Abstract
We have optimized the display of the B domain of staphylococcal protein A on the surface of Lactococcus lactis. The maximum binding capacity was estimated at 0.146 μg of antibody per 108 cells and was sustained at 86% after treatment with simulated gastric juice. A tumor necrosis factor alpha (TNF-α)-binding affibody was also displayed and bound TNF-α, which could be useful in the treatment of inflammatory bowel disease.
Lactic acid bacteria (LAB) have received considerable attention in recent years, based on their record of safe usage as a constituent of fermented foods and their health-promoting effects as probiotics (5). Recombinant LAB could also be used in therapy, with most applications aimed at the delivery of antigens or therapeutic proteins to human mucosal surfaces (27). Another potential application of recombinant LAB involves surface attachment of affinity molecules, such as antibodies, single-chain variable fragments (scFv), or specific oligosaccharides, which can target pathogens, toxins, or inflammatory mediators in the human intestine (9, 22).
We describe the surface display of two types of affinity molecule, the B domain and the tumor necrosis factor alpha (TNF-α)-binding affibody, on a model LAB, Lactococcus lactis. The B domain, which is one out of five antibody-binding domains of staphylococcal protein A (15), was already used as a model protein (2). The surface display of the B domain was reported for Escherichia coli in a biosensor application (8), for Saccharomyces cerevisiae as a whole-cell immunoadsorbent (16), and for certain viruses for specific cell targeting (20).
The affinity of the B domain or its synthetic homologue, the Z domain (17), for the antibody Fc region has been redirected to several other proteins by randomization of amino acids involved in the interaction, using the genetic combinatorial library and phage display (18). The variants of the Z domain were termed “affibodies” and were directed against various proteins (reviewed in reference 19) and also against TNF-α (7).
TNF-α is well established as a proinflammatory cytokine in the pathology of inflammatory bowel disease (IBD), and monoclonal antibodies against TNF-α are routinely used in parenteral therapy (23) but can have systemic side effects. The abundant presence of TNF-α in the stool samples of IBD patients (4) and the successful treatment of experimental colitis in rats by oral administration of avian IgY (28) justify the oral delivery of an agent with the capability of removing TNF-α in IBD. We have therefore replaced the B domain in our surface-displayed fusion protein with an affibody against TNF-α (7) as a second type of binding molecule. LAB with surface-displayed affibody against TNF-α could be used to bind TNF-α in the intestine, with the potential for use in the treatment of IBD. This novel approach could also overcome the problems with the gastrointestinal stability of antibodies.
Preparation of gene constructs.
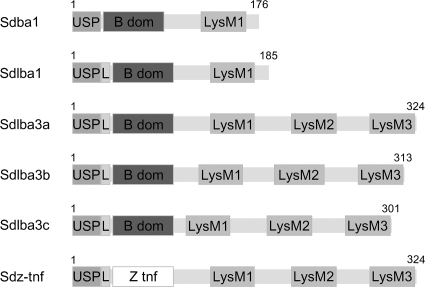
Several gene constructs were prepared as described in supplemental material to give a fusion protein with optimal surface binding capabilities (Fig. 1). Each gene construct is composed of three functional parts: a signal sequence for secretion to the growth medium (derived from the Usp45 protein [26] and strengthened by a synthetic propeptide [12]), an affinity domain (the B domain or the TNF-α-binding affibody), and a variant of a peptidoglycan binding domain of AcmA (1, 21) for surface attachment. Noncovalent binding via AcmA was chosen, because covalent surface binding using a sortase motif was described to be less effective in a previous report of surface display of staphylococcal nuclease (6).
FIG. 1.
Gene constructs for lactococcal surface display.
Fusion protein expression.
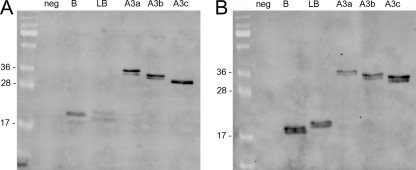
Nisin-controlled expression (13) was used to express the fusion proteins. Overnight cultures of Lactococcus lactis harboring the appropriate plasmid were diluted (1:100) in 10 ml of fresh GM-17 medium, grown to an optical density (A600) of 0.5 to 0.8, and induced with 25 ng/ml nisin (Fluka). Three hours after induction, the culture was split in two. Five milliliters was stored at 4°C for flow cytometry and fluorescence microscopy, and the remaining 5 ml was used for SDS-PAGE and Western blot analysis, as described in the supplemental material. All fusion proteins were detected by fluorescein isothiocyanate (FITC)-conjugated anti-protein A antibody (Abcam) in both cell lysate and growth medium, and their observed and calculated molecular masses corresponded (Sdba1, 18.4 kDa; Sdlba1, 19.4 kDa; Sdlba3a, 34.0 kDa; Sdlba3b, 32.9 kDa; and Sdlba3c, 31.8 kDa) (Fig. 2). The concentration of fusion proteins in the growth medium was comparable to that in the cell lysate, which contained cytoplasmic and surface-bound fusion proteins. This shows that not all secreted fusion proteins bind to the surface, possibly due to increased production and secretion rates in comparison to the rate of binding. Two bands of similar sizes were detected with each fusion protein, which corresponded to fusion proteins with and without a secretion signal and with a calculated molecular mass difference of 2.9 kDa. The processing of secretion signals was evaluated with N-terminal sequencing. The growth medium fractions of the Sdba1 and Sdlba1 proteins were concentrated 10-fold on an Amicon Ultra-4 (3-kDa cutoff; Millipore), separated on SDS-PAGE gel, blotted to a polyvinylidene difluoride (PVDF) membrane, and prepared for use with an Applied Biosystems 492 protein sequencer. The determined sequences GSADN and LEISS correspond to the N-terminal sequences of Sdba1 and Sdlba1, respectively, without the Usp45 signal. The HtrA protease-deficient strain was tested as a reported strategy for increasing recombinant protein expression (14), and a small increase in the expression level was indeed observed (data not shown).
FIG. 2.
Expression of fusion proteins for the surface display of the B domain in L. lactis NZ9000. (A) Cell lysate. (B) Growth medium. neg, pNZ8148; B, pSDBA1; LB, pSDLBA1; A3a, pSDLBA3a; A3b, pSDLBA3b; A3c, pSDLBA3c.
Surface display of the B domain.
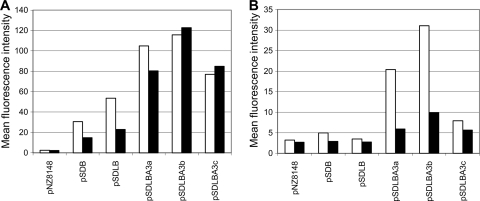
The quantity and functionality of the surface-displayed B domain were characterized by flow cytometry. Ten microliters of cell cultures (1 × 107 cells/ml) was added to 500 μl of Tris-buffered saline (TBS) and pelleted. Cells were resuspended in 500 μl of TBS containing 1 μg of specific FITC-conjugated anti-protein A antibody (Abcam) or 2 μg of nonspecific Alexa Fluor 488-conjugated rabbit anti-mouse antibody (Invitrogen) and incubated for 2 h at room temperature with constant shaking. Cells were then washed three times with 200 μl 0.1% TBS-Tween (TBST) and resuspended in 500 μl of TBS. Stained cells were analyzed with a FACSCalibur (Becton Dickinson, Inc.) flow cytometer. At least 100,000 bacterial cells were counted for each sample. The results are presented as mean fluorescence intensity (MFI) values for lactococcal cells, which correlate with the amount of surface-bound fusion protein. Surface display of B domain was detected for all gene constructs by specific antibody, while the functionality of the displayed B domain was confirmed by the ability to bind nonspecific antibody via the Fc region (Fig. 3). However, significant differences between the constructs were observed. Inclusion of a synthetic propeptide and, especially, the use of three peptidoglycan binding domains contributed to the increased surface display and functionality of the B domain, in accordance with previous observations (25). The expression of Sdlba3b resulted in the highest-level responses with both specific and nonspecific antibody in both bacterial strains. A more detailed comparison between pNZ8148 (control)- and pSDLBA3b-containing cells stained with specific (Fig. 4A) and nonspecific (Fig. 4B) antibody is shown. A distinct shift in fluorescence intensity between controls (gray) and Sdlba3b-expressing cells (black line) can be observed in both cases. The importance of the length of the spacer between the displayed protein and the cell wall anchor was demonstrated, with the Sdlba3b construct being superior to Sdlba3a and Sdlba3c. The different spacers may affect the spatial orientation of the displayed protein, affecting its binding ability, or may affect the expression levels of the fusion proteins, which differ slightly (Fig. 2). The results for the NZ9000 strain were superior to those for NZ9000ΔhtrA, despite the higher protein expression levels in NZ9000ΔhtrA. The functionality of the displayed B domain in the NZ9000ΔhtrA strain was impaired, as shown by a decrease in nonspecific-antibody binding (Fig. 3B). This could be explained by the reported disturbance of protein secretion in the absence of HtrA (24).
FIG. 3.
Flow cytometry of lactococcal cells harboring different plasmids, stained with specific (white bars) or nonspecific antibody (black bars). (A) NZ9000 strain; (B) NZ9000ΔhtrA strain.
FIG. 4.
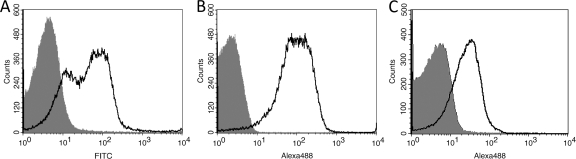
Shift in fluorescence intensity. (A) Control and Sdlba3b-expressing cells detected with specific FITC-conjugated antibody. MFI (control) = 2.61, MFI (sample) = 115.70. (B) Control and Sdlba3b-expressing cells detected with nonspecific Alexa Fluor 488-conjugated antibody MFI (control) = 2.26, MFI (sample) = 150.71. (C) Control and Sdz-tnf-expressing cells detected with Alexa Fluor 488-conjugated TNF-α. MFI (control) = 4.83, MFI (sample) = 31.67.
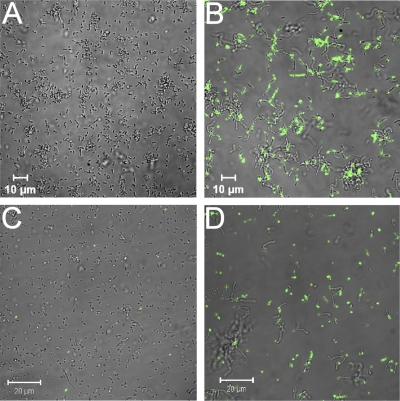
Fluorescence microscopy was used to confirm the results of flow cytometry. The staining protocol was similar, except that the starting volume of cell cultures was 20 μl and the final volume of TBS for resuspension 200 μl. Fluorescence microscopy was performed using a Carl Zeiss LSM 510 confocal microscope. Alexa Fluor 488 was excited with an argon laser (488 nm), and emission was filtered using a narrow-band 505- to 530-nm filter. Sdlba3b-expressing cells showed stronger binding of antibody and greater intensity of fluorescence than control cells (Fig. 5), which again confirmed the surface display and functionality of the B domain.
FIG. 5.
Control (A) and Sdlba3b-expressing (B) cells detected with nonspecific Alexa Fluor 488-conjugated antibody. Control (C) and Sdz-tnf-expressing (D) cells detected with Alexa Fluor 488-conjugated TNF-α.
Binding capacity.
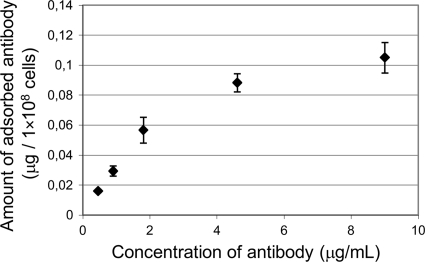
The capacity of B domain-displaying cells to bind nonspecific antibody was quantified by a spectroscopic method as described in the supplemental material. The amount of antibody adsorbed on 1 × 108 cells was plotted against the concentration of antibody and was observed to be concentration dependent (Fig. 6). The Langmuir adsorption isotherm (11) was assumed for the binding of antibody. The Langmuir linear regression method (10) {CAb/Γ = (CAb/Γmax) + [1/(K × Γmax)]} was used to fit the Langmuir equation to the data and yielded the equation CAb/Γ = (6,837 × CAb) + 22,688 (r2 = 0.9897). The maximal amount of adsorbed antibody (Γmax) was calculated to be 0.146 μg per 1 × 108 cells, which corresponds to approximately 4,500 molecules per cell. This is less than the amount reported for yeast (16); however, the smaller cell size and the different display mechanism have to be considered. Nevertheless, the capacity to bind microgram quantities of target proteins by commonly used bacterial doses (1 × 109 to 1 × 1010) could well suggest physiological significance.
FIG. 6.
Adsorption of nonspecific antibody to B domain displaying cells.
Gastric resistance.
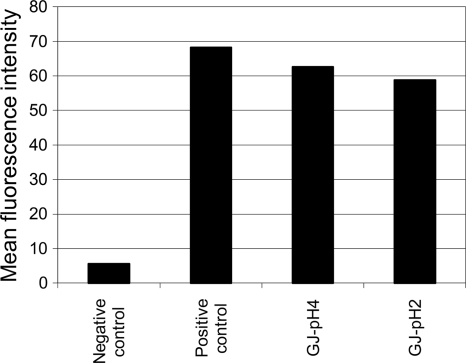
The harsh gastric conditions are the biggest hurdle in the oral delivery of proteins. The stability and functionality of the surface-displayed B domain were tested against simulated gastric juice (GJ), which was prepared as described in reference 3, using 3 mg/ml pepsin (Sigma- Aldrich), and the pH was adjusted to 2.0 or 4.0. Five hundred microliters of pNZ8148 (negative-control)- or pSDLBA3b-containing cells was mixed with either 500 μl of TBS buffer (pH 7.0; positive control) or 500 μl of simulated GJ and incubated at room temperature for 30 min. Fifty microliters of the mixture was stained with nonspecific antibody and analyzed by flow cytometry as described above. The reduction of MFI in samples exposed to simulated gastric juice was less than 14%, even after 30 min at pH 2.0 (Fig. 7). This indicates that L. lactis with the surface-displayed B domain or its homologues would retain considerable functionality after oral administration.
FIG. 7.
Flow cytometric analysis of binding of nonspecific antibody to pSDLBA3b-containing cells after the incubation of cells at pH 7.0 (positive control) or under simulated gastric conditions at pH 4.0 (GJ-pH 4) or pH 2.0 (GJ-pH 2). pNZ8148-containing cells were used as a negative control.
Display of affibody and binding of TNF-α.
The TNF-α-binding affibody gene (sdz-tnf) (see Table S1 in the supplemental material) was based on the Z00185 polypeptide (7) and was displayed in a manner analogous to that observed for the B domain. The surface display of the TNF-α-binding affibody was characterized by its ability to bind recombinant TNF-α, which was labeled using an Alexa Fluor 488 microscale protein-labeling kit (Invitrogen). Flow cytometry of pNZ8148-containing (Fig. 4C, gray [control]) and pSDZ-TNF-containing (black line) cells, which were stained with fluorescently labeled TNF-α, showed a distinct shift in fluorescence intensity. Increased fluorescence of Sdz-tnf-expressing cells was also observed with the use of fluorescence microscopy (Fig. 5). We have therefore demonstrated that L. lactis with surface-displayed TNF-α-binding affibody was capable of binding fluorescently labeled TNF-α. The binding of TNF-α by recombinant L. lactis could be used for the removal of TNF-α from the human intestine, which should be useful in the alleviation of the symptoms of IBD. Besides TNF-α-binding affibody, other affibodies could be applied in a similar fashion, yielding novel possible applications.
Supplementary Material
Acknowledgments
This work was supported by Slovenian Research Agency grant no. P4-0127.
We are grateful to Igor Mierau for providing the NZ9000ΔhtrA strain, to Vladka Gaberc Porekar for providing TNF-α, and to Roger Pain for critical reading of the manuscript.
Footnotes
Published ahead of print on 27 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bosma, T., R. Kanninga, J. Neef, S. A. Audouy, M. L. van Roosmalen, A. Steen, G. Buist, J. Kok, O. P. Kuipers, G. Robillard, and K. Leenhouts. 2006. Novel surface display system for proteins on non-genetically modified gram-positive bacteria. Appl. Environ. Microbiol. 72:880-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bratkovic, T., A. Berlec, T. Popovic, M. Lunder, S. Kreft, U. Urleb, and B. Strukelj. 2006. Engineered staphylococcal protein A's IgG-binding domain with cathepsin L inhibitory activity. Biochem. Biophys. Res. Commun. 349:449-453. [DOI] [PubMed] [Google Scholar]
- 3.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 1998. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 84:759-768. [DOI] [PubMed] [Google Scholar]
- 4.de Silva, D. G., L. N. Mendis, N. Sheron, G. J. Alexander, D. C. Candy, H. Chart, and B. Rowe. 1992. TNF alpha in stool as marker of intestinal inflammation. Lancet 340:372. [DOI] [PubMed] [Google Scholar]
- 5.de Vrese, M., and J. Schrezenmeir. 2008. Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 111:1-66. [DOI] [PubMed] [Google Scholar]
- 6.Dieye, Y., S. Usai, F. Clier, A. Gruss, and J. C. Piard. 2001. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 183:4157-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonsson, A., H. Wallberg, N. Herne, S. Stahl, and F. Y. Frejd. 2009. Generation of tumour-necrosis-factor-alpha-specific affibody molecules capable of blocking receptor binding in vitro. Biotechnol. Appl. Biochem. 54:93-103. [DOI] [PubMed] [Google Scholar]
- 8.Jose, J., J. W. Chung, B. J. Jeon, R. M. Maas, C. H. Nam, and J. C. Pyun. 2009. Escherichia coli with autodisplayed Z-domain of protein A for signal amplification of SPR biosensor. Biosens. Bioelectron. 24:1324-1329. [DOI] [PubMed] [Google Scholar]
- 9.Kruger, C., Y. Hu, Q. Pan, H. Marcotte, A. Hultberg, D. Delwar, P. J. van Dalen, P. H. Pouwels, R. J. Leer, C. G. Kelly, C. van Dollenweerd, J. K. Ma, and L. Hammarstrom. 2002. In situ delivery of passive immunity by lactobacilli producing single-chain antibodies. Nat. Biotechnol. 20:702-706. [DOI] [PubMed] [Google Scholar]
- 10.Langmuir, I. 1918. The adsorption of gases on plane surfaces of glass, mica and platinum. J. Am. Chem. Soc. 40:1361-1403. [Google Scholar]
- 11.Langmuir, I. 1916. The constitution and fundamental properties of solids and liquids. J. Am. Chem. Soc. 38:2221-2295. [Google Scholar]
- 12.Le Loir, Y., S. Nouaille, J. Commissaire, L. Bretigny, A. Gruss, and P. Langella. 2001. Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 67:4119-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mierau, I., and M. Kleerebezem. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705-717. [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi, A., I. Poquet, V. Azevedo, J. Commissaire, L. Bermudez-Humaran, E. Domakova, Y. Le Loir, S. C. Oliveira, A. Gruss, and P. Langella. 2002. Controlled production of stable heterologous proteins in Lactococcus lactis. Appl. Environ. Microbiol. 68:3141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moks, T., L. Abrahmsen, B. Nilsson, U. Hellman, J. Sjoquist, and M. Uhlen. 1986. Staphylococcal protein A consists of five IgG-binding domains. Eur. J. Biochem. 156:637-643. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura, Y., S. Shibasaki, M. Ueda, A. Tanaka, H. Fukuda, and A. Kondo. 2001. Development of novel whole-cell immunoadsorbents by yeast surface display of the IgG-binding domain. Appl. Microbiol. Biotechnol. 57:500-505. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson, B., T. Moks, B. Jansson, L. Abrahmsen, A. Elmblad, E. Holmgren, C. Henrichson, T. A. Jones, and M. Uhlen. 1987. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1:107-113. [DOI] [PubMed] [Google Scholar]
- 18.Nord, K., E. Gunneriusson, J. Ringdahl, S. Stahl, M. Uhlen, and P. A. Nygren. 1997. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat. Biotechnol. 15:772-777. [DOI] [PubMed] [Google Scholar]
- 19.Nygren, P. A. 2008. Alternative binding proteins: affibody binding proteins developed from a small three-helix bundle scaffold. FEBS J. 275:2668-2676. [DOI] [PubMed] [Google Scholar]
- 20.Ohno, K., K. Sawai, Y. Iijima, B. Levin, and D. Meruelo. 1997. Cell-specific targeting of Sindbis virus vectors displaying IgG-binding domains of protein A. Nat. Biotechnol. 15:763-767. [DOI] [PubMed] [Google Scholar]
- 21.Okano, K., Q. Zhang, S. Kimura, J. Narita, T. Tanaka, H. Fukuda, and A. Kondo. 2008. System using tandem repeats of the cA peptidoglycan-binding domain from Lactococcus lactis for display of both N- and C-terminal fusions on cell surfaces of lactic acid bacteria. Appl. Environ. Microbiol. 74:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton, A. W., R. Morona, and J. C. Paton. 2006. Designer probiotics for prevention of enteric infections. Nat. Rev. Microbiol. 4:193-200. [DOI] [PubMed] [Google Scholar]
- 23.Sandborn, W. J., and S. B. Hanauer. 1999. Antitumor necrosis factor therapy for inflammatory bowel disease: a review of agents, pharmacology, clinical results, and safety. Inflamm. Bowel Dis. 5:119-133. [DOI] [PubMed] [Google Scholar]
- 24.Sriraman, K., and G. Jayaraman. 2008. HtrA is essential for efficient secretion of recombinant proteins by Lactococcus lactis. Appl. Environ. Microbiol. 74:7442-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steen, A., G. Buist, G. J. Horsburgh, G. Venema, O. P. Kuipers, S. J. Foster, and J. Kok. 2005. AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 272:2854-2868. [DOI] [PubMed] [Google Scholar]
- 26.van Asseldonk, M., W. M. de Vos, and G. Simons. 1993. Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and a heterologous alpha-amylase. Mol. Gen. Genet. 240:428-434. [DOI] [PubMed] [Google Scholar]
- 27.Wells, J. M., and A. Mercenier. 2008. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 6:349-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Worledge, K. L., R. Godiska, T. A. Barrett, and J. A. Kink. 2000. Oral administration of avian tumor necrosis factor antibodies effectively treats experimental colitis in rats. Dig. Dis. Sci. 45:2298-2305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.