15,300 ± 2,307/5,483 ± 361 | 2.79 | 0.017 |
| l-Alanine | 2TMS | 223.1267 | 5.97 | 1,053 | 9,963 ± 1,767/36,233 ± 13,799 | 0.27 | 0.100 |
| 3TMS | 305.1663 | 8.76 | 1,279 | 5,900 ± 335/96,467 ± 20,766 | 0.06 | 0.017 | |
| Phosphate | 3TMS | 314.0959 | 7.85 | 1,236 | 53,233 ± 28,407/170,000 ± 17,578 | 0.31 | 0.003 |
| l-Proline | 2TMS | 259.1424 | 8.15 | 1,250 | 5,400 ± 1,542/63,433 ± 7,250 | 0.09 | 0.003 |
| Succinic acid | 2TMS | 262.1057 | 8.32 | 1,258 | 49,300 ± 4,078/81,567 ± 15,815 | 0.60 | 0.048 |
| Fumaric acid | 2TMS | 260.0900 | 8.68 | 1,275 | 3,907 ± 457/7,900 ± 123 | 0.49 | 0.006 |
| Malic acid | 2TMS | 350.1401 | 9.96 | 1,441 | 3,880 ± 98/17,833 ± 981 | 0.22 | 0.001 |
| Glucosef | 5TMS, 1MeOxg | 569.2876 | 13.44 | 1,852 | 433,667 ± 12,423/3,757 ± 45 | 115.44 | 0.0003 |
| Maltoseh | 8TMS, 1MeOx | 947.459 | 16.38 | 2,659 | 3,553 ± 983/127 ± 27 | 28.05 | 0.027 |
| Trehalosei | 8TMS | 918.4324 | 18.49 | 2,661 | 433,333 ± 26,312/476,000 ± 17,349 | 0.91 | 0.158 |
RT, retention time during gas chromatography.
RI, retention index calculated using alkane standards (C10 to C40).
Integrated area of chromatographic peak under Pi-limiting conditions versus Pi-excess conditions. Values are means and standard deviations from triplicate experiments.
Determined by Student's t test.
TMS, trimethylsilyl.
Glucose levels are relatively uncertain due to possible contamination by glucose of the medium during the extraction process; Pi-limited cells retain high glucose concentrations in the medium after 24 h of cultivation.
MeOX, methoxyamine.
Maltose was detected only by the EI mode due to its higher molecular mass and was extracted with cold methanol.
Trehalose data were included despite a P value of >0.05, as trehalose can serve as a precursor for maltose by the action of trehalose synthase (TreS).
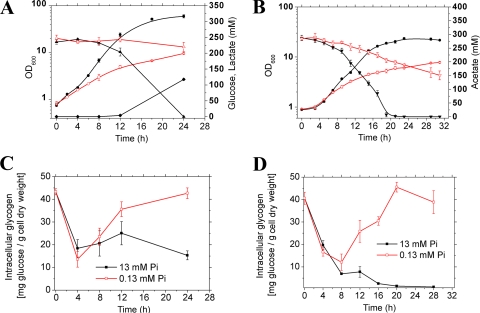
FIG. 2.
Growth (OD600, circles), carbon source consumption (triangles), and cellular glycogen pools (squares) are shown for C. glutamicum cultivated in CGXII minimal medium with 222 mM glucose (A and C) or with 300 mM potassium acetate (B and D) either under Pi excess or Pi limitation. The inoculum was precultivated twice in the same medium under Pi limitation. Panel A also shows lactate formation (rhombic symbols), which did not occur during growth on acetate (B). Mean values and standard deviations of triplicate cultures are shown.
The metabolites that showed the most greatly increased levels in Pi-limited cells were glucose (115-fold) and maltose (28-fold). Whereas the ratio measured for glucose might be caused by contamination through the high concentration of external glucose that was still present in the medium of the Pi-limited cultures after 24 h, the increased pool of maltose cannot be explained in this way. As outlined below, it is related to glycogen metabolism.
Phosphorus-containing-metabolite profiling of C. glutamicum using 31P-NMR spectroscopy.
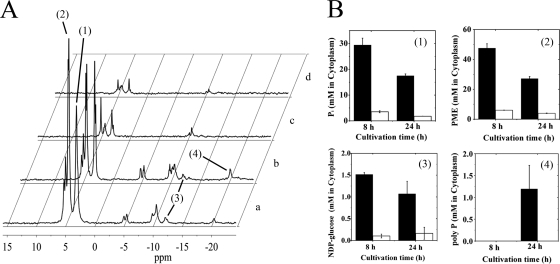
To complement GC-MS analysis, in vivo 31P-NMR was applied to measure different intracellular phosphorus-containing metabolites in C. glutamicum cells cultivated under either Pi excess or limitation. The results are summarized in Fig. 3. The cytoplasmic Pi concentration after 24 h of growth was calculated to be 17.5 ± 0.72 mM under Pi excess and 1.8 ± 0.01 mM under Pi limitation. This difference (9.7-fold) is larger than the one determined by GC-MS (3-fold) but reflects the native situation accurately. The concentration of phosphomonoesters was also found to be much lower in Pi-starved cells (4.0 ± 0.16 mM after 24 h) than in Pi-excess cells (27.1 ± 1.4 mM after 24 h). Similarly, the concentration of NDP-glucose was about 6-fold lower in Pi-starved cells (0.16 ± 0.13 mM after 24 h) than Pi-excess cells (1.07 ± 0.28 mM after 24 h). Polyphosphate was detected in stationary-phase cells grown under Pi excess but not in cells grown under Pi limitation, as expected.
FIG. 3.
In vivo 31P-NMR spectrum (A) and measurements to determine the cytoplasmic concentrations of intracellular Pi, phosphate monoesters (PME), NDP-glucose, and polyphosphate (polyP) in cells cultivated for 8 h (a) and 24 h (b) in CGXII glucose minimal medium with 13 mM Pi (black bars) or for 8 h (c) and 24 h (d) in CGXII glucose minimal medium with 0.13 mM Pi (white bars) (B). Signals representing intracellular Pi (1), phosphate monoesters (2), NDP-glucose (3), and polyphosphate (4) are marked in the 31P-NMR spectra, which were recorded using a Varian Inova 400 MHz spectrometer operating at a 31P frequency of 161.985 MHz, as described in Materials and Methods. The experiment was performed twice with comparable results.
Influence of the transcriptional regulator SugR on the glucose uptake rate under Pi limitation.
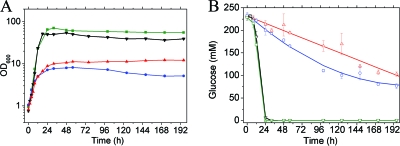
The DeoR-type transcriptional regulator SugR represses genes of the phosphoenolpyruvate phosphotransferase system (PTS), genes of several glycolytic enzymes, and a variety of further metabolic genes (8, 9, 11, 40-42). The repressing function of SugR has been reported to be relieved by several sugar phosphates, i.e., fructose 6-phosphate, fructose 1-phosphate, glucose 6-phosphate, and fructose 1,6-bisphosphate (9, 11, 42). The levels of these metabolites are high when cells grow on sugars, such as glucose or fructose, and low when the cells grow on gluconeogenic carbon sources, such as acetate. As the level of Pi monoesters in glucose-grown cells was found to be much lower in Pi-starved cells, the question of whether the low glucose uptake rates observed for cells growing under Pi limitation [46 nmol min−1 (mg of protein)−1 compared to 192 nmol min−1 (mg of protein)−1 under Pi excess] might be related to the activity of SugR arose.
To test this possibility, we analyzed growth and glucose consumption of a ΔsugR mutant of C. glutamicum (9) under Pi excess and Pi starvation. As shown in Fig. 4, the ΔsugR mutant grew similarly to the wild type under Pi limitation, but to a lower optical density. Under Pi excess, on the other hand, the ΔsugR mutant reached a higher optical density than the wild type. Under Pi excess, the glucose consumption rate of the ΔsugR mutant was comparable to that of the wild type, whereas it was 2.5-fold higher (114 nmol min−1 [mg of protein]−1) under Pi limitation (the period from 8 h to 24 h after start of the cultivation was used for calculation). This supports the assumption that the low glucose consumption rate under Pi limitation is at least partially due to repression of the PTS genes for glucose uptake (ptsG, ptsI, and ptsH) and of glycolytic genes by SugR, due to low levels of its effector metabolites. The major reason for the low glucose consumption rate under Pi limitation might, however, be a low rate of phosphoenolpyruvate (PEP) formation.
FIG. 4.
Growth of (A) and glucose consumption by (B) of C. glutamicum wild type (black and red) and the ΔsugR mutant (green and blue) in CGXII minimal medium with 4% (wt/vol) glucose and either 13 mM Pi (black and green) or 0.13 mM Pi (red and blue). Mean values and standard deviations of triplicate cultures are shown.
Influence of Pi limitation on the glycogen pool of C. glutamicum.
C. glutamicum cells growing on glucose, fructose, or sucrose accumulate glycogen up to 90 mg per g of cells (dry weight) in the early exponential growth phase and degrade the polymer when the sugar becomes limiting. In contrast, only marginal amounts of glycogen are formed in cells growing on the gluconeogenic substrates acetate or lactate (36). Recent studies revealed a close connection between maltose and glycogen metabolism (38). The finding of a greatly increased maltose pool in Pi-limited cells prompted us to also measure the glycogen content of cells. As shown in Fig. 2, there was a significant discrepancy between results obtained under Pi-limiting and Pi-excess conditions. Comparable to previously published data (36), cells grown on glucose under Pi excess accumulated glycogen up to 30 mg glucose per g of cells (dry weight) in the early exponential growth phase and then started to degrade it before reaching the stationary phase. In contrast, Pi-limited cells accumulated glycogen up to 24 h to a level of about 45 mg glucose per g of cells (dry weight). Even more surprising was the observation that cells grown with acetate as the sole carbon source also formed glycogen under Pi limitation up to levels comparable to that of glucose-grown cells (Fig. 2C and D). In agreement with previous results (36), no glycogen was formed by acetate-grown cells under Pi excess (Fig. 2D). The high glycogen level that was measured at time zero in all cultures resulted from the precultivation of the inoculum under Pi-limiting conditions. Based on these results, Pi limitation causes glycogen accumulation in C. glutamicum.
The results described above raised the question of whether also other types of growth limitations have an influence on glycogen accumulation. Therefore, the glycogen content was measured in C. glutamicum cells cultivated either under nitrogen excess and nitrogen limitation or under iron excess and iron limitation. Both types of stresses have been studied in the past, and key players involved in the adaptation to these stresses have been identified (for reviews, see references 4 and 10). As shown in Fig. S4 in the supplemental material, these limitations did not cause an accumulation of glycogen in the stationary phase during growth on glucose or acetate. This indicates that glycogen accumulation is one of the specific responses of the cell to Pi limitation.
Influence of Pi limitation on the expression of pgm and glgC.
Glycogen synthesis in C. glutamicum involves four enzymes, i.e., phosphoglucomutase (pgm), catalyzing the conversion of glucose 6-phosphate to glucose 1-phosphate, ADP-glucose pyrophosphorylase (glgC), which converts glucose 1-phosphate and ATP to ADP-glucose and pyrophosphate, glycogen synthase (glgA), converting [α-1,4-glucan]n and ADP-glucose to [α-1,4-glucan]n + 1 and ADP, and branching enzyme (glgB), which introduces α-1,6-glycosidic bonds into linear α-1,4-glucans. To test the influence of Pi limitation on the expression of pgm and glgC, the corresponding promoter regions were cloned into the promoter probe vector pET2 containing a promoterless chloramphenicol acetyltransferase reporter gene, and the resulting plasmids pET2-pgm and pET2-glgC were transferred into C. glutamicum wild type. As shown in Table 3, expression of the phosphoglucomutase gene pgm was 6-fold higher in cells grown for 24 h under Pi limitation than in cells grown for 24 h under Pi excess. In the case of the ADP-glucose pyrophosphorylase gene glgC the expression level was 3-fold higher under Pi limitation than Pi excess. These results indicate that genes of the glycogen synthesis pathway are activated or derepressed under Pi limitation, and increased levels of the two enzyme activities could be at least partially responsible for the increased flux of glucose 6-phosphate into the glycogen pathway and reduced fluxes into glycolysis and the pentose phosphate pathway.
TABLE 3.
Influence of Pi concentration on the expression of the pgm and glgC genes in C. glutamicum
| Growth condition | CAT activitya (nmol min−1 mg protein−1) |
|
|---|---|---|
| C. glutamicum pET2-pgm | C. glutamicum pET2-glgC | |
| Pi excess (13 mM) | 31 ± 5 | 195 ± 37 |
| Pi limitation (0.13 mM) | 189 ± 37 | 583 ± 6 |
Strains were cultivated for 24 h in CGXII medium with 222 mM glucose under either Pi excess or Pi limitation. Chloramphenicol acetyltransferase activities were determined using cell-free extracts. Means and standard deviations derived from three independent cultivations are given.
In silico simulation of the phosphate starvation response using a genome-scale model of C. glutamicum.
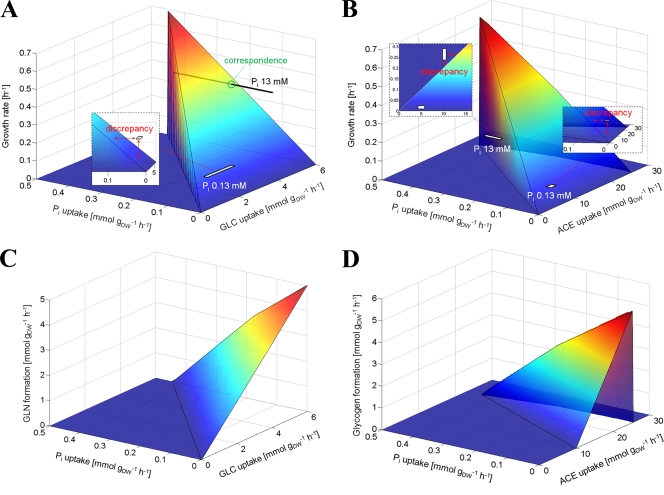
In order to study the influence of Pi limitation on metabolism in silico by flux balance analysis (FBA), a genome-scale model of C. glutamicum (20) was expanded by including two reactions required for glycogen synthesis (ADP-glucose pyrophosphorylase and glycogen synthase) and one reaction responsible for glycogen degradation, which represents both glycogen phosphorylase and maltodextrin phosphorylase. The resulting optimal phenotypes for growth under variation of glucose and Pi uptake are shown in Fig. 5A. As expected, under the precondition of a sufficient glucose uptake rate (>4 mmol [g of cells (dry weight)]−1 h−1), the growth rate is linearly dependent on the Pi uptake rate. However, there is a discrepancy between the simulated maximal growth rate and the experimentally determined growth rate under Pi-limiting conditions, the latter being located at a point where no steady-state flux solution of the network exists. To reach the experimentally observed growth rate of about 0.16 h−1 under Pi limitation in the simulation, glucose and phosphate uptake rates have to be significantly higher (>2 mmol g of cells (dry weight)−1 h−1 and >0.1 mmol g of cells (dry weight)−1 h−1, respectively). A similar observation was made for the results obtained for the acetate-grown cells. Here discrepancies were found for both phosphate-limiting and phosphate-excess conditions (Fig. 5B). As expected, simulations showed that increasing the glucose or acetate uptake rate at low phosphate uptake rates would not lead to higher cellular growth.
FIG. 5.
Simulated phenotypes under growth optimization of the genome-scale metabolic network model. In silico solutions of growth rates and glycogen (GLN) formation under variation of Pi uptake in combination with either glucose (GLC) uptake (A and C) or acetate (ACE) uptake (B and D) form three-dimensional surfaces in each case. For comparison, measured growth rates for Pi-limited and -excess cultures, including experimental errors, are mapped as light gray rectangles.
Since a strong influence of Pi limitation on the glycogen pool was observed (see above), the influence of varying glucose and phosphate uptake rates on glycogen formation was tested in silico. As shown in Fig. 5C, an increased rate of glycogen synthesis was predicted at low Pi uptake and high glucose uptake rates. The same behavior was also observed when acetate instead of glucose was used as carbon source (Fig. 5D). These predictions are in agreement with the experimental results.
DISCUSSION
Previous studies on the response of bacteria to Pi-limiting conditions focused mainly on gene expression, regulators, and enzymes involved in the Pi starvation response. In the work presented here, the influence of Pi limitation on metabolite levels was analyzed by GC-MS using C. glutamicum as a model organism. An important result was the detection of greatly elevated maltose levels under Pi limitation, which raises the question of how this disaccharide is formed in cells growing on glucose. The only pathway that has been described in literature for C. glutamicum is the conversion of trehalose to maltose by trehalose synthase (TreS). TreS was shown to be the only enzyme present in C. glutamicum capable of converting trehalose to maltose and vice versa, and evidence that TreS is mainly responsible for trehalose degradation was presented (48). Trehalose, which serves as a stress protection compound and as a prerequisite for mycolate production, is synthesized either from UDP-glucose and glucose 6-phosphate via the OtsA-OtsB pathway or from malto-oligosaccharides or α-1,4-glucans via the TreY-TreZ pathway (43, 48). The cytoplasmic trehalose level as determined by GC-MS was much higher than the maltose level (by a factor of 103 to 104) and was only slightly decreased under Pi-limitation compared to Pi-excess.
Besides TreS, one alternative enzyme candidate could also play a role in maltose formation. The protein encoded by cg1012 shows sequence similarity to the E. coli maltodextrin glucosidase MalZ. MalZ removes glucose residues from the reducing end of maltodextrins, which are composed of more than two glucose residues (i.e., maltotriose, maltotetraose, etc.), and forms maltose as an end product (5). It is not yet clear whether this enzyme activity is present in C. glutamicum (38), but it would offer an alternative explanation for the high internal glucose level of Pi-limited cells (besides the possibility that it is due to contamination from residual extracellular glucose).
Recent studies have indicated that C. glutamicum catabolizes maltose in the same way as E. coli (2) by MalQ (cg2523), a maltodextrin glucanotransferase (also called amylomaltase) which forms from any maltodextrin, including maltose, larger maltodextrins, and glucose. Glucose can then be phosphorylated either by an ATP-dependent glucokinase (30) or by a polyphosphate/NTP-dependent glucokinase (25) to glucose 6-phosphate and catabolized, whereas the maltodextrins are degraded by maltodextrin phosphorylase (MalP) to glucose 1-phosphate, which is converted to glucose 6-phosphate by phosphoglucomutase (38). In Mycobacterium tuberculosis and M. smegmatis, maltose formed from trehalose by TreS is incorporated into glycogen by the consecutive action of the maltose kinase Pep1 and the maltosyltransferase GlgE (7, 17). Genes encoding homologs of Pep1 (cg2530) and GlgE (cg1382) were also identified in C. glutamicum but have not yet been characterized (G. M. Seibold, unpublished data).
In a previous study, the presence of maltose in cells of C. glutamicum cultivated on glucose under Pi excess was reported (39). Thus, maltose might be a regular metabolite in C. glutamicum not only during growth on maltose. The question of whether the increased maltose pool observed under Pi-limitation is caused by an increased synthesis or by a decreased degradation or both cannot be answered at the moment.
Based on the close connection of maltose and glycogen metabolism, we found that Pi starvation also had a strong influence on the cellular glycogen pool. Whereas under Pi excess, glycogen is formed in the early exponential phase and then degraded again, Pi-starved cells form a glycogen pool in the exponential phase but also retain it in the stationary phase, irrespective of whether glucose or acetate was used as the carbon source. The large glycogen pool under Pi starvation could be due to increased synthesis, as suggested by the increased expression of pgm and glgC. The regulators responsible for this increased expression are not yet known. The global transcriptional regulator RamA was recently shown to function as an activator of glgC (37), but current knowledge suggests that RamA responds to a metabolite involved in acetate catabolism rather than to Pi. Besides an increased glycogen synthesis rate, a decreased glycogen degradation rate could also be responsible for the altered glycogen pool under Pi limitation. Glycogen degradation in C. glutamicum involves glycogen phosphorylase (GlgP), which phosphorolytically cleaves α-1,4-glycosidic bonds at the nonreducing ends of glycogen and forms glucose 1-phosphate and phosphorylase-limited dextrins. The debranching enzyme (GlgX) converts these dextrins to linear maltodextrins (2 < n < 20, where n is the number of glucose molecules), which are then further degraded by MalP to glucose 1-phosphate (38). As GlgP and MalP both require Pi, their activity might be limited at reduced cytoplasmic Pi concentrations.
The in silico simulation data based on a stoichiometric genome-scale metabolic model (20) that was modified to include glycogen synthesis and degradation reactions predicted increased glycogen formation in exponentially growing cells under Pi limitation with glucose or acetate as the carbon source. However, the model was not able to correctly predict the experimentally determined growth rates under Pi starvation. Reasons for the observed discrepancies could be a lack of qualitative information and/or quantitative accuracy of the model, which mainly includes all biomass-related reactions but only rough estimations of its stoichiometric coefficients. Furthermore, the model does not incorporate any kind of Pi-dependent regulation of central metabolism and storage pool metabolism (glycogen, polyphosphate). An inclusion of regulation would necessitate the formulation of mechanistic models, which is currently impossible due to the lack of quantitative knowledge of regulatory and metabolic processes. Therefore, the results from our stoichiometric analysis should be regarded as a first step in simulating the complex metabolism of carbohydrate storage pools like glycogen.
Central carbon metabolism and energy metabolism are inevitably connected to the availability of Pi, as key enzymatic reactions require Pi as substrate, such as glyceraldehyde 3-phosphate dehydrogenase and F1Fo-ATP synthase. In addition, many reactions require ATP or ADP as substrates, such as phosphofructokinase, 3-phosphoglycerate kinase, and pyruvate kinase. As the cytoplasmic concentrations of Pi, ADP, and ATP are lower under Pi-limiting conditions, a reduced enzyme activity and consequently a reduced glycolytic flux can be envisaged, resulting in a reduced PEP synthesis rate and thus a reduced glucose consumption rate. In this study, another consequence of Pi limitation was found, namely, an altered glycogen metabolism resulting in an increased and more stable glycogen pool. To our knowledge, such a link has not yet been described. Further studies are required to elucidate the molecular details of this connection.
Supplementary Material
Acknowledgments
This work was financially supported by the German Ministry of Education and Research (BMBF) within the program “GenoMik-Plus” (grant 0313805D to M.B.).
We thank Volker Wendisch and Verena Engels for providing the C. glutamicum ΔsugR strain and Katharina Nöh for valuable contributions at the beginning of this project.
Footnotes
Published ahead of print on 27 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Becker, S. A., A. M. Feist, M. L. Mo, G. Hannum, B. O. Palsson, and M. J. Herrgard. 2007. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox. Nat. Protocols 2:727-738. [DOI] [PubMed] [Google Scholar]
- 2.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkovski, A. (ed.). 2008. Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 4.Burkovski, A. 2007. Nitrogen control in Corynebacterium glutamicum: proteins, mechanisms, signals. J. Microbiol. Biotechnol. 17:187-194. [PubMed] [Google Scholar]
- 5.Dippel, R., and W. Boos. 2005. The maltodextrin system of Escherichia coli: metabolism and transport. J. Bacteriol. 187:8322-8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggeling, L., and M. Bott (ed.). 2005. Handbook of Corynebacterium glutamicum. CRC Press, Taylor & Francis Group, Boca Raton, Florida.
- 7.Elbein, A. D., I. Pastuszak, A. J. Tackett, T. Wilson, and Y. T. Pan. 2010. Last step in the conversion of trehalose to glycogen: a mycobacterial enzyme that transfers maltose from maltose 1-phosphate to glycogen. J. Biol. Chem. 285:9803-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engels, V., S. N. Lindner, and V. F. Wendisch. 2008. The global repressor SugR controls expression of genes of glycolysis and of the l-lactate dehydrogenase LdhA in Corynebacterium glutamicum. J. Bacteriol. 190:8033-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engels, V., and V. F. Wendisch. 2007. The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 189:2955-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frunzke, J., and M. Bott. 2008. Regulation of iron homeostasis in Corynebacterium glutamicum, p. 241-266. In A. Burkovski (ed.), Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 11.Gaigalat, L., J. P. Schlüter, M. Hartmann, S. Mormann, A. Tauch, A. Pühler, and J. Kalinowski. 2007. The DeoR-type transcriptional regulator SugR acts as a repressor for genes encoding the phosphoenolpyruvate: phosphotransferase system (PTS) in Corynebacterium glutamicum. BMC Mol. Biol. 8:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermann, T. 2003. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 104:155-172. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh, Y. J., and B. L. Wanner. 2010. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulett, F. M. 2002. The Pho regulon, p. 193-201. In J. A. Sonenshein and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 15.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabus, A., A. Niebisch, and M. Bott. 2007. Role of cytochrome bd oxidase from Corynebacterium glutamicum in growth and lysine production. Appl. Environ. Microbiol. 73:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalscheuer, R., K. Syson, U. Veeraraghavan, B. Weinrick, K. E. Biermann, Z. Liu, J. C. Sacchettini, G. Besra, S. Bornemann, and W. R. Jacobs. 2010. Self-poisoning of Mycobacterium tuberculosis by targeting GlgE in an α-glucan pathway. Nat. Chem. Biol. 6:376-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Studies on amino acid fermentation. Part I. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3:193-205. [PubMed] [Google Scholar]
- 20.Kjeldsen, K. R., and J. Nielsen. 2009. In silico genome-scale reconstruction and validation of the Corynebacterium glutamicum metabolic network. Biotechnol. Bioeng. 102:583-597. [DOI] [PubMed] [Google Scholar]
- 21.Kocan, M., S. Schaffer, T. Ishige, U. Sorger-Herrmann, V. F. Wendisch, and M. Bott. 2006. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J. Bacteriol. 188:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert, C., D. Weuster-Botz, R. Weichenhain, E. W. Kreutz, A. A. De Graaf, and S. M. Schoberth. 2002. Monitoring of inorganic polyphosphate dynamics in Corynebacterium glutamicum using a novel oxygen sparger for real time P-31 in vivo NMR. Acta Biotechnol. 22:245-260. [Google Scholar]
- 23.Lee, S. H., H. M. Woo, B. H. Jung, J. G. Lee, O. S. Kwon, H. S. Pyo, M. H. Choi, and B. C. Chung. 2007. Metabolomic approach to evaluate the toxicological effects of nonylphenol with rat urine. Anal. Chem. 79:6102-6110. [DOI] [PubMed] [Google Scholar]
- 24.Liebl, W. 2005. Corynebacterium taxonomy, p. 9-34. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Taylor & Francis Group, Boca Raton, FL.
- 25.Lindner, S. N., S. Knebel, S. R. Pallerla, S. M. Schoberth, and V. F. Wendisch. 2010. Cg2091 encodes a polyphosphate/ATP-dependent glucokinase of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. doi: 10.1007/s00253-010-2568-5. [DOI] [PubMed]
- 26.Lindner, S. N., S. Knebel, H. Wesseling, S. M. Schoberth, and V. F. Wendisch. 2009. Exopolyphosphatases Ppx1 and Ppx2 from Corynebacterium glutamicum. Appl. Environ. Microbiol. 75:3161-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindner, S. N., H. Niederholtmeyer, K. Schmitz, S. M. Schoberth, and V. F. Wendisch. 2010. Polyphosphate/ATP-dependent NAD kinase of Corynebacterium glutamicum: biochemical properties and impact of ppnK overexpression on lysine production. Appl. Microbiol. Biotechnol. doi: 10.1007/s00253-010-2481-y. [DOI] [PubMed]
- 28.Lindner, S. N., D. Vidaurre, S. Willbold, S. M. Schoberth, and V. F. Wendisch. 2007. NCgl2620 encodes a class II polyphosphate kinase in Corynebacterium glutamicum. Appl. Environ. Microbiol. 73:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pallerla, S. R., S. Knebel, T. Polen, P. Klauth, J. Hollender, V. F. Wendisch, and S. M. Schoberth. 2005. Formation of volutin granules in Corynebacterium glutamicum. FEMS Microbiol. Lett. 243:133-140. [DOI] [PubMed] [Google Scholar]
- 30.Park, S. Y., H. K. Kim, S. K. Yoo, T. K. Oh, and J. K. Lee. 2000. Characterization of glk, a gene coding for glucose kinase of Corynebacterium glutamicum. FEMS Microbiol. Lett. 188:209-215. [DOI] [PubMed] [Google Scholar]
- 31.Parrou, J. L., and J. Francois. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248:186-188. [DOI] [PubMed] [Google Scholar]
- 32.Price, N. D., J. L. Reed, and B. O. Palsson. 2004. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nature Rev. Microbiol. 2:886-897. [DOI] [PubMed] [Google Scholar]
- 33.Rittmann, D., U. Sorger-Hermann, and V. F. Wendisch. 2005. Phosphate starvation-inducible gene ushA encodes a 5′ nucleotidase required for growth of Corynebacterium glutamicum on nucleotides as the phosphorus source. Appl. Environ. Microbiol. 71:4339-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaaf, S., and M. Bott. 2007. Target genes and DNA-binding sites of the response regulator PhoR from Corynebacterium glutamicum. J. Bacteriol. 189:5002-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seibold, G., M. Auchter, S. Berens, J. Kalinowski, and B. J. Eikmanns. 2006. Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J. Biotechnol. 124:381-391. [DOI] [PubMed] [Google Scholar]
- 36.Seibold, G., S. Dempf, J. Schreiner, and B. J. Eikmanns. 2007. Glycogen formation in Corynebacterium glutamicum and role of ADP-glucose pyrophosphorylase. Microbiology 153:1275-1285. [DOI] [PubMed] [Google Scholar]
- 37.Seibold, G. M., C. T. Hagmann, M. Schietzel, D. Emer, M. Auchter, J. Schreiner, and B. J. Eikmanns. 2010. The transcriptional regulators RamA and RamB are involved in the regulation of glycogen synthesis in Corynebacterium glutamicum. Microbiology 156:1256-1263. [DOI] [PubMed] [Google Scholar]
- 38.Seibold, G. M., M. Wurst, and B. J. Eikmanns. 2009. Roles of maltodextrin and glycogen phosphorylases in maltose utilization and glycogen metabolism in Corynebacterium glutamicum. Microbiology 155:347-358. [DOI] [PubMed] [Google Scholar]
- 39.Strelkov, S., M. von Elstermann, and D. Schomburg. 2004. Comprehensive analysis of metabolites in Corynebacterium glutamicum by gas chromatography/mass spectrometry. Biol. Chem. 385:853-861. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka, Y., H. Teramoto, M. Inui, and H. Yukawa. 2008. Regulation of expression of general components of the phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS) by the global regulator SugR in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 78:309-318. [DOI] [PubMed] [Google Scholar]
- 41.Toyoda, K., H. Teramoto, M. Inui, and H. Yukawa. 2008. Expression of the gapA gene encoding glyceraldehyde 3-phosphate dehydrogenase of Corynebacterium glutamicum is regulated by the global regulator SugR. Appl. Microbiol. Biotechnol. 81:291-301. [DOI] [PubMed] [Google Scholar]
- 42.Toyoda, K., H. Teramoto, M. Inui, and H. Yukawa. 2009. Molecular mechanism of SugR-mediated sugar-dependent expression of the ldhA gene encoding L-lactate dehydrogenase in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 83:315-327. [DOI] [PubMed] [Google Scholar]
- 43.Tzvetkov, M., C. Klopprogge, O. Zelder, and W. Liebl. 2003. Genetic dissection of trehalose biosynthesis in Corynebacterium glutamicum: inactivation of trehalose production leads to impaired growth and an altered cell wall lipid composition. Microbiology 149:1659-1673. [DOI] [PubMed] [Google Scholar]
- 44.Vasicova, P., Z. Abrhamova, J. Nesvera, M. Patek, H. Sahm, and B. Eikmanns. 1998. Integrative and autonomously replicating vectors for analysis of promoters in Corynebacterium glutamicum. Biotechnol. Techniques 12:743-746. [Google Scholar]
- 45.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 46.Wendisch, V. F., and M. Bott. 2005. Phosphorus metabolism, p. 377-396. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL.
- 47.Wold, S., M. Sjöström, and L. Eriksson. 2001. PLS-regression: a basic tool of chemometrics. Chemom. Intell. Lab. Syst. 58:109-130. [Google Scholar]
- 48.Wolf, A., R. Krämer, and S. Morbach. 2003. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol. Microbiol. 49:1119-1134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.