Abstract
At present there is little quantitative information on the identity and composition of bacterial populations in the rumen microbial community. Quantitative fluorescence in situ hybridization using newly designed oligonucleotide probes was applied to identify the microbial populations in liquid and solid fractions of rumen digesta from cows fed barley silage or grass hay diets with or without flaxseed. Bacteroidetes, Firmicutes, and Proteobacteria were abundant in both fractions, constituting 31.8 to 87.3% of the total cell numbers. They belong mainly to the order Bacteroidales (0.1 to 19.2%), hybridizing with probe BAC1080; the families Lachnospiraceae (9.3 to 25.5%) and Ruminococcaceae (5.5 to 23.8%), hybridizing with LAC435 and RUM831, respectively; and the classes Deltaproteobacteria (5.8 to 28.3%) and Gammaproteobacteria (1.2 to 8.2%). All were more abundant in the rumen communities of cows fed diets containing silage (75.2 to 87.3%) than in those of cows fed diets containing hay (31.8 to 49.5%). The addition of flaxseed reduced their abundance in the rumens of cows fed silage-based diets (to 45.2 to 58.7%) but did not change markedly their abundance in the rumens of cows fed hay-based diets (31.8 to 49.5%). Fibrolytic species, including Fibrobacter succinogenes and Ruminococcus spp., and archaeal methanogens accounted for only a small proportion (0.4 to 2.1% and 0.2 to 0.6%, respectively) of total cell numbers. Depending on diet, between 37.0 and 91.6% of microbial cells specifically hybridized with the probes used in this study, allowing them to be identified in situ. The identities of other microbial populations (8.4 to 63.0%) remain unknown.
The rumen is an anaerobic ecosystem used by herbivores to convert fibrous plant material into fermentation products that are in turn used as energy by the host. Fibrolytic degradation is accomplished by a complex microbial community which includes specialized fungi, protozoa, and bacteria (14). More than 200 bacterial species (5) have been isolated from rumen, and many of these have been phylogenetically and physiologically characterized. Several of these, including Fibrobacter succinogenes, Ruminococcus albus, and Ruminococcus flavefaciens, have the ability to hydrolyze cellulose in axenic culture (24). Despite the presence of these fibrolytic populations, a large portion of the fiber in low-quality forage diets passes through the rumen undigested. In the rumen, fibrolytic bacteria do not digest plant cell walls in isolation but rather interact with a consortium of bacteria (18). Although culture-dependent studies have improved our understanding of rumen microbiology, the importance of the isolates to the structure and function of the rumen microbial community, with the possible exception of the fibrolytic strains, is still unknown. Expanding our knowledge of the structure and function of the rumen microbial community may provide insights into approaches to improve the efficiency of fiber digestion and biofuel production (14).
To provide a high-resolution view of the population structure of the rumen bacterial community, we used quantitative fluorescence in situ hybridization (qFISH) to investigate the composition and distribution of bacterial populations associated with the liquid and solid rumen contents from 12 ruminally cannulated Holstein dairy cows (3 cows were used for each diet) fed (for at least 21 days) grass hay or barley silage diets with or without flaxseed (Table 1). Six new 16S rRNA-targeted FISH probes (Table 2) for not only the fibrolytic groups but also other unclassified bacterial groups in the rumen were designed, using ARB software (17), against the rumen 16S rRNA gene sequences (data not shown) retrieved from the Ribosomal Database Project (RDP) database (6). The new probes target Bacteroidales-related clones (probe BAC1080) (phylum Bacteroidetes), Lachnospiraceae- and Ruminococcaceae-related clones (probes LAC435 and RUM831, respectively) (phylum Firmicutes), Butyrivibrio fibrisolvens-related clones (probe BFI826), and R. albus- and R. flavefaciens-related clones (probes RAL1436 and RFL155, respectively).
TABLE 1.
Composition of diets used in this study
| Ingredient | Diet composition (% dry weight) |
|||
|---|---|---|---|---|
| Hay-based diet | Hay and flaxseed diet | Silage-based diet | Silage and flaxseed diet | |
| Alfalfa grass hay (chopped) | 47.5 | 47.5 | 0 | 0 |
| Barley silage | 0 | 0 | 47.5 | 47.5 |
| Steamed rolled barley grain | 47.5 | 32.5 | 47.5 | 32.5 |
| Ground flaxseeds | 0 | 15 | 0 | 15 |
| Other | 5 | 5 | 5 | 5 |
TABLE 2.
Oligonucleotide probes and their target populations used in this study for FISH analyses
| Probe namea | Target rRNA | Designed target(s) | % FAb | Reference |
|---|---|---|---|---|
| EUB338 (00159) | 16S | Domain Bacteria | 0-50 | 16 |
| EUB338II (00160) | 16S | Phylum Planctomycetes | 0-50 | 16 |
| EUB338III (00161) | 16S | Phylum Verrucomicrobia | 0-50 | 16 |
| NONEUB (00243) | 16S | Control probe complementary to EUB338 | 0-50 | 16 |
| ALF968 (00021) | 16S | Class Alphaproteobacteria, phylum Proteobacteria | 20 | 16 |
| BET42a (00034) | 23S | Class Betaproteobacteria, phylum Proteobacteria | 35 | 16 |
| GAM42a (00174) | 23S | Class Gammaproteobacteria, phylum Proteobacteria | 35 | 16 |
| SRB385 (00300) | 16S | Class Deltaproteobacteria, phylum Proteobacteria | 35 | 16 |
| SRB385Db (00301) | 16S | Class Deltaproteobacteria, phylum Proteobacteria | 35 | 16 |
| HGC69a (00182) | 23S | Phylum Actinobacteria | 25 | 16 |
| GNSB941 (00718) | 16S | Phylum Chloroflexi | 35 | 16 |
| CFX1223 (00719) | 16S | Phylum Chloroflexi | 35 | 16 |
| SPIRO1400 (01004) | 16S | Subgroup of family Spirochaetaceae | 20 | 16 |
| TM7-905 (00600) | 16S | Candidate phylum TM7 | 20 | 16 |
| LGC354A (00195) | 16S | Phylum Firmicutes | 35 | 16 |
| LGC354B (00196) | 16S | Phylum Firmicutes | 35 | 16 |
| LGC354C (00197) | 16S | Phylum Firmicutes | 35 | 16 |
| RUM831 | 16S | Rumen clones in family Ruminococcaceae, phylum Firmicutes | 35 | This study |
| RAL1436 | 16S | Ruminococcus albus-related clones, phylum Firmicutes | 20 | This study |
| RFL155 | 16S | Ruminococcus flavefaciens-related clones, phylum Firmicutes | 45 | This study |
| LAC435 | 16S | Clones in family Lachnospiraceae, phylum Firmicutes | 35 | This study |
| BFI826 | 16S | Butyrivibrio fibrisolvens-related clones, phylum Firmicutes | 35 | This study |
| BAC1080 | 16S | Clones in order Bacteroidales, phylum Bacteroidetes | 20 | This study |
| Fibr225 (00005) | 16S | Fibrobacter succinogenes-related clones, phylum Fibrobacteres | 20c | 16 |
| ARCH915 (00027) | 16S | Domain Archaea | 20 | 16 |
The numbers in parentheses after the probe names represent the probe accession numbers in probeBase (16).
FA, formamide concentration used in the FISH buffer.
The optimum formamide concentration for the probe was determined in this study.
The optimal formamide concentrations (OFC) of the new probes used in FISH were assessed in different ways. Probes RUM831 and BAC1080 were assessed by using pure cultures of Ruminococcus and Prevotella strains with zero and one mismatch (Fig. 1) to the probes. The OFC of probes LAC435 and BFI826 were assessed using Clone-FISH (21) with zero and one mismatch 16S rRNA clone (Fig. 1) by following the procedure described previously (9, 10). The highest formamide concentration (tested in 5% stepwise increases) at which a clear fluorescent signal was observed with the reference bacterium or competent cells with zero mismatches after FISH probing, but not with bacteria or competent cells with one mismatch, was selected. The OFC of probes FIB225 (designed by Stahl et al. [23]), RFL155, and RAL1436 were assessed using only pure cultures of F. succinogenes, R. flavefaciens, and R. albus, respectively, all having perfect matches to each probe (Fig. 1). The highest formamide concentration (tested in 5% stepwise increases) at which a clear fluorescent signal was observed with the reference bacterium after FISH probing was selected. These probes were employed with other available probes (Table 2) chosen from probeBase (16) based on the alignment and classification of the 16S rRNA gene sequences retrieved from rumen communities.
FIG. 1.
Alignments of the probe sequences and their target sites and sequences of corresponding sites in reference bacteria or clones. The probe names in parentheses after the abbreviated names are according to Oligonucleotide Probe Database nomenclature (2). Only the nucleotides that are different from target sequences are shown. E, empty space; R., Ruminococcus; P., Prevotella; F., Fibrobacter.
The digest samples from the top, bottom, and middle of the rumen were collected through a cannula, thoroughly mixed, and fractioned as liquid fraction (LiqF) and solid fraction (SolF). On-site, about 100 ml was transferred to a heavy-wall 250-ml beaker and squeezed using a Bodum coffee maker plunger (Bodum Inc., Triengen, Switzerland). The extruded liquid samples (containing the planktonic cells) were fixed in ethanol and paraformaldehyde (PFA) for FISH probing (3). The remaining liquid was discarded, and the squeezed particulate samples (used to collect particulate-attached cells) were washed with 100 ml phosphate buffer (5.23 g/liter K2HPO4, 2.27 g/liter KH2PO4, 3.00 g/liter NaHCO3, and 20 ml/liter 2.5% cysteine HCl) by stirring gently with a spatula, followed by squeezing again and decanting. Washed particulate samples (5 g) were then fixed for FISH as described above.
After fixation, the particulate samples plus the fixation solution were transferred into a stomacher bag and “stomached” (Stomacher 400 Circulator, Seaward England) at 230 rpm for 6 min. Treated samples were then transferred into a clean 250-ml beaker and squeezed again. Microscopic examination of the squeezed residues after DAPI (4′,6-diamidino-2-phenylindole) staining (100 μl [0.003 mg/ml] for 10 min) showed only a few bacterial cells attached on the plant fibers, indicating that most bacterial cells had been “stomached” into the liquid (data not shown). To recover cells, filtrates were centrifuged (5,000 × g), and the cell pellet was washed three times with phosphate buffer before being used for FISH probing. On the day of sampling, each cow was sampled twice, at 1100 h and 1600 h. The liquid FISH samples obtained from the 3 cows fed with the same diet (at two different sampling times) were mixed, as were the particulate FISH samples, and used in qFISH analysis. FISH was carried out according to Amann (3). FISH was carried out on glass coverslips (24 by 60 mm) coated with gelatin (9). DAPI staining of biomass samples was carried out after FISH probing. FISH and DAPI images were captured with a Zeiss epifluorescence microscope (Zeiss PM III) equipped with a Canon 5D Mark II camera. Raw images captured randomly were transferred into gray TIF images and sharpened in Adobe Photoshop CS3. Cells stained with DAPI and hybridized to the probes were enumerated using the function provided in ImageJ (1). The percent compositions of these probe-defined groups (against all DAPI-stained cells in the same microscopic field) in the different fractions of rumen contents from cows fed different diets are presented in Table 3.
TABLE 3.
Distribution and composition of FISH probe-defined groups in rumen microbial communities in cows fed with different diets
| Probe-defined microbial group | Composition (mean value [%] ± SD)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| Hay-based diet |
Hay and flaxseed diet |
Silage-based diet |
Silage and flaxseed diet |
|||||
| LiqF | SolF | LiqF | SolF | LiqF | SolF | LiqF | SolF | |
| BAC1080 | 9.6 ± 1.33 | 0.1 ± 0.02 | 19.2 ± 3.71 | 4.2 ± 0.72 | 14.2 ± 3.11 | 18.8 ± 3.88 | 14.4 ± 2.89 | 16.7 ± 4.33 |
| ALF968 | 0.2 ± 0.02 | 0.2 ± 0.02 | 0.2 ± 0.03 | 0.2 ± 0.04 | 0.7 ± 0.14 | 1.5 ± 0.41 | 0.1 ± 0.01 | 0.1 ± 0.01 |
| BET42a | 0 | 0 | 0.6 ± 0.01 | 1.2 ± 0.27 | 0.1 ± 0.01 | <0.1 | 0.4 ± 0.06 | 0.2 ± 0.04 |
| GAM42a | 3.2 ± 0.53 | 4.4 ± 0.57 | 4.2 ± 0.76 | 4.5 ± 0.67 | 2.0 ± 0.32 | 1.2 ± 0.23 | 8.2 ± 1.23 | 5.3 ± 0.95 |
| SRBmix | 5.8 ± 0.88 | 11.6 ± 2.43 | 9.0 ± 1.52 | 10.1 ± 2.56 | 28.3 ± 4.43 | 23.3 ± 4.54 | 7.7 ± 0.78 | 13.2 ± 2.22 |
| CHLmix | 1.7 ± 0.27 | 0 | 0.5 ± 0.01 | 0 ± 0 | 0.2 ± 0.02 | 0.4 ± 0.07 | 0.1 ± 0.01 | 0.1 ± 0.02 |
| SPIRO1400 | 0.5 ± 0.09 | 1.9 ± 0.32 | 1.7 ± 0.33 | 2.0 ± 0.21 | 1.4 ± 0.31 | 1.9 ± 0.33 | 0.4 ± 0.03 | 0.4 ± 0.07 |
| TM7-905 | 0.6 ± 0.08 | 0.8 ± 0.07 | 0.5 ± 0.01 | 0.1 ± 0.03 | 1.5 ± 0.23 | 0.2 ± 0.02 | 0.6 ± 0.02 | 0.3 ± 0.08 |
| HGC69a | 1.3 ± 0.28 | 2.1 ± 0.31 | 0.3 ± 0.06 | 0.3 ± 0.05 | 0.4 ± 0.03 | 0.1 ± 0.02 | 0.5 ± 0.09 | 0.2 ± 0.02 |
| RUM831 | 5.5 ± 0.13 | 5.7 ± 0.89 | 5.8 ± 0.73 | 8.9 ± 1.32 | 18.0 ± 4.13 | 23.8 ± 3.11 | 5.6 ± 1.14 | 7.4 ± 1.32 |
| RAL1436 | 0.4 ± 0.06 | 0.3 ± 0.03 | 0.2 ± 0.06 | 0.2 ± 0.03 | 0.3 ± 0.05 | 0.6 ± 0.09 | 0.7 ± 0.13 | 0.6 ± 0.12 |
| RFL155 | 0.7 ± 0.11 | 0.2 ± 0.03 | 0.3 ± 0.07 | 0.7 ± 0.19 | 0.1 ± 0.01 | 0.8 ± 0.11 | 0.5 ± 0.06 | 1.2 ± 0.34 |
| LAC435 | 25.5 ± 3.98 | 10.0 ± 1.51 | 9.6 ± 1.31 | 11.7 ± 1.67 | 12.6 ± 2.56 | 20.2 ± 3.23 | 9.3 ± 1.51 | 16.1 ± 3.31 |
| BFI826 | 0.3 ± 0.06 | 0.4 ± 0.05 | 0.4 ± 0.06 | 0.7 ± 0.12 | 0.5 ± 0.05 | 0.3 ± 0.08 | 2.4 ± 0.37 | 0.2 ± 0.02 |
| Fibr225 | 0 | 0 | 0.2 ± 0.04 | 0.1 ± 0.02 | 0.8 ± 0.14 | 0.7 ± 0.14 | 0.4 ± 0.11 | 0.1 ± 0.04 |
| ARCH915 | 0.3 ± 0.08 | 0.2 ± 0.07 | 0.6 ± 0.01 | 0.3 ± 0.07 | 0.6 ± 0.09 | 0.1 ± 0.02 | 0.4 ± 0.05 | 0.4 ± 0.06 |
| Total hybridizedb | 54.1 | 37 | 52.4 | 43.7 | 80.9 | 91.6 | 48 | 60.7 |
| Otherc | 45.9 | 63 | 47.6 | 56.3 | 19.1 | 8.4 | 52 | 39.3 |
The two numbers represent the mean value (%) and the standard deviation of individual probe-defined microbial groups in a specified rumen digest fraction, which were calculated based on 3 mean values, each consisting of 20 enumerations.
The numbers represent the sum of percentages of all individual probe-defined microbial groups in a specified rumen digest fraction. The percentages obtained with FISH probes RAL1436, RFL155, and BFI826 were not counted in the sum because the bacterial cells hybridizing with the former two probes also hybridized with RUM831, and the bacterial cells hybridizing with the last probe also hybridized with probe LAC435.
The numbers represent the percentages of microorganisms which were not identified by FISH in a specified rumen digest fraction.
We provided quantitative data by using qFISH to show that Bacteroidetes, Firmicutes, and Proteobacteria were abundant in both the LiqF and the SolF, constituting 31.8 to 87.3% of the total cell numbers. These FISH data add weight to the view that Firmicutes and Bacteroidetes might be dominant in rumens, as suggested previously from their high ratios retrieved from 16S rRNA clone libraries (e.g., see references 12, 26, and 27). However, information emerging from 16S rRNA gene clone library data cannot be used to reach conclusions on the quantitative composition of the rumen bacterial community. Bacteria may have 1 to 14 copies of rRNA genes, and several biases are known to be associated with their PCR amplification (8).
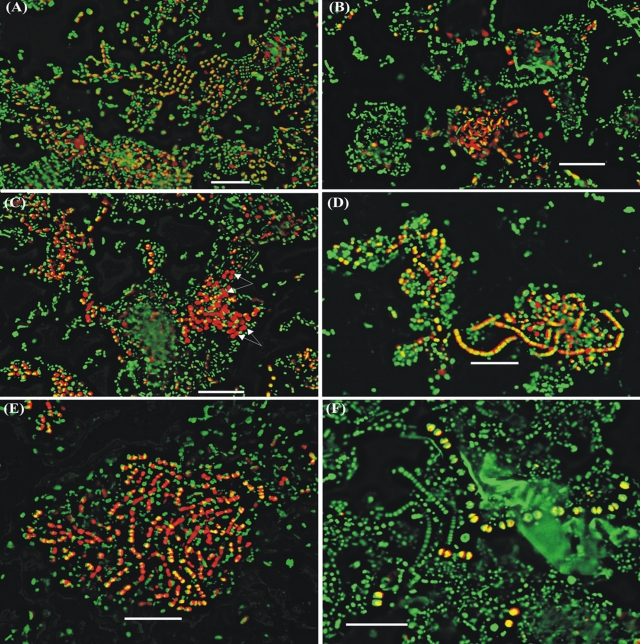
These 3 dominant bacterial groups have been identified at a high-resolution level. They belong mainly to the order Bacteroidales (0.1 to 19.2%), hybridizing with probe BAC1080 (Fig. 2 A); the families Lachnospiraceae (9.3 to 25.5%) and Ruminococcaceae (5.5 to 23.8%), hybridizing with LAC435 (Fig. 2E) and RUM831 (Fig. 2D), respectively; and the classes Deltaproteobacteria (5.8 to 28.3%) and Gammaproteobacteria (1.2 to 8.2%), hybridizing with SRBmix (equal moles of SRB385 and SRB385Db) (Fig. 2C) and GAM42a (Fig. 2B), respectively. All were more abundant in the microbial communities in the rumens of cows fed diets containing silage (75.2 to 87.3%) than in those in the rumens of cows fed diets containing hay (31.8 to 49.5%). These results show how diets containing different forages (hay or silage) may influence the distribution of the microbial populations, which is in line with data by Tajima et al. (25). We also found in this study that the addition of flaxseed (to inhibit methane emission) reduced their abundance in the rumens of cows fed silage-based diets (to 45.2 to 58.7%) but did not change markedly their abundance in the rumens of cows fed hay-based diets (31.8 to 49.5%), suggesting that adding flaxseed to these diets also affected rumen microbial community composition, although the extent of its influence reflected the forage used, being more profound with a silage-based diet than when hay was used.
FIG. 2.
Images of digest samples from the rumens of cows fed hay- or silage-based diets with and without flaxseed after color combination. Images from probes are labeled in red, and those from DAPI staining are in green. The yellow (combination of red and green), including those partly colored cells in panels A to F, hybridized with probes BAC1080, GAM42a, SRBmix, RUM831, LAC435, and ARCH915, respectively. A few cells (arrows) hybridizing with SRBmix (C) were not stained by DAPI. Bars, 10 μm.
We also present evidence here to suggest that Proteobacteria are common members of the microbial community, with sulfur-reducing bacteria (SRB) belonging to Deltaproteobacteria in particular being readily detected (up to 28% of the total cells) in both the LiqF and the SolF of rumen contents from cows fed the four different diets examined here. SRB have seldom been retrieved in clone libraries obtained from rumen samples. Lin et al. (15) have estimated SRB abundance in the rumen using DNA hybridization and concluded that they were of minor importance (0.7 to 0.8% of the total rRNA). Our estimates are much higher than those for every diet regime examined, possibly reflecting the coverage of the probes used in the two different studies. The probe mixture SRBmix used here targets most members of the Deltaproteobacteria, while those of Lin et al. (15) covered mainly members of the Desulfobacteraceae, Desulfovibrionaceae, and Desulfobulbaceae. We also recognized that the probe mixture SRBmix perfectly matched with the 16S rRNA genes of some bacteria other than SRB in Deltaproteobacteria. The possibility of overestimation of SRB cannot be ruled out. Interestingly, our data suggest that Gammaproteobacteria were abundant in some of the rumen communities we examined by FISH, comprising 1.2 to 8.2% of total cells.
The other unexpected finding was that the fibrolytic bacteria and archaeal methanogens accounted for only a minor fraction of the communities. Of the three characterized fibrolytic bacterial species, F. succinogenes was not detected in the rumen digesta from cattle fed the hay-based diet but was present in the remainder of the diets. In contrast, R. albus and R. flavefaciens were present in both the LiqF and the SolF of the rumen digesta from cows fed all four diets. Although the importance of these bacteria within the rumen microbial community cannot be denied, these three populations accounted for only 0.7 to 2.1% of the total microbial cells. This numerical range compares well with that determined previously for F. succinogenes (0.1 to 6.9% of total rRNA) (4, 23) and Ruminococcus spp. (1.5 to 2.9% of total rRNA) (11), considering that different animals and diets were used in those studies and that different specificities of the probes and different detection methods were used. However, this is much lower than the 9% (of total rRNA) detected by Michalet-Doreau et al. (19) in their work. The abundance of fibrolytic B. fibrisolvens-related species was also low, being present at <1% in all fractions, except in the LiqF in cows fed the mixture of silage and flaxseed, where they contributed 2.4% of total cells.
Methanogens hybridized to ARCH915 (Fig. 2F) were present (0.1 to 0.6%) in all rumen samples examined by FISH, which is close to or within the range (0.3 to 3.3%) estimated in other studies (15, 22). Interestingly, no marked difference in abundance of the methanogens could be seen between the samples from the rumens of cows fed diets with flaxseed and those from the rumens of cows fed diets without flaxseed, although it has been reported (7) that the addition of fatty acids could decrease methane production in the rumen. This may be due to the presence of methanogens with different activities in different rumen samples or the inability of probe ARCH915 to hybridize to all methanogens in the rumen samples examined here.
Bacteria belonging to Chloroflexi, TM7, Spirochetes, and Actinobacteria hybridizing with CHLmix, TM7-905, SPRO1400, and HGC69a, respectively, accounted for only a minor fraction of the total cell numbers observed. In most cases, their abundances in each fraction did not change markedly with diet, always being present in small numbers (0 to 1%), suggesting that they have a minor role there. This conclusion, however, has to be confirmed since many (8.4 to 63.0%, depending on diet) of the bacteria could not be identified in the rumens of cows fed with all diets except the silage-based diet (Table 3).
FISH with the probes designed in this study failed to identify all of the bacterial cells. This is because the probes do not target all rumen 16S rRNA gene sequences and/or the true extent of rumen biodiversity has not been revealed from cloning analyses. This indicates that our current understanding of the quantitative composition of the rumen microbial community is far from complete. Moreover, no physiological data were generated in this study to suggest what the role(s) of most of the dominant populations (except the SRB hybridized with probe SRBmix) identified by FISH might be, meaning that it is still not possible to link their abundance to their in situ function. Furthermore, each FISH-probed population probably includes bacteria with different phenotypes. Clearly, much needs to be done before the structure and function of the rumen microbial community are fully understood.
FISH is a useful tool in the investigation of microbial composition in complex ecosystems (3). However, FISH probes targeting rumen bacterial populations are limited. By comparison with other culture-independent methods, e.g., quantitative PCR, FISH has several advantages (8). In particular, in combination with histochemical staining methods (20) and microautoradiography (MAR-FISH) (13), the in situ ecophysiology of a targeted population can be determined under specified electron acceptor conditions. These techniques may provide important clues as to the functional role of microbial populations within complex communities, like that of the rumen. The possession of the FISH probes described in this paper could allow such studies to be undertaken in herbivore rumens.
Acknowledgments
Financial support for this study was provided by the Alberta Agricultural Research Institute and the Canadian Triticale Biorefinery Initiative through the Agricultural Bioproducts Initiative Program of Agriculture and Agri-Food Canada. Financial support for Y.K. was through a Peer Review Project of Agriculture and Agri-Food Canada.
We thank Jay Yanke at Lethbridge Research Centre (LRC) Culture Collection for providing fresh bacterial cultures.
Footnotes
Published ahead of print on 27 August 2010.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I. 1995. In situ hybridization of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecological manual. Kluwer Academic Publications, London, United Kingdom.
- 4.Briesacher, S. L., T. May, K. N. Grigsby, M. S. Kerley, R. V. Anthony, and J. A. Paterson. 1992. Use of DNA probes to monitor nutritional effects on ruminal prokaryotes and Fibrobacter succinogenes S85. J. Anim. Sci. 70:289-295. [DOI] [PubMed] [Google Scholar]
- 5.Bryant, M. P. 1959. Bacterial species of the rumen. Bacteriol. Rev. 23:125-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohme, F., A. Machmuller, A. Wasserfallen, and M. Kreuzer. 2000. Comparative efficiency of various fats rich in medium-chain fatty acids to suppress rumen methanogenesis as measured with RUSITEC. Can. J. Agric. Sci. 80:473-482. [Google Scholar]
- 8.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 9.Kong, Y. H., J. L. Nielsen, and P. H. Nielsen. 2005. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 71:4076-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong, Y. H., Y. Xia, J. L. Nielsen, and P. H. Nielsen. 2007. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plants. Microbiology 153:4061-4073. [DOI] [PubMed] [Google Scholar]
- 11.Krause, D. O., B. P. Dalrymple, W. J. Smith, R. I. Mackie, and C. S. McSweeney. 1999. 16S rDNA sequencing of Ruminococcus albus and Ruminococcus flavefaciens: design of a signature probe and its application in adult sheep. Microbiology 145:1797-1807. [DOI] [PubMed] [Google Scholar]
- 12.Larue, R., Z. Yu, V. A. Parisi, A. R. Egan, and M. Morrison. 2005. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 7:530-543. [DOI] [PubMed] [Google Scholar]
- 13.Lee, M., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescence in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leschine, S. B. 1995. Cellulose degradation in anaerobic environments. Annu. Rev. Microbial. 49:399-426. [DOI] [PubMed] [Google Scholar]
- 15.Lin, C. Z., L. Raskin, and D. A. Stahl. 1997. Microbial community structure in gastrointestinal tracts of domestic animals: comparative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol. 22:281-294. [Google Scholar]
- 16.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAllister, T. A., and K. J. Cheng. 1996. Microbial strategies in the ruminal digestion of cereal grains. Anim. Feed Sci. Technol. 62:29-36. [Google Scholar]
- 19.Michalet-Doreau, B., I. Fernandez, C. Peyron, L. Millet, and G. Fonty. 2001. Fibrolytic activities and cellulolytic bacterial community structure in the solid and liquid phases of rumen contents. Reprod. Nutr. Dev. 41:187-194. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen, J. L., K. Kragelund, and P. H. Nielsen. 2010. Ecophysiological analysis of microorganisms in complex microbial systems by combination of fluorescence in situ hybridization with extracellular staining techniques, p. 117-128. In S. P. Cummings (ed.), Bioremediation: methods and protocols. Humana Press Inc., Portland, OR. [DOI] [PubMed]
- 21.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 22.Sharp, R., C. J. Ziemer, M. D. Stern, and D. A. Stahl. 1998. Taxon-specific associations between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiol. 26:71-78. [Google Scholar]
- 23.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart, C. S., H. J. Flint, and M. P. Bryant. 1980. The rumen bacteria, p. 10-46. In P. N. Hobson and C. S. Stewart (ed.), The rumen microbial ecosystem. Blackie Academic & Professional, London, United Kingdom.
- 25.Tajima, K., R. I. Aminov, T. Nagamine, H. Matsui, M. Nakamura, and Y. Benno. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tajima, K., I. Nonaka, K. Higuchi, N. Takusari, M. Kurihara, A. Takenaka, M. Mitsumori, H. Kajikawa, and R. I. Aminov. 2007. Influence of high temperature and humidity on rumen bacterial diversity in Holstein heifers. Anaerobe 13:57-64. [DOI] [PubMed] [Google Scholar]
- 27.Whitford, M. F., R. J. Forster, C. E. Beard, J. Gong, and R. M. Teather. 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4:153-163. [DOI] [PubMed] [Google Scholar]