Abstract
The study site located in the Mediterranean Sea was visited eight times in 2005 and 2006 to collect samples from the epipelagic (5 m), mesopelagic (200 m, 600 m), and bathypelagic (1,000 m, 2,000 m) zones. Randomly amplified polymorphic DNA PCR (RAPD-PCR) analysis was used to obtain fingerprints from microbial and viral size fractions using two different primers each. Depending on the primer used, the number of bands in the water column varied between 12 to 24 and 6 to 19 for the microbial size fraction and between 16 to 26 and 8 to 22 for the viral size fraction. The majority of sequences from the microbial fraction was related to Alphaproteobacteria, Cyanobacteria, Gammaproteobacteria, Firmicutes, and Eukaryota. Only 9% of sequences obtained from the viral fraction were of identifiable viral origin; however, 76% of sequences had no close relatives in the nr database of GenBank. Only 20.1% of complete phage genomes tested in silico resulted in potential RAPD-PCR products, and only 12% of these were targeted by both primers. Also, in silico analysis indicated that RAPD-PCR profiles obtained by the two different primers are largely representative of two different subsets of the viral community. Also, correlation analyses and Mantel tests indicate that the links between changes in the microbial and viral community were strongest in the bathypelagic. Thus, these results suggest a strong codevelopment of virus and host communities in deep waters. The data also indicate that virus communities in the bathypelagic zone can exhibit substantial temporal dynamics.
Viruses comprise the most abundant biological entities in the ocean (29) and play an integral part in global geochemical cycles (12, 35). Also, viruses have been proposed to influence the community composition of their hosts by selectively infecting the winners in the competition for nutrients (32, 33, 38). Indeed, the viral influence on prokaryotic community composition has been demonstrated in several studies (see, e.g., references 5, 26, and 41).
The community composition of prokaryotes or eukaryotic single-celled organisms can be investigated by PCR-based fingerprinting techniques based on small subunit rRNA genes or their flanking regions. However, viruses lack such conserved genes, and consequently obtaining genetic data on the virus community in a water sample is challenging. Several authors have used primers specific for certain virus groups, e.g., Phycodnaviridae (8, 27) or picornaviruslike viruses (11). Undoubtedly, metagenomics currently offers the most comprehensive genetic data set on an entire viral community (see, for example, references 2, 4, and 6) and has become accessible to most research groups due to ever decreasing costs. Nevertheless, the challenge of adequately analyzing the large data sets yielded by metagenomics remains.
Recently, Winget and Wommack (37) demonstrated the use of randomly amplified polymorphic DNA PCR (RAPD-PCR) to assess viral community composition. These authors showed that the banding patterns obtained by RAPD-PCR from viral communities are highly reproducible, making RAPD-PCR a valuable high-throughput and low-cost technique to assess viral community composition on a routine basis. Other studies have used RAPD-PCR for strain typing of closely related viruses (9) and to study the benthic viral community of the Chesapeake Bay (13), the pelagic viral community during an iron-induced phytoplankton bloom in the Southern Ocean (34), and virus-host interactions at hydrothermal vents (36). Since RAPD-PCR is a relatively novel fingerprinting approach to study complex viral communities, one may still ask the question of how to best interpret such results in an ecological context. To address this, we performed RAPD-PCR analysis on viral and microbial communities (prokaryotes and single-celled planktonic eukaryotes) at a station in the northwestern Mediterranean Sea to determine their temporal and depth variation. The results were coupled with in silico RAPD-PCR analysis of available whole viral genomes and sequence analysis of the DNA sequences obtained from selected RAPD-PCR bands. Finally, the RAPD-PCR banding patterns were used to relate changes in the viral and microbial communities with each other and with other microbial and environmental parameters to better understand the mechanism driving temporal and depth variation of viral and microbial communities at the study site.
MATERIALS AND METHODS
Study site and sampling.
Sampling was conducted at the Dynamique des Flux Atmosphériques en Mediterranée (DYFAMED) site located in the Ligurian Basin of the Mediterranean Sea (43°25′N, 07°52′E). The site was visited eight times in 2005 and 2006 (27 September 2005, 25 October 2005, 19 December 2005, 7 February 2006, 7 March 2006, 2 April 2006, 6 May 2006, and 30 June 2006). The sampling scheme covered depths of 5, 200, 600, 1,000, and 2,000 m. Due to differences in abundances of viruses and microbes over depth, we sampled 12 liters from a depth of 5 m, 24 liters from a depth of 200 m, and 36 liters from a depth of 600 to 2,000 m. Sampling was performed using 12-liter Niskin bottles mounted on a carousel sampler (SBE32; Sea-Bird Electronics). The samples were amended with sodium azide (final concentration, 0.005%) to prevent changes in microbial and viral community composition and stored at 4 to 10°C until further processing within 24 to 36 h after sampling. Details on the structuring of the water column (salinity, temperature, potential density), the bacterial and archaeal community composition analyzed by denaturing gradient gel electrophoresis (DGGE) and catalyzed reporter deposition fluorescence in situ hybridization, and the standing stocks of prokaryotes and viruses are reported elsewhere (39, 40).
RAPD-PCR analyses.
Microorganisms and viruses were concentrated by using sequential tangential-flow ultrafiltration (Vivaflow 200, 0.22-μm pore size and 100-kDa molecular mass cutoff, PES; Vivasciences) until a final volume of 30 ml for each concentrate was reached. The microorganisms in the >0.22-μm fraction were harvested by low-speed centrifugation (3,500 × g, 4°C, 15 min), resuspended in 1 ml of filtered and autoclaved seawater, pelleted again, and resuspended in 45 μl of filtered and autoclaved seawater. Viruses in the 100-kDa, 0.22-μm fraction were harvested by ultracentrifugation at 105 × g and 4°C for 5 h (L8-55M, rotor SW28 at 27.5 × 103 rpm; Beckman Coulter) and resuspended overnight in 1 ml of MSM buffer (450 mM NaCl, 50 mM MgSO4, 50 mM Tris-HCl [pH 8.0], 0.005% [wt/vol] glycerol). Subsequently, the virus concentrates were centrifuged at 105 × g and 4°C for 4 h (L8-55M, rotor SW55 at 32.5 × 103 rpm; Beckman Coulter) and resuspended overnight in 45 μl of MSM buffer. The microbial and viral concentrates were checked for contamination by flow-cytometry (FACSCalibur; BD Biosciences) according to previously published protocols (7, 19).
The following protocol is frequently used to prepare samples for pulsed-field gel electrophoresis and may not be necessary for RAPD-PCR (37). Agarose plugs were prepared by mixing either microbial or viral concentrates with 45 μl of molten 2% low-melting-point agarose (catalog no. 162-0137; Bio-Rad) in 1× TBE buffer (90 mM Tris-HCl [pH 8.3], 90 mM boric acid, 2 mM EDTA) using plug molds. Plugs from the >0.22-μm fraction were submerged in 1 ml of lysozyme buffer (10 mM Tris-HCl [pH 7.2], 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, 1 mg of lysozyme [Sigma, catalog no. L7651] ml−1) and incubated at 37°C for 1 h. Subsequently, plugs were washed in 50 ml of wash buffer (20 mM Tris [pH 8.0], 50 mM EDTA) for 10 min and incubated in 1 ml of proteinase K buffer (100 mM EDTA [pH 8.0], 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1 mg of proteinase K [Sigma, catalog no. 82456] ml−1) at 50°C for 14 h. Finally, the plugs were washed four times in 50 ml of wash buffer for 1 h each with the washing buffer for the second step containing 1 mM phenylmethylsulfonyl fluoride to inactivate any residual proteinase K. Treatment of the plugs from the viral concentrates followed the same procedure, excluding the lysozyme step. Plugs were stored in 15 ml of wash buffer at 4°C until analysis. The amount of molten agarose containing the template DNA used in RAPD-PCR was adjusted to obtain adequate levels of PCR products (≥100 ng μl−1) and varied between 2 to 5 μl. For each microbial and viral concentrate two PCRs using either primer CRA-22 (5′-CCG CAG CCA A-3′) or OPA-13 (5′-CAG CAC CCA C-3′ 42]; MWG Biotech) were performed, including negative controls containing no template DNA. PCR chemicals were from MBI Fermentas, and each 50-μl PCR mixture contained 5 μl of 10× Taq buffer (100 mM Tris-HCl [pH 8.8], 500 mM KCl, 0.8% Nonidet P-40), 4 μl of 25 mM MgCl2, 1.25 μl of 10 mM deoxynucleoside triphosphate mix, 0.5 μl of 100 μM primer, and 0.25 μl of a 5-U μl−1 Taq polymerase solution. The cycling protocol was as previously described (42), except that the final elongation step lasted 30 min (15). PCR fragments were cleaned and concentrated by using a QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions, resulting in a final volume of 28 ml in elution buffer (Qiagen). The PCR products including the negative controls were initially checked and quantified on agarose gels using a molecular mass standard. For high-resolution separation of the RAPD-PCR products, 200 ng of the products per sample was separated electrophoretically on 2.5% standard agarose gels in 1× TBE run at 5 V cm−1 for 140 min. The gels were stained with SYBR Gold (1:10,000 dilution of stock solution; Invitrogen/Molecular Probes) for 30 min, and images were obtained by using a Gel Doc EQ (Bio-Rad) gel documentation system. The images were analyzed for the number of bands per sample (presence versus absence). The band patterns were translated into a binary data matrix for further statistical analysis as previously described (20) (see Fig. S1 to S4 in the supplemental material).
In silico RAPD-PCR analysis of complete phage genomes.
To determine whether the primers CRA-22 and OPA-13 target the same or different groups of viruses, we performed in silico RAPD-PCR analysis using 424 complete phage genomes retrieved from GenBank. The tool PrimerSearch (part of the EMBOSS toolkit; version 6.0) was used to obtain the number and size of potential PCR products for each of the complete phage genomes tested. A 10% mismatch between primers and target sequences was allowed, corresponding to 1 mismatched nucleotide per primer. Only PCR products in the size range of 20 to 3,000 bp were taken into account for further analysis.
Sequencing and phylogenetic analysis.
Selected RAPD-PCR bands were excised from the gels, and the nucleic acids were extracted by using a QIAquick gel extraction kit (catalog no. 28706; Qiagen). The extracted fragments were reamplified and reanalyzed as described above using original samples as standards to confirm correct placement of the excised bands on the gels. Reamplified RAPD-PCR fragments were cloned and sequenced by a commercial sequencing service (MWG Biotech). DNA sequences obtained from RAPD-PCR fragments were analyzed by searching the nr and env_nt databases of GenBank using tblastx (searching a translated nucleotide database using a translated nucleotide query [1]). Hits were assumed to be significant at expectation values of ≤10−3.
Statistical analyses.
All statistical analyses were performed in Mathematica (version 7.0.1; Wolfram Research). Spearman rank correlation coefficients were used to evaluate the relationships between parameters. Only statistically significant (P ≤ 0.05) and relevant (−0.5 > r > 0.5) correlations were taken into account. Differences in the average values between two parameters were tested by using a Student t test. Samples where grouped according to depth layers (epipelagic, 5 m; mesopelagic, 200 and 600 m; bathypelagic, 1,000 and 2,000 m), and an analysis of variance (ANOVA) was performed to test for differences between these depth layers. Mantel tests (18) were used to link changes in the banding patterns of the microbial and viral size fraction, as determined by RAPD-PCR with each other and with changes in bacterial community composition (39). For this purpose, the banding patterns determined by RAPD-PCR and for the bacterial community by DGGE (39) were converted into dissimilarity matrices by calculating Jaccard's dissimilarity coefficient. The Mantel statistic (rm) was calculated as the Spearman rank correlation coefficient between the upper nondiagonal values of the dissimilarity matrices being compared. Probability values for the null hypothesis of the Mantel tests were calculated by using permutation tests (17) based on 100 random permutations, sufficient to obtain P values accurately to two decimal places. The results of the statistical tests were assumed to be significant at P values of ≤0.05.
Nucleotide sequence accession numbers.
In all, 84 distinct nucleotide sequences (39 from the >0.22-μm fraction and 45 from the viral size fraction) were obtained from RAPD-PCR fragments. The sequences were deposited into GenBank under accession numbers ET671171 to ET671254.
RESULTS
Variability in microbial and viral RAPD-PCR data.
The >0.22-μm size fraction contained mainly prokaryotes and single-celled eukaryotic organisms, whereas the 100-kDa to 0.22-μm size fraction exclusively contained viruses, as validated by flow cytometry. Considering that RAPD-PCR data for the microbial and viral size fraction obtained by the primers CRA-22 and OPA-13 constitute two sets of discriminatory parameters (the presence or absence of bands), it is legitimate to concatenate the data sets of the two primers. Henceforth, data obtained by RAPD-PCR analysis from the >0.22-μm size fraction will be indicated by MCRA (primer CRA-22), MOPA (primer OPA-13), and MICRO (data for both primers combined). Likewise, data obtained by RAPD-PCR analysis from the 100-kDa to 0.22-μm size fraction will be designated VCRA (primer CRA-22), VOPA (primer OPA-13), and VIR (data for both primers combined).
The number of bands in the water column for MCRA, MOPA, and MICRO varied between 12 and 24, 6 and 19, and 21 and 41, respectively (Table 1). For VCRA, VOPA, and VIR, the number of bands in the water column varied between 16 and 26, 8 and 22, and 26 and 46, respectively (Table 1). The number of bands in VCRA, VOPA, and VIR was significantly higher than in MCRA, MOPA, and MICRO (Student t test: VCRA-MCRA, P = 0.0038; VOPA-MOPA, P < 0.0001; and VIR-MICRO, P < 0.0001). The number of bands detected in MCRA, MOPA, and MICRO was significantly lower in the epipelagic compared to the meso- and bathypelagic (ANOVA: MCRA, F ratio = 7.98, P = 0.0017; MOPA, F ratio = 7.96, P = 0.0018; and MICRO, F-ratio = 18.59, P < 0.0001). However, the number of bands in VCRA, VOPA, and VIR did not differ between depth-layers (ANOVA: VCRA, F ratio = 1.53, P = 0.2394; VOPA, F ratio = 0.81, P = 0.4562; and MICRO, F ratio = 1.47, P < 0.2520). In the water column and the three depth layers, the number of bands for MCRA was significantly higher than for MOPA (Student t test: in every case, P < 0.0001). Similarly, the number of bands for VCRA was significantly higher than for VOPA in the water column and in the meso- and bathypelagic (Student t test: water column, P < 0.0001; mesopelagic, P = 0.0082; and bathypelagic, P = 0.0096) but not in the epipelagic zone (Student t test: P = 0.1717).
TABLE 1.
Number of bands in RAPD-PCR banding patternsa
| Fraction | Parameter |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avg (SD) |
Range (no. of bands) |
No. of samples |
||||||||||
| Total | Epi. | Meso. | Bathy. | Total | Epi. | Meso. | Bathy. | Total | Epi. | Meso. | Bathy. | |
| MCRA | 19 (2.4) | 17 (2.5) | 19 (1.7) | 20 (2.4) | 12-24 | 12-19 | 16-21 | 16-24 | 40 | 8 | 16 | 16 |
| MOPA | 13 (3.1) | 11 (1.9) | 13 (3.2) | 14 (3.0) | 6-19 | 8-14 | 6-19 | 9-19 | 39 | 8 | 16 | 15 |
| MICRO | 32 (4.7) | 27 (3.6) | 32 (4.3) | 34 (4.0) | 21-41 | 21-33 | 24-40 | 27-41 | 39 | 8 | 16 | 15 |
| VCRA | 21 (2.9) | 20 (3.2) | 20 (3.0) | 22 (2.7) | 16-26 | 17-25 | 16-25 | 18-26 | 30 | 6 | 12 | 12 |
| VOPA | 18 (2.8) | 17 (4.2) | 17 (2.0) | 18 (2.7) | 8-22 | 8-20 | 13-21 | 13-22 | 34 | 7 | 13 | 14 |
| VIR | 38 (5.0) | 37 (6.5) | 38 (4.4) | 39.8 (4.8) | 26-46 | 26-44 | 29-43 | 31-46 | 30 | 6 | 12 | 12 |
The table gives the number of bands for the microbial (MCRA, MOPA, and MICRO) and viral (VCRA, VOPA, and VIR) size fractions obtained by RAPD-PCR analysis for the entire water column and in the three depth layers (i.e., the epi-, meso-, and bathypelagic zones [Epi., Meso., and Bathy., respectively]). The averages (rounded to integers), standard deviations, ranges, and numbers of samples are given.
Relationships of microbial and viral RAPD-PCR data with other parameters.
Data on bacterial community composition and richness determined by DGGE analysis of PCR-amplified fragments of the 16S rRNA gene and prokaryotic and viral abundance were reported previously (39, 40). Here, we use these data to determine their relationships to the microbial and viral RAPD-PCR data. Physicochemical parameters (salinity, temperature, and potential density) were only occasionally and weakly related to changes in the number of bands or changes in microbial and viral community composition (data not shown).
Changes in the number of bands.
In the water column, the number of bands in VCRA, VOPA, and VIR correlated negatively with prokaryotic abundance (Table 2). Also, VIR correlated negatively with bacterial richness (Table 2). However, the number of bands in MICRO and VIR did not correlate with each other, nor did MCRA and MOPA correlate with VCRA or VOPA in the entire water column (Table 3). In the epipelagic zone, MOPA correlated positively with VIR. In the mesopelagic zone, VCRA, VOPA, and VIR correlated negatively with prokaryotic abundance (Table 2). Also, in the mesopelagic zone, VCRA correlated positively with MOPA and MICRO (Table 3). In the bathypelagic zone, prokaryotic abundance correlated positively with MOPA and MICRO (Table 2) and negatively with VOPA and VIR (Table 3). Also, in the bathypelagic zone, VOPA and VIR correlated negatively with viral abundance and bacterial richness (Table 2). A common feature for all depth layers was that all correlations between prokaryotic abundance and VCRA, VOPA, and VIR were negative (Table 2).
TABLE 2.
Spearman rank correlation coefficients between the number of bands detected in RAPD-PCR data and other biological parameters for the entire water column and for the epi-, meso-, and bathpelagic zonesa
| Zone | Parameter | Spearman rank coefficient (r) |
|||||
|---|---|---|---|---|---|---|---|
| MCRA | MOPA | MICRO | VCRA | VOPA | VIR | ||
| Water column | Prokaryotic abundance | -0.60 | -0.56 | -0.67 | |||
| Viral abundance | |||||||
| Richness-Bacteria | -0.54 | ||||||
| Epipelagic | Prokaryotic abundance | ||||||
| Viral abundance | |||||||
| Richness-Bacteria | |||||||
| Mesopelagic | Prokaryotic abundance | 0.65 | -0.62 | -0.69 | |||
| Viral abundance | |||||||
| Richness-Bacteria | |||||||
| Bathypelagic | Prokaryotic abundance | 0.57 | 0.58 | -0.64 | -0.69 | ||
| Viral abundance | -0.73 | -0.60 | |||||
| Richness-Bacteria | -0.59 | -0.76 | |||||
The table gives the Spearman rank correlation coefficients between the number of bands detected in microbial (MCRA, MOPA, and MICRO) and viral (VCRA, VOPA, and VIR) RAPD-PCR data. Additionally, correlations with bacterial richness (based on DGGE analysis [39]) and prokaryotic and viral abundance (40) are given. Only statistically significant (P ≤ 0.05) and relevant (−0.5 > r > 0.5) correlation coefficients are given.
TABLE 3.
Spearman rank correlation coefficients between the number of bands detected in RAPD-PCR data for the entire water column and in epi-, meso-, and bathypelagic zonesa
| Zone | Parameter | Spearman rank coefficient (r) |
|||
|---|---|---|---|---|---|
| MCRA | MOPA | MICRO | VCRA | ||
| Water column | MOPA | ||||
| VCRA | |||||
| VOPA | |||||
| VIR | |||||
| Epipelagic | MOPA | ||||
| VCRA | |||||
| VOPA | |||||
| VIR | 0.82 | ||||
| Mesopelagic | MOPA | ||||
| VCRA | 0.60 | 0.59 | |||
| VOPA | |||||
| VIR | |||||
| Bathypelagic | MOPA | ||||
| VCRA | |||||
| VOPA | -0.59 | ||||
| VIR | -0.64 | ||||
The table gives the Spearman rank correlation coefficients between the number of bands detected in microbial (MCRA, MOPA, and MICRO) and viral (VCRA, VOPA, and VIR) RAPD-PCR data. Only statistically significant (P ≤ 0.05) and relevant (−0.5 > r > 0.5) correlation coefficients are given.
Changes in the banding patterns.
Schematic representations of the banding patterns for MCRA, MOPA, VCRA, and VOPA can be found in Fig. S5 to S8 in the supplemental material. In the entire water column, changes in MICRO were linked to changes in the banding patterns of VIR (Table 4). Also, changes in the bacterial community composition were linked to changes in MOPA and VCRA. In the epipelagic zone, changes in the bacterial community composition determined by DGGE (39) were linked to changes in MCRA and MICRO, and changes in VCRA were linked to changes in MOPA and MICRO. In the mesopelagic zone, changes in the bacterial community were linked to changes in the banding patterns of MOPA. In the bathypelagic zone, the banding patterns in VCRA changed with bacterial community composition. Also, in the bathypelagic zone, changes in the banding patterns of VOPA and VIR were linked to changes in MCRA and MICRO. The most interesting finding was that links between changes in the microbial and viral compartments were most pronounced in the bathypelagic zone.
TABLE 4.
Mantel tests for the entire water column and in epi-, meso-, and bathypelagic zonesa
| Zone | Parameter | Mantel test (rm) |
|||||
|---|---|---|---|---|---|---|---|
| MCRA | MOPA | MICRO | VCRA | VOPA | VIR | ||
| Water column | MOPA | ||||||
| VCRA | |||||||
| VOPA | |||||||
| VIR | 0.53 | 0.56 | |||||
| DGGE-Bacteria | 0.53 | 0.53 | |||||
| Epipelagic | MOPA | ||||||
| VCRA | 0.59 | 0.55 | |||||
| VOPA | 0.51 | ||||||
| VIR | |||||||
| DGGE-Bacteria | 0.61 | 0.57 | |||||
| Mesopelagic | MOPA | ||||||
| VCRA | |||||||
| VOPA | |||||||
| VIR | |||||||
| DGGE-Bacteria | 0.57 | ||||||
| Bathypelagic | MOPA | ||||||
| VCRA | |||||||
| VOPA | 0.64 | 0.62 | |||||
| VIR | 0.59 | 0.53 | |||||
| DGGE-Bacteria | 0.61 | ||||||
Results of the Mantel tests performed to detect relationships between changes in microbial (MCRA, MOPA, and MICRO) and viral (VCRA, VOPA, and VIR) fingerprints obtained by RAPD-PCR data and bacterial community composition (fingerprints of DGGE-Bacteria [39]). Only statistically significant (P ≤ 0.05) and relevant (rm > 0.5) Mantel statistics are given.
In silico RAPD-PCR analysis of viral target sequences.
Of a total of 424 complete phage genomes tested, primer CRA-22 gave potential PCR products for 49 (11.6% of the total) and primer OPA-13 for 36 (8.5% of the total) viral genomes, allowing for one mismatched nucleotide between primer and target. Thus, both primers targeted 85 virus genomes (20.1% of the total). Under stringent conditions, no potential PCR product was obtained for the primer CRA-22, and only two virus genomes resulted in PCR products with the primer OPA-13. From these 85 virus genomes, only 10 (12%) were targeted by both primers. In summary, 67 to 74% of the successfully targeted phage genomes yielded one potential RAPD-PCR fragment, while 20 to 25% yielded two fragments, and 6 to 8% yielded three or more fragments (Table 5).
TABLE 5.
Insilico RAPD-PCR analysisa
| Primer | % Virus genomes (no. of genomes) |
|||
|---|---|---|---|---|
| One band | Two bands | ≥3 bands | Total | |
| CRA-22 | 73.5 (36) | 20.4 (10) | 6.1 (3) | 100 (49) |
| OPA-13 | 66.7 (24) | 25.0 (9) | 8.3 (3) | 100 (36) |
The table gives the percentages and numbers of virus genomes that resulted in 1, 2, and ≥3 PCR products using the primers CRA-22 and OPA-13.
Sequence analyses of RAPD-PCR fragments.
We obtained 16 nucleotide sequences from MCRA, 23 from MOPA, 23 from VCRA, and 22 from VOPA, totaling 84 distinct nucleotide sequences (see Tables S1 to S4 and Fig. S5 to S8 in the supplemental material). The sequences varied in length between 170 to 570 bp for MCRA, 238 to 864 bp for MOPA, 173 to 1,040 bp for VCRA, and 185 to 700 bp for VOPA. The GC content averaged 46.4% for MCRA, 49.0% for MOPA, 47.9% for MICRO, 41.8% for VCRA, 44.0% for VOPA, and 42.9% for VIR. Also, the GC content of MICRO was significantly higher than of VIR (Student t test: P = 0.0015). For MCRA, MOPA, MICRO, VCRA, VOPA, and VIR the sequence length did not correlate with the GC content of the sequences (in every case −0.5 ≤ r ≤ 0.5).
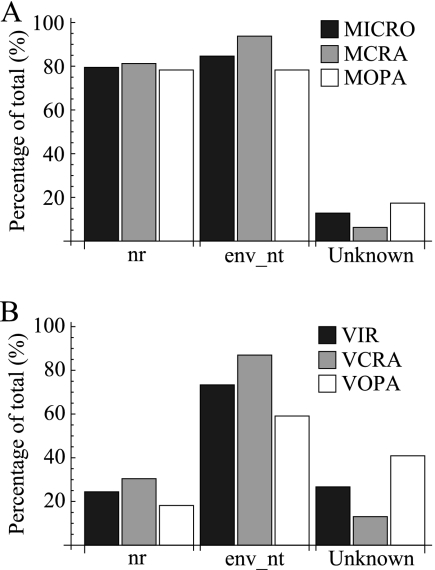
Between 79 and 81% of the microbial nucleic acid sequences had significant hits in the nr database of GenBank and between 78 and 94% in the env_nt database (Fig. 1 A). Only between 6 and 17% of the microbial sequences had no significant hits in either database (Fig. 1B). In contrast, only 18 to 30% of viral sequences had known relatives in the nr database, whereas, 59 to 87% had significant hits in the env_nt database (Fig. 1B). The percentage of viral sequences with no hits in either database ranged between 13 and 41% (Fig. 1B).
FIG. 1.
Percentage of significant hits in GenBank. The figure shows the percentage of significant hits in the nr and env_nt databases of GenBank for the microbial (A) and viral (B) sequences obtained from RAPD-PCR bands. Also, the percentage of sequences for which no close relatives could be found in either database is shown.
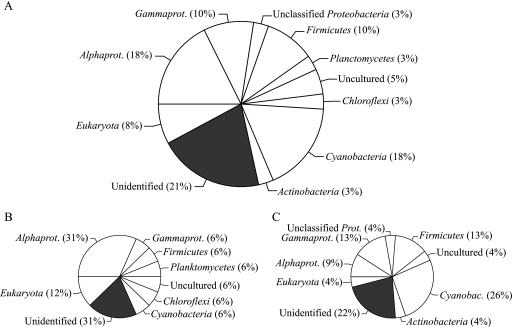
Phylogenetic analyses revealed that 18% of the sequences obtained from MICRO could each be attributed to Alphaproteobacteria and Cyanobacteria and that 21% of the sequences had no significant hit in the nr database of GenBank (Fig. 2 A). Other significant fractions of the sequences had close relatives among Gammaproteobacteria (10%), Firmicutes (10%), and Eukaryota (8%). The picture was different when we analyzed MCRA and MOPA separately (Fig. 2B and C). On the one hand, MCRA was dominated by Alphaproteobacteria (31%), unidentified sequences (31%), and Eukaryota (12%; Fig. 2B). On the other hand, Cyanobacteria (26%), unidentified sequences (22%), Gammaproteobacteria (13%), and Firmicutes (13%) constituted the largest fractions in MOPA. Interestingly, no sequences related to Archaea were obtained from MCRA or MOPA (Fig. 2, see Tables S1 and S2 in the supplemental material).
FIG. 2.
Taxonomic breakdown of significant hits to nucleic acid sequences obtained from microbial concentrates. The pie chart shows the number of significant hits in each of the phylogenetic groups (as a percentage of the total) based on comparing the nucleic acid sequences obtained from MICRO (A), MCRA (B), and MOPA (C) to the nr database of GenBank using tblastx.
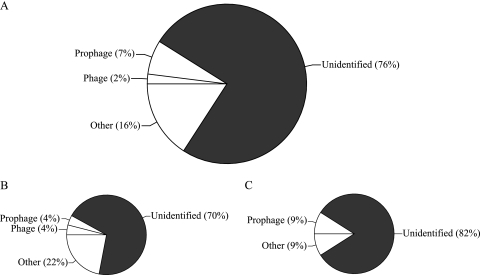
Only small fractions of sequences in VIR (9%), VCRA (8%), and VOPA (9%) were associated with known viruses (Fig. 3). Other sequences in VIR (16%), VCRA (22%), and VOPA (9%) had significant hits in the groups Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Cyanobacteria, Euryarchaeota, and Eukaryota (Fig. 3, see Tables S3 and S4 in the supplemental material).
FIG. 3.
Taxonomic breakdown of significant hits to nucleic acid sequences obtained from viral concentrates. The pie chart shows the number of significant hits to phage- and prophage-related sequences, as well as other phage-unrelated sequences (as a percentage of the total) based on comparing the nucleic acid sequences obtained from VIR (A), VCRA (B), and VOPA (C) to the nr database of GenBank using tblastx. The category “other” corresponds to significant hits in the groups Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Cyanobacteria, Euryarchaeota, and Eukaryota.
DISCUSSION
Critical evaluation of the approach.
The preparation of microbial and viral concentrates is particularly crucial when using RAPD-PCR, because the primers are not specific for any particular group of organisms or viruses (13, 37). In the present study, there is evidence that the microbial and viral concentrates, and thus subsequent analyses, were not affected by cross-contamination. (i) Flow cytometric analysis of the concentrates did not indicate cross-contamination. (ii) Sequences detected in MCRA and MOPA were not detected in VCRA or VOPA (see Tables S1 to S4 in the supplemental material). (iii) A majority of sequences obtained from VCRA and VOPA had no significant hit in the nr database of GenBank, whereas the majority of sequences from MCRA and MOPA had close relatives in this database (Fig. 1). In summary, these results indicate that the filtration and subsequent cleaning procedure, consisting of several washing steps, to obtain the microbial and viral concentrates was successful.
The majority of cloned and subsequently sequenced microbial or viral RAPD-PCR bands obtained from either primer yielded one nucleic acid sequence. However, occasionally, we obtained more than one distinct nucleic acid sequence from a single cloned band regardless of which primer was used (see Fig. S1 to S4 in the supplemental material). Similar results have been reported previously (13, 37) and in the case of benthic viral assemblages in an estuary have been interpreted as evidence for similar genes being widely distributed (13). Thus, the actual number of different viral and microbial target sequences appears to be higher than what can be distinguished on the agarose gels. These results indicate a limitation of the method that is similar to other nucleic acid based fingerprinting methods such as DGGE analysis (see also reference 23).
Interpreting viral RAPD-PCR profiles.
The use of RAPD-PCR to obtain fingerprints of viral communities is a relatively new approach (13, 34, 36, 37), but it was used previously to obtain nucleic acid probes specific for virus populations (42). Although other authors demonstrated the usefulness and reliability of RAPD-PCR analysis to obtain fingerprints of viral communities (13, 37), questions remain on the interpretation of the results in an ecological context. To get a better idea of what the targets are and what the differences between PCR products obtained by the primers CRA-22 and OPA-13 might be, we performed in silico analyses using complete phage genomes retrieved from GenBank. Although the results need to be interpreted carefully due to the small sample size (424 complete phage genomes tested), the results reveal a number of interesting trends. First, when stringent conditions were chosen, i.e., no mismatches were allowed, we obtained no PCR products for the primer CRA-22, and only two virus genomes gave PCR products when the primer OPA-13 was used. Even under less stringent conditions allowing for one mismatched nucleotide between primer and target sequence, only 20.1% of the virus genomes tested yielded potential PCR fragments. There are two possible, but not mutually exclusive conclusions from these results. (i) Even if both primers are used to obtain viral fingerprints, the results will only be based on a relatively small subset of the viruses in the sample. (ii) The currently available complete phage genomes in the nr database of GenBank are indeed not representative for marine pelagic virus communities. The relatively small percentage of identifiable RAPD-PCR sequences compared to the nr database of GenBank versus the much higher number of significant hits in the env_nt database (Fig. 1) would argue for the latter conclusion (see also reference 2).
The finding that only 12% of the successfully targeted phage genomes gave potential PCR fragments with both primers, suggests that the profiles obtained by CRA-22 and OPA-13 are largely representative of two different subsets of the viral community. This conclusion is further supported by the lack of a correlation in the number of bands detected for VCRA and VOPA in the entire water column and the three depth layers (Table 3). Also, changes in the banding patterns in VCRA and VOPA were not linked in the entire water column and the meso- and bathypelagic (Table 4), further supporting the idea that the two primers target different groups of the viral community in the investigated environments.
This brings up the question of how representative RAPD-PCR data are for any given viral community. It has been argued that conventional PCR-based profiling techniques such as DGGE or terminal restriction fragment length polymorphism analysis target the more abundant phylotypes of the community; however, the exact relative contribution of each of the signals cannot be given with certainty due to biases introduced by the PCR amplification (3, 25, 30). The results of PCR-based fingerprinting techniques of prokaryotic communities using the 16S rRNA gene as a target are sometimes interpreted as a measure of apparent richness, although a lot more phylotypes might be present in any one sample based on massively parallel tag sequencing (28). In addition, the number of 16S rRNA genes per prokaryotic genome is not constant and can vary widely between different phylotypes adding to the complication of interpreting such results quantitatively (10, 16). This also suggests that in the case of conventional prokaryotic fingerprints (e.g., DGGE), the number of bands does not necessarily equal the number of dominant phylotypes and that a certain amount of variation is inherently present in such data (23). Thus, this issue is also pertaining to profiling techniques targeting specific genes such as the 16S rRNA gene, and yet those techniques have proven wildly successful. Since the kinetics of PCR amplification in RAPD-PCR are the same as in the analysis of PCR-amplified fragments of the 16S rRNA gene, the same arguments hold. According to our in silico analysis conducted with complete phage genomes, the most important choice to make in RAPD-PCR analysis of viral communities is which primer to use. Based on our data, the primer CRA-22 resulted in a higher number of bands than the primer OPA-13 for the viral and the microbial fraction (Table 1). Thus, primer CRA-22 appears to be more discriminative than OPA-13 in the investigated environment.
Interpretation of microbial RAPD-PCR profiles.
Viral infection is a stochastic process depending on the abundance of hosts (21). Thus, it is reasonable to assume and supported by observations (7, 22) that the majority of double-stranded DNA viruses found in the water column infect the most abundant groups of organisms, prokaryotes and single-celled eukaryotes (e.g., flagellates). RAPD-PCR analysis of viral communities does not distinguish between different virus types. Thus, to obtain data on the relationship between a grand majority of the virus community and their hosts, it is helpful to study the communities of prokaryotes and single-celled eukaryotes, preferably using similar methods.
A direct comparison of the results of the bacterial DGGE data by Winter et al. (39) with the microbial RAPD-PCR data is problematic because the latter also targets Archaea and Eukaryota. Thus, the broader set of target organisms may explain why the number of bands detected in MCRA, MOPA, and MICRO did not correlate with bacterial richness as detected by DGGE in the water column or the three depth layers (Table 2).
Another interesting finding is that the number of viral RAPD-PCR bands exceeded the number of bands obtained from the microbial size fraction for each primer alone and in combination. The most parsimonious explanation for this result is that the number of different primer target sites in a virus community is higher compared to the corresponding microbial community. This leads to two conclusions: (i) the number of different virus types in the water column is higher than the corresponding number of different microbial hosts (see, for example, reference 14) and, (ii) assuming that the primary sequence of nucleotides in a given DNA molecule is random, the chance of a given 10-mer to finding a priming site is 1:410. However, DNA sequences are not random, and our results may lead to the conclusion that the diversity of priming sites for the primers used in the present study is higher in viral than in microbial genomes despite the fact that genomes of prokaryotes and single-celled eukaryotes are generally larger than those of viruses. Based on our data, neither of the two conclusions can be completely dismissed or verified; however, it is likely a combination of the two that led to our results.
The number of bands detected in MCRA and MOPA were not correlated with each other, either in the water column or in the three depth layers (Table 3). Similarly, changes in the banding patterns of MCRA and MOPA were not linked (Table 4). This suggests that data obtained from MCRA and MOPA appear to be representative of two different subgroups of the microbial community, similarly to data obtained from VCRA and VOPA. Nevertheless, the nucleic acid sequences obtained from MCRA and MOPA were mostly affiliated to the same broad groups, with the exceptions of Planktomycetes, Chloroflexi, Actinobacteria, and some unclassified Proteobacteria (Fig. 2B and C). Thus, the differences appear to be largely due to lower level taxonomic ranks compared to the ones analyzed here. A previous study conducted at the same site revealed that, on average, archaeal cells comprised 2.7, 13.2, and 8.0% of DAPI (4′,6′-diamidino-2-phenylindole)-stained cells in the epi-, meso-, and bathypelagic zones, respectively (39). However, the lack of sequences affiliated with Archaea as obtained from MCRA and MOPA does not contradict previously published results, since we did not obtain sequences from all bands detected on the gels and, thus we might have missed the archaeal phylotypes.
Prokaryotes and single-celled eukaryotes as viral hosts.
Based on previously published data from the same sampling site and period (39, 40), prokaryotic abundance and bacterial richness (as detected by DGGE analysis) were positively correlated in the bathypelagic zone but not so in the epi- or mesopelagic. This is similar to the concomitant increase in prokaryotic abundance and the number of bands in MOPA and MICRO in the bathypelagic (Table 2), where the decline of eukaryotic cells with depth was much stronger than that of prokaryotic cells (31, 40). This suggests that the overwhelming majority of viruses in the bathypelagic zone target prokaryotes. Further support comes from the finding that in the epipelagic zone, where the abundance of single-celled eukaryotes was much higher than in the meso- and bathypelagic zones, no correlation between viral RAPD-PCR data and bacterial richness, as well as prokaryotic and viral abundance, was found (Table 2).
Prokaryotic and viral abundances were tightly linked in the entire water column and in the three depth layers at the study site, where an increase in the abundance of prokaryotes and viruses in the bathypelagic zone occurred during winter and spring (February to June [40]). This largely overlaps with the period when the complete breakdown of the thermal stratification resulted in a uniform water column with the potential for deep vertical mixing (39). Winter et al. (39) argue that environmental changes in the bathypelagic zone during the nonstratified period from December to April (e.g., enhanced availability of nutrients and dissolved organic carbon) resulted in increased prokaryotic growth and viral production at depth. Thus, the bathypelagic zone at the sampling station during the nonstratified period was characterized by enhanced prokaryotic and viral abundance (40), concomitant with enhanced bacterial richness (39) and an increased number of bands in MOPA and MICRO (Table 2, see Fig. S5 and S6 in the supplemental material). Surprisingly, a simultaneous decrease in the number of bands in VOPA and VIR occurred (Table 3, see Fig. S7 and S8 in the supplemental material). There are two non-mutually exclusive interpretations of these results. (i) The virus community changed in a way that precludes an increasing number of virus types to be detected by RAPD-PCR because the primers used did not target these new virus types. (ii) The increase in viral abundance is mainly due to a small number of virus types that outcompete less-adapted viruses with respect to the limiting resource, i.e., prokaryotic hosts in the bathypelagic zone. Thus, the results could be indicative of a virus bloom dominated by a small number of virus types. Either way, the data clearly show a substantial change in the virus community that occurred concomitantly with changes in the microbial and bacterial community, particularly from April to June. The data also indicate that virus communities in the bathypelagic zone exhibited substantial temporal dynamics, as has been shown previously for prokaryotic communities at the study site (39).
For the entire water column, our data indicate that changes in the community composition of the microbial compartment were linked to changes in the virus community, as well as to changes in the bacterial community (Table 4). However, contrary to our expectations, the strongest indications for links between changes in the microbial compartment, as well as in the bacterial community, with changes in the viral community were detected in the bathypelagic zone (Table 4). Thus, given the scarcity of eukaryotic predators able to influence prokaryotic community composition (24) in deep waters at the study site (31), our data suggest that viral lysis was a stronger factor than protozoan grazing for influencing microbial community composition. This also indicates that viral lysis is the biggest source of mortality for prokaryotes in the bathypelagic zone.
Williamson et al. (36) showed that lysogenic viruses predominate at deep-sea diffuse-flow hydrothermal vents. Although we also found some prophage-related sequences in the viral size fraction, the majority of sequences in the present study could not be identified (Fig. 3). However, the links between changes in the viral and microbial community, as well as in the bacterial community (Table 4), detected in this study are more suggestive of lytic virus production (either by lytic viruses or temperate viruses directly entering the lytic cycle). Similar results could also be caused by prophage induction; however, one would need to assume relative constant induction of prophage throughout our study period.
Summary and conclusions.
In summary, RAPD-PCR data obtained using either primer CRA-22 or OPA-13 are largely representative of two different subsets of the viral community. Depending on the primer used, the number of nucleic acid sequences with significant hits in the env_nt database of GenBank was only slightly lower for the viral compared to the microbial community; however, substantially fewer viral sequences than microbial sequences also had close relatives in the nr database of GenBank, with few sequences being identifiable as of viral origin. These results suggest that the viral sequences currently available in the nr database of GenBank do not appear to be representative of the diversity of marine viruses.
Our RAPD-PCR data indicate particularly strong ties between changes in the microbial and viral community in the bathypelagic zone at the study site. Further evidence for these striking temporal dynamics at depth is provided by DGGE analysis of the bacterial community (39), as well as changes in prokaryotic and viral abundance (40). The low number of potential eukaryotic hosts in the bathypelagic zone at the study site (31) and the strong links between changes in the microbial and viral RAPD-PCR data indicate that viruses are the main source of mortality for prokaryotes in the bathypelagic zone at the study site. Also, the data indicate that virus communities in the bathypelagic zone can exhibit substantial temporal dynamics.
Supplementary Material
Acknowledgments
We thank Jean-Claude Marty, Jacques Chiaverini, Floriane Girard, and Stéphane Gouy for organizing the cruises to the DYFAMED site. We acknowledge the captains and crews of RV Tethys II for their assistance at sea and M.-E. Kerros for technical assistance. We thank C. A. Suttle for suggesting the in silico nucleic acid sequence analysis to evaluate the RAPD-PCR approach for viral communities. The constructive comments of three anonymous reviewers are also greatly appreciated.
This study was financially supported by a Marie Curie postdoctoral fellowship from the European Commission to C.W. (project ILVIROMAB, no. 007712). The cruises were financed by INSU-CNRS (Institut National des Sciences de l'Univers-Centre National de la Recherche Scientifique). Additional support came from the project ANR-AQUAPHAGE (ANR 07 BDIV 015-06; coordinated by M.G.W.) of the French Science Ministry.
Footnotes
Published ahead of print 20 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, S., P. Böger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assay in analysis of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bench, S. R., T. E. Hanson, K. E. Williamson, D. Ghosh, M. Radosovich, K. Wang, and K. E. Wommack. 2007. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 73:7629-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvier, T., and P. A. del Giorgio. 2007. Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ. Microbiol. 9:287-297. [DOI] [PubMed] [Google Scholar]
- 6.Breitbart, M., L. Wegley, S. Leeds, T. Schoenfeld, and F. Rohwer. 2004. Phage community dynamics in hot springs. Appl. Environ. Microbiol. 70:1633-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brussaard, C. P. D. 2004. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70:1506-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, F., C. A. Suttle, and S. M. Short. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comeau, A. M., S. M. Short, and C. A. Suttle. 2004. The use of degenerate-primed random amplification of polymorphic DNA (DP-RAPD) for strain-typing and inferring the genetic similarity among closely related viruses. J. Virol. Methods 118:95-100. [DOI] [PubMed] [Google Scholar]
- 10.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques 34:790-802. [DOI] [PubMed] [Google Scholar]
- 11.Culley, A. I., A. S. Lang, and C. A. Suttle. 2003. High diversity of unknown picorna-like viruses in the sea. Nature 424:1054-1057. [DOI] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 13.Helton, R. R., and K. E. Wommack. 2009. Seasonal dynamics and metagenomic characterization of estuarine viriobenthos assemblages by randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 75:2259-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmfeldt, K., M. Middelboe, O. Nybroe, and L. Riemann. 2007. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl. Environ. Microbiol. 73:6730-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janse, I., J. Bok, and G. Zwart. 2004. A simple remedy against artificial double bands in denaturing gradient gel electrophoresis. J. Microb. Methods 57:279-281. [DOI] [PubMed] [Google Scholar]
- 16.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legendre, P., and L. Legendre. 1998. Numerical ecology. Developments in environmental modeling, 2nd ed. Elsevier Science B.V., Amsterdam, Netherlands.
- 18.Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209-220. [PubMed] [Google Scholar]
- 19.Marie, D., F. Partensky, D. Vaulot, and C. P. D. Brussaard. 1999. Enumeration of phytoplankton, bacteria, and viruses in marine samples, p. 11.11.1-11.11.15. In J. P. Robinson, Z. Darzynkiewicz, P. N. Dean, A. Orfao, P. S. Rabinovitch, C. C. Stewart, H. J. Tanke, and L. L. Wheeless (ed.), Current protocols in cytometry. John Wiley & Sons, Inc., New York, NY. [DOI] [PubMed]
- 20.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray, A. G., and G. A. Jackson. 1992. Viral dynamics: a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Prog. Ser. 89:103-116. [Google Scholar]
- 22.Payet, J. P., and C. A. Suttle. 2008. Physical and biological correlates of virus dynamics in the southern Beaufort Sea and Amundsen Gulf. J. Mar. Sys. 74:933-945. [Google Scholar]
- 23.Pei, A. Y., W. E. Oberdorf, C. W. Nossa, A. Agarwal, P. Chokshi, E. A. Gerz, Z. Jin, P. Lee, L. Yang, M. Poles, S. M. Brown, S. Sotero, T. DeSantis, E. Brodie, K. Nelson, and Z. Pei. 2010. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl. Environ. Microbiol. 76:3886-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pernthaler, J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3:537-546. [DOI] [PubMed] [Google Scholar]
- 25.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwalbach, M. S., I. Hewson, and J. A. Fuhrman. 2004. Viral effects on bacterial community composition in marine plankton microcosms. Aquat. Microb. Ecol. 34:117-127. [Google Scholar]
- 27.Short, S. M., and C. A. Suttle. 2000. Denaturing gradient gel electrophoresis resolves virus sequences amplified with degenerate primers. Biotechniques 28:20-26. [DOI] [PubMed] [Google Scholar]
- 28.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the unexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka, T., and F. Rassoulzadegan. 2002. Full-depth profile (0-2000m) of bacteria, heterotrophic nanoflagellates and ciliates in the NW Mediterranean Sea: vertical partitioning of microbe trophic structures. Deep Sea Res. II 49:2093-2107. [Google Scholar]
- 32.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 33.Thingstad, T. F., and R. Lignell. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13:19-27. [Google Scholar]
- 34.Weinbauer, M. G., J.-M. Arrieta, C. Griebler, and G. J. Herndl. 2009. Enhanced viral production and infection of bacterioplankton during an iron-induced phytoplankton bloom in the Southern Ocean. Limnol. Oceanogr. 54:774-784. [Google Scholar]
- 35.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781-788. [Google Scholar]
- 36.Williamson, S. J., S. C. Cary, K. E. Williamson, R. R. Helton, S. R. Bench, D. Winget, and K. E. Wommack. 2008. Lysogenic virus-host interactions predominate at deep-sea diffuse-flow hydrothermal vents. ISME J. 2:1112-1121. [DOI] [PubMed] [Google Scholar]
- 37.Winget, D. M., and K. E. Wommack. 2008. Randomly amplified polymorphic DNA PCR as a tool for assessment of marine viral richness. Appl. Environ. Microbiol. 74:2612-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winter, C., T. Bouvier, M. G. Weinbauer, and T. F. Thingstad. 2010. Trade-offs between competition and defense specialists among unicellular planktonic organisms: the “killing the winner” hypothesis revisited. Microb. Mol. Biol. Rev. 74:42-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter, C., M.-E. Kerros, and M. G. Weinbauer. 2009. Seasonal changes of bacterial and archaeal communities in the dark ocean: evidence from the Mediterranean Sea. Limnol. Oceanogr. 54:160-170. [Google Scholar]
- 40.Winter, C., M. E. Kerros, and M. G. Weinbauer. 2009. Seasonal and depth-related dynamics of prokaryotes and viruses in surface and deep waters of the northwestern Mediterranean Sea. Deep Sea Res. I 56:1972-1982. [Google Scholar]
- 41.Winter, C., A. Smit, G. J. Herndl, and M. G. Weinbauer. 2004. Impact of virioplankton on archaeal and bacterial community richness as assessed in seawater batch cultures. Appl. Environ. Microbiol. 70:804-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wommack, K. E., J. Ravel, R. T. Hill, and R. R. Colwell. 1999. Hybridization analysis of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 65:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.