Abstract
Nontyphoidal salmonellae are major causes of food-borne disease worldwide. In Uruguay, Salmonella enterica serovar Enteritidis was the most commonly isolated serovar throughout the last decade, with a marked epidemic period between 1995 and 2004. In a previous study, we conducted comparative genomics of 29 epidemic-spanning S. Enteritidis field isolates, and here we evaluated the pathogenic potential of the same set of isolates using several phenotypic assays. The sample included 15 isolates from human gastroenteritis, 5 from invasive disease, and 9 from nonhuman sources. Contrary to the genetic homogeneity previously observed, we found great phenotypic variability among these isolates. One-third of them were defective in at least one assay, namely, 10 isolates were defective in motility, 8 in invasion of Caco-2 cells, and 10 in survival in egg albumen. Twelve isolates were tested for invasiveness in 3-day-old chickens, and five of these were significantly less invasive than the reference strain. The two oldest preepidemic isolates were reduced in fitness in all assays, providing a plausible explanation for the previous negligible incidence of S. Enteritidis in Uruguay and supporting the view that the introduction or emergence of a more virulent strain was responsible for the marked rise of this serovar. Further, we found differences in fitness among the isolates which depended on the source of isolation. A total of 1 out of 14 isolates from human gastroenteritis, but 6 out of 13 isolates from other sources, was impaired in at least two assays, suggesting enhanced fitness among strains able to cause intestinal disease in humans.
Salmonella enterica subsp. enterica serovar Enteritidis (S. Enteritidis) is a major cause of food-borne disease worldwide. It represents an important public health problem with marked economic impact (3, 18). It is usually acquired by ingestion of contaminated water or food, and poultry-derived products, particularly eggs, are the major source of human infection (22, 45).
During the 1980s, S. Enteritidis emerged as a major concern for food safety in Europe and the Americas, and by 1990 in the United States and 1993 in Europe, it became the most frequently isolated Salmonella serovar (48). Data obtained from the WHO Global Salmonella Survey program (http://www.who.int/salmsurv/links/en/) confirmed that between 2000 and 2004, S. Enteritidis was the most frequently isolated serovar of S. enterica from human sources worldwide. In Uruguay, S. Enteritidis was rarely isolated until 1994, but in 1995, a first outbreak occurred, initiating an epidemic of human infection that lasted until 2004 (4, 6). In the period from 1997 to 2004, S. Enteritidis was the most frequently identified serovar in Uruguay, accounting for more than 50% of all strains received each year at the National Salmonella Center (NSC; Institute of Hygiene, Uruguay) and for more than 85% of the strains isolated from humans (6). In 2009, S. Enteritidis reemerged as the main cause of salmonellosis in Uruguay, outnumbering S. Typhimurium as the most frequent serovar isolated from humans (NSC unpublished data).
The factors responsible for this S. Enteritidis pandemic are still unclear, but several facts have been proposed to contribute, including changes in farming practices, the existence of a rodent reservoir, eradication of competing S. Gallinarum strains by vaccination, and evolution of the pathogen to become more virulent (9, 48). In this respect, it has been suggested that the rapid spread of S. Enteritidis throughout Europe and the United States might indicate the emergence of a new, more virulent strain of the bacterium due to the acquisition of the ability to colonize and persist in poultry (37).
S. Enteritidis can asymptomatically colonize the gastrointestinal tract and deeper organs of chickens, including the ovary and oviduct, through which it can be transmitted to the forming eggs (23, 26). An important epidemiological characteristic of this serovar is its capacity to persist in the hostile environment of albumen in laid eggs, and it is postulated that this ability is essential for its eventual transmission to humans (8, 21, 26, 27). In addition, survival of S. Enteritidis in a nonhost environment, such as soil or fresh fruits and vegetables, has also been reported, which may certainly influence the epidemiology of this pathogen (10, 11, 35).
In the human host, S. Enteritidis colonizes the small intestine, and most commonly, it causes a self-limiting gastroenteritis characterized by mucosal inflammation and diarrhea (29). Many reports using both animal and cell culture models indicate that the ability to invade and survive in epithelial cells is important for Salmonella pathogenesis (17, 38, 42, 53). Upon interaction with enterocytes, Salmonella induces a proinflammatory response characterized by basal secretion of interleukin-8 (IL-8; CXCL8), CCL20 (MIP3a), and various other proinflammatory chemokines that recruit neutrophils and dendritic cells into the subepithelial compartment (13, 34, 41, 51), ultimately leading to diarrhea.
Previously, we reported the genetic diversity of a comprehensive set of S. Enteritidis field isolates recovered from different sources and isolated before, during, and after the epidemic in Uruguay (5). Using molecular typing and microarray-based comparative genomic hybridization (CGH) methods, we found a major genomic profile for the strains circulating in the country during the epidemic, which is remarkably similar to that of fully sequenced S. Enteritidis PT4 P125109, a United Kingdom clinical isolate that is highly virulent in chickens. Significantly, the most divergent strains were the two oldest preepidemic isolates, isolated more than 5 years before the beginning of the epidemic (Table 1) (5). Further, preliminary cell invasion assays showed a degree of variation between isolates that was greater than would be expected given the typing and CGH analysis.
TABLE 1.
Summary of the isolation periods, origins, and CGH and virulence plasmid patterns for the 29 Uruguayan S. Enteritidis isolates used in this study
| Isolatea | Period of isolation | Sourceb | CGHc | SVPd |
|---|---|---|---|---|
| 31/88 | Preepidemic | Gastroenteritis | 75 | + |
| 08/89 | Invasive disease | 74 | + | |
| 53/94 | Food | 39 | + | |
| 57/94 | Food | 1 | + | |
| 47/95 | Epidemic | Gastroenteritis | 2 | + |
| 51/95 | Gastroenteritis | 0 | + | |
| 108/95 | Gastroenteritis | 0 | + | |
| 49/98 | Food | 9 | + | |
| 80/98 | Invasive disease | 1 | + | |
| 100/99 | Gastroenteritis | 0 | + | |
| 130/99 | Gastroenteritis | 0 | − | |
| 132/99 | Invasive disease | 0 | + | |
| 206/99 | Food | 45 | − | |
| 32/00 | Animal | 8 | + | |
| 125/00 | Gastroenteritis | 0 | + | |
| 48/01 | Food | 0 | + | |
| 251/01 | Egg | 0 | + | |
| 254/01 | Egg | 0 | + | |
| 8/02 | Gastroenteritis | 0 | + | |
| 32/02 | Egg | 1 | − | |
| 65/02 | Gastroenteritis | 0 | + | |
| 77/02 | Gastroenteritis | 2 | + | |
| 199/02 | Invasive disease | 0 | − | |
| 214/02 | Invasive disease | 0 | − | |
| 47/03 | Gastroenteritis | 6 | + | |
| 106/04 | Postepidemic | Gastroenteritis | 0 | + |
| 10/05 | Gastroenteritis | 2 | + | |
| 92/05 | Gastroenteritis | 1 | + | |
| 93/05 | Gastroenteritis | 0 | + |
Strain designations adhere to the following rule: number of isolate/year of isolation.
Food refers to any product for human consumption (e.g., cake or sandwich), with the exception of eggs.
The number of chromosomal genes of difference between the test isolate and reference strain, according to CGH analysis (5). The total number of genes analyzed was 5,863.
SVP, Salmonella virulence plasmid; +, virulence plasmid present; −, virulence plasmid absent.
In the present study, we examined the pathogenic potential of this set of isolates by assessing their ability to attach, invade, and elicit a proinflammatory transcriptional response in cultured human intestinal epithelial cells. Furthermore, virulence after oral inoculation into newly hatched chickens and the capacity to survive in egg albumen were also assessed. For comparison, S. Enteritidis PT4 P125109 was included in all the studies as a reference.
MATERIALS AND METHODS
Bacterial strains, tissue culture cells, and culture conditions.
A total of 29 S. Enteritidis isolates collected in Uruguay over the last 2 decades (1988 to 2005) were studied. Comprehensive information about these isolates has been previously reported (5). The sample included isolates from human (gastroenteritis and systemic infections) and nonhuman sources and covered the different epidemiological periods (before, during, and after the epidemic of S. Enteritidis in Uruguay) as well as all isolates showing variability in our previously reported genotyping analysis (Table 1). All isolates were confirmed biochemically and serologically at the NSC and were stored at −80°C in Luria-Bertani (LB) broth containing 25% glycerol. For ease of interpretation of the figures, the isolates were classified by source, while the two oldest preepidemic isolates were placed in a different group due to their unique genetic lineage.
S. Enteritidis PT4 P12519 (NCTC 13349, here referred to as PT4) was obtained from the Wellcome Trust Sanger Institute (Hinxton, United Kingdom). S. Enteritidis M1511 is a PT4 derivative with a deleted invC gene (Kanr), constructed by P22 transduction of invC::aphT from S. Enteritidis SB566 (14), and was kindly provided by Wolf Dietrich Hardt (Institut für Mikrobiologie, Zürich, Switzerland). Another PT4 derivative, a fliC knockout mutant, was constructed by P22 transduction of a fliC::aphT cassette from strain SEFK32 (47). The absence of motility in this strain was confirmed by plating in soft agar and microscopic visualization.
S. enterica serovar Gallinarum strain 09 was also obtained from the Sanger Institute and was used as a negative control in motility assays and as a positive control in egg albumen survival assays.
Strains were grown aerobically in an orbital shaking incubator (200 rpm) at 37°C in LB broth, with 80 μg/ml of kanamycin added when required.
The human colon carcinoma (Caco-2) cell line was obtained from the American Type Culture Collection. Caco-2 cells were maintained in minimal essential medium with Earle's salts (high glucose, 4.5 g/liter), supplemented with 4 mM l-glutamine and 20% fetal calf serum at 37°C in 5% CO2, at up to 80% confluence.
Motility tests.
For motility testing, 2 μl of overnight cultures of bacteria grown in LB broth were spotted onto the surface of an LB plate containing 0.3% agar (soft agar) and incubated for 6 h at 37°C before the diameter of the halo of growth was measured. Results were expressed as the percent values of the diameter of growth for each isolate, considering PT4 as 100%. The assays were repeated at least twice, and the results were reproducible. Results were confirmed by phase-contrast microscope visualization of mid-log-phase bacterial cultures grown in LB broth.
Caco-2 adhesion and invasion assays.
Assays measuring bacterial invasion into Caco-2 cells were carried out as previously described (5), with small modifications; namely, a short centrifugation step (5 min, 200 g/960 rpm in a Sorvall Legend RT centrifuge, rotor 6445, at room temperature) was carried out immediately upon bacterial inoculation to counteract defects in motility. Each strain was evaluated in two independent experiments, and each time tested at least in duplicate. Invasion levels were expressed as the percentages of recovered intracellular bacteria related to the inoculum. For statistical analysis, the isolates were classified positive for invasion if the percentage of the inoculum recovered from the intracellular environment was higher than 0.11% (30% of the invasion value obtained for PT4) and negative if it was less than or equal to 0.11%. This cutoff was established based on our previous report in which isolates differing significantly (P < 0.01) from PT4 in cell invasiveness showed less than or equal to 30% of its invasion level (5). The strain M1511 (ΔinvC) used as a control for the noninvasive strain showed a percentage of inoculum that equaled 0.03% (data not shown).
For the adhesion assays, replica plates of the invasion assays were processed as described previously, but instead of addition of medium containing gentamicin, the cells were gently washed 4 times with warm phosphate-buffered saline (PBS) and directly lysed with 0.1% Triton X-100 for bacterial counting. The number of CFU obtained in the adhesion assays comprises the adherent plus the intracellular bacteria.
To polarize Caco-2 cells, 5 × 104 cells were seeded in Costar transwells (6.5-mm diameter, 4-μm pore size, polycarbonate) in 24-well plates and grown for 19 days, changing the culture medium every other day, until transepithelial electrical resistance (TEER) stabilization (TEER > 200 Ω·cm2).
Quantitative real-time PCR.
Caco-2 cells were infected as done previously for the invasion assays, but after 1.5 h of incubation with culture medium supplemented with 100 μg/ml of gentamicin, the antibiotic concentration was changed to 10 μg/ml, and the cells were incubated for an additional 1.5 h. Thus, at 3 h postinfection, the cells were washed 3 times with PBS and lysed with Trizol (Invitrogen). Total RNA extraction was carried out as indicated by the supplier. A total of 500 ng of the resulting total RNA was reverse transcribed using Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) and random primers in a 20-μl reaction, and 2 μl of a 1/16 dilution of this reaction was used for real-time PCR using Sybr green (QuantiTect; Qiagen) in a Corbett thermocycler. Primer sequences used are as follows: 18S-F, 5′-ACATCCAAGGAAGGCAGCAG-3′; 18S-R, 5′-TTTTCGTCACTACCTCCCCG-3′; CCL20-F, 5′-CGAATCAGAAGCAGCAAGCA-3′; CCL20-R, 5′-CTTTGTGTGAAAGATGATAGCAT-3′; IL-8-F, 5′-CAGCTCTGTGTGAAGGTGCAG-3′; IL-8-R, 5′-TTATGAATTCTCAGCCCTCTTC-3′; TNF-α-F, 5′-GCTCCAGGCGGTGCTTGTTCC-3′; and TNF-α-R, 5′-AGGTCCCTGGGGAACTCTTCC-3′.
The cycling programs used were as follows: for 18S RNA and tumor necrosis factor alpha (TNF-α), 15 min at 95°C and 40 cycles of 15 s at 94°C, 30 s at 60°C, and 30 s at 72°C, and for CCL20 and IL-8, 15 min at 95°C and 40 cycles of 15 s at 94°C, 30 s at 54°C, and 30 s at 72°C. We used the comparative threshold cycle (CT) method for relative mRNA quantitation. Briefly, the CT obtained for each gene was normalized by the CT obtained for 18S RNA in each sample, giving the ΔCT (ΔCT = CT for 18S RNA − CT for the sample gene). The corresponding 2−ΔCT value was calculated for each gene in each sample, and the 2−ΔCT values were compared to the 2−ΔCT value obtained for the same gene in the uninfected sample to obtain the fold variation in mRNA abundance of the sample related to the uninfected control (fold variation = 2−ΔCTinfected/2−ΔCTuninfected). Each isolate was assayed in duplicate. Nonreverse-transcribed controls rendered no detectable CT values or were amplified at least 10 cycles later than the corresponding reverse-transcribed samples.
Bacterial survival in egg albumen.
The ability of different bacterial isolates to survive in egg albumen was quantified essentially as described by Lu et al. (31). Organic, antibiotic-free chicken eggs from a local farm were disinfected by immersion in 70% ethanol, dried, and cracked into a sterile container. Egg albumen from 10 eggs was pooled and beaten for 3 min. All bacterial strains were grown in LB broth overnight at 37°C with shaking before being used in assays. An overnight culture of bacteria was added to 4 ml of an albumen pool in a Falcon tube to a final concentration of 1 × 103 to 2 × 103 CFU/ml and thoroughly mixed with a vortex. An aliquot was removed immediately after inoculation, diluted in PBS, and plated on LB agar for bacterial enumeration, considering the value of the initial inoculum as 100%. The tubes were incubated at 37°C, and at different time points (0.5, 2, 3, and 6 days), aliquots were removed for plating on LB agar. Surviving bacteria were enumerated and related to the initial values to obtain the percentages of survival. Since different batches of eggs vary in their bactericidal activity, the time course of survival of the isolates in egg albumen varies slightly in different assays. Each assay was repeated twice in duplicate, and data obtained from one representative assay at the most discriminative time point (2 days postinoculation [p.i.]) are presented. An isolate was classified positive for survival in egg albumen if the percentage of the inoculum obtained after 2 days of incubation was >30% and negative if it was ≤30%.
Chicken virulence assays.
Salmonella-free 3-day-old Lohmann white chickens obtained from a local farm were inoculated orally with each S. Enteritidis isolate at 1 × 109 to 2 × 109 CFU in 200 μl. For inoculation, bacteria were grown overnight with shaking at 37°C in 10 ml of LB medium and resuspended in 6 ml of sterile PBS. The animals were provided with feed and water ad libitum and maintained in isolated cages. At 7 days postinoculation, the chickens were euthanized, and the spleens and ovaries-oviducts were removed aseptically. Spleens were homogenized in 2.5 ml sterile water, and the number of CFU/spleen was quantified by plating serial dilutions of homogenates in LB agar. For negative samples, 1 ml of the original samples was enriched in tetrathionate broth (Oxoid) for 24 h at 37°C before being streaked on selective Salmonella-Shigella (SS) agar plates. Suspected Salmonella colonies were further verified by culture on lysine iron agar and phenyl alanine agar. Samples showing confirmed Salmonella colonies were considered positive, and 3 CFU/spleen was assigned for counting purposes. Ovaries-oviducts were directly enriched in tetrathionate broth and processed as described for enrichment from spleens. Ovary-oviduct samples showing confirmed Salmonella colonies were considered positive. Each isolate was tested in two independent experiments with at least 5 chickens per isolate each time. Control groups of chickens inoculated with sterile PBS were processed as done previously, and samples of organ homogenates were plated in SS plates after enrichment in tetrathionate broth. All animal manipulations were performed according to the National Ethical Committee for Animal Experimentation requirements.
Statistical analysis.
For analysis of the frequency of positive strains in Caco-2 or egg survival assays, we used Fisher's exact test (GraphPad Prism software), considering α of <0.05. For analysis of differences in the Caco-2 transcriptional response to the infection or invasiveness in chickens, we used one-way analysis of variance (ANOVA) with Dunnett's multiple comparison test (GraphPad Prism software), considering the difference statistically significant if the P value was <0.05.
RESULTS
Caco-2 cell adhesion and invasion tests.
To study the interaction of S. Enteritidis isolates and human intestinal epithelium, we used the enterocyte-like Caco-2 cell line cocultured with bacteria (7, 12, 16, 28). Motility is an important factor influencing invasion (30, 40, 46, 47); thus, we first tested the motility of the strains in soft agar (Fig. 1 A). A total of 10 out of 29 isolates showed markedly decreased motility compared to that of PT4 isolates (≤20% of the motility of PT4). Two of these were the old preepidemic strains, and of the other eight motility-impaired isolates, only two were recovered from gastroenteritis cases.
FIG. 1.
(A) Motility of S. Enteritidis isolates. The dashed line indicates the 20% cutoff below which an isolate is assigned a motility-impaired phenotype. S. Gal, S. Gallinarum strain. A representative experiment is shown. (B) Caco-2 adhesion and invasion assays. Total associated bacteria (adhesion) or intracellular bacteria (invasion) were quantified. The dashed line indicates the 0.11% cutoff used as a threshold to differentiate invasion-negative from invasion-positive isolates. Data shown are means ± standard errors. (C) Frequency of invasion-positive S. Enteritidis isolated from human gastroenteritis compared to the frequency of isolates obtained from other sources, i.e., isolates obtained from human systemic disease and nonhuman sources. The P value obtained using Fisher's exact test is shown.
Due to the observed differences in motility, the plates used in the Caco-2 assays were subjected to a short centrifugation step immediately upon inoculation to counteract the motility defects and synchronize bacterium-cell contact. When this step was included, we found that all isolates exhibited levels of adhesion to the cells comparable to those of the reference strain, with values varying between 0.99 and 3.45% of the added inoculum (mean ± standard deviation [SD] = 2.12 ± 0.56) (Fig. 1B). However, the invasion assays revealed a higher heterogeneity of behaviors, with the number of intracellular bacteria varying between 0.02 and 1.10% of the initial inoculum (mean ± SD = 0.27 ± 0.23), confirming our previously reported results (5). Most of the isolates with defective motility also exhibited diminished invasion capacity despite the fact that the centrifugation step significantly increased their invasiveness. Thus, 8 of 9 strains that we previously showed as having impaired abilities to invade Caco-2 cells without centrifugation (5) remained hypoinvasive here in the centrifugation-assisted assays (≤30% of the invasiveness of PT4) (Fig. 1B). Some hypoinvasive isolates were assayed for invasion into polarized Caco-2 cells, but their levels of invasion remained low (data not shown).
We classified the isolates as positive or negative for invasion according to the percentages of the inoculum recovered from the intracellular environment (see Materials and Methods), and for statistical analysis, we grouped them by their source of isolation (gastroenteritis versus other origin). The subset of isolates recovered from human gastroenteritis cases during the epidemic or postepidemic periods displayed significantly better fitness than the isolates from all other sources (P of 0.033 in Fisher's exact test) (Fig. 1C).
Transcriptional response of Caco-2 cells to S. Enteritidis infection.
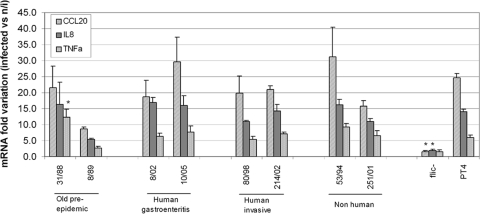
We selected 8 isolates covering all sources and periods of isolation to analyze their ability to elicit a proinflammatory response from Caco-2 cells. For this, we measured the IL-8, CCL20, and TNF-α mRNA levels from infected Caco-2 cells in comparison with those from uninfected cells (1, 52). It has been reported that the response of epithelial cells to Salmonella infection is mediated mainly through Toll-like receptor 5 activation by flagellin (20, 41, 44, 52); thus, we used a fliC::aphT PT4-derived strain as a negative control. Gene expression above the negative-control level was clearly induced in Caco-2 cells upon infection with all the isolates (Fig. 2). No significant differences were observed in the transcriptional responses elicited by the individual isolates compared with that elicited by PT4, except TNF-α expression levels were significantly higher (P < 0.05 in one-way ANOVA compared with that of PT4) after infection with the preepidemic isolate 31/88.
FIG. 2.
Quantitative real-time PCR of CCL20, IL-8, and TNF-α genes from Caco-2 cells infected with S. Enteritidis isolates. Averages ± standard errors are shown. n/i, not infected; fliC−, cells infected with the fliC::aphT PT4-derived strain; *, significant difference related to cells infected with PT4 shown by one-way ANOVA (P < 0.05).
Survival of S. Enteritidis isolates in egg albumen.
Egg albumen can repress the proliferation of S. Enteritidis due to iron restriction, high pH, and enzymatic activities that destabilize the bacterial cell wall (25). It is postulated that survival in egg albumen is essential for S. Enteritidis transmission to humans (21). Thus, we decided to evaluate if there were differences among the isolates in their capacity to survive in egg albumen. As shown in Fig. 3 A, a great diversity of phenotypes was found. Both of the preepidemic isolates showed diminished survival, and survival levels were markedly different between isolates derived from human and nonhuman sources. Our results show that 2 out of 16 human isolates and 6 out of 9 nonhuman isolates were negative for survival (P of 0.01 in Fisher's exact test) (Fig. 3B).
FIG. 3.
(A) S. Enteritidis survival in egg albumen at 2 days postinoculation. Values are expressed as average percentages of initial inocula ± standard errors. The dashed line indicates the 30% cutoff used as a threshold to differentiate survival-negative from survival-positive isolates. (B) Frequency of survival-positive S. Enteritidis isolates obtained from human sources compared to that of isolates obtained from nonhuman sources. The P value obtained using Fisher's exact test is shown.
Three particular isolates were most drastically affected in their capacity to survive (less than 3% of the initial inoculum), namely, the oldest preepidemic isolate (31/88), one isolate obtained from eggs during the peak of the epidemic (254/01), and a human gastroenteritis isolate (47/03) (Fig. 3A). Conversely, several other isolates obtained from different periods and sources showed markedly increased survival in egg albumen compared to that of PT4. We found no clear correlation between the motility of the isolates and their capacity to survive in egg albumen. In fact, only 4 out of 10 isolates with impaired survival in egg albumen were also impaired in motility. Furthermore, the chicken-adapted serovar S. Gallinarum, which is nonmotile, showed a high level of survival (90% of the initial inoculum at 48 hours p.i.) in egg albumen.
In vivo virulence assays in chickens.
As the ability to invade deeper organs in chickens is important for S. Enteritidis transmission to humans, we assessed 12 isolates selected for spleen and ovary-oviduct colonization after oral inoculation into newly hatched chickens (Fig. 4).
FIG. 4.
S. Enteritidis colonization of chicken spleens at 7 days postinoculation. Each symbol represents an individual animal from two independent experiments. The corresponding median for each group is represented with a solid line. *, significant difference compared to that of PT4 shown by one-way ANOVA (P < 0.05).
Consistent with previous reports (2), the PT4 reference strain showed a very high level of invasiveness in this virulence model. All isolates were capable of invading chicken spleens, but several isolates showed significantly lower spleen invasiveness compared to that of PT4 (P < 0.05), namely, the oldest preepidemic isolate (31/88), both isolates obtained from invasive disease (199/02 and 214/02), and two isolates obtained from nonhuman sources (53/94 and 251/01) (Fig. 4). Regarding the colonization of the ovaries-oviducts, we found that all isolates that showed significantly lower invasiveness into the spleens also showed low levels of ovary-oviduct colonization (Table 2). In addition, some isolates that colonized the spleen well were nevertheless poor colonizers of the reproductive organs (isolates 8/02 and 206/99). No clear signs of disease or mortality were observed in any group of animals.
TABLE 2.
S. Enteritidis colonization of chicks' ovaries-oviductsd
| Isolate | Colonizationa | Proportionb |
|---|---|---|
| 31/88c | 0/15 | 0 |
| 08/89 | 2/10 | 0.20 |
| 8/02 | 0/13 | 0 |
| 10/05 | 7/20 | 0.35 |
| 93/05 | 2/19 | 0.11 |
| 199/02c | 2/20 | 0.10 |
| 214/02c | 1/14 | 0.07 |
| 53/94c | 1/18 | 0.06 |
| 206/99 | 1/19 | 0.05 |
| 251/01c | 2/24 | 0.08 |
| 254/01 | 6/19 | 0.32 |
| 32/02 | 2/15 | 0.13 |
| PT4 | 9/33 | 0.27 |
Results expressed as the number of chicks that were Salmonella colonized in their ovaries-oviducts/total number of chicks analyzed for each isolate.
Results expressed as the proportion of Salmonella-positive animals from the total number of animals tested for each isolate.
Isolates showing significant difference of spleen invasiveness compared to that of PT4.
Animals shown in Fig. 4 were analyzed for the presence of Salmonella in enriched homogenates of ovaries-oviducts.
DISCUSSION
S. Enteritidis emerged as a major etiological agent for food-borne disease in Uruguay during the 1990s, and for several years, it was the most frequently isolated serovar of Salmonella in the country. A similar phenomenon occurred in Europe and the United States about a decade earlier. The factors responsible for this worldwide epidemic are still undefined, but evolution of the virulence of the pathogen has been proposed to contribute. In this study, a set of S. Enteritidis isolates from Uruguay during the last 2 decades, and covering different epidemiological situations and diverse sources of isolation, were assayed in several in vitro or in vivo tests for evaluation of their virulence-associated phenotypes. The main findings are as follows: (i) a great heterogeneity of phenotypes was found, which is markedly different from the high homogeneity previously found in the same set of isolates by genomic comparison; (ii) the preepidemic isolates showed low levels of fitness in most of the assays evaluated; and (iii) the subset of isolates from human gastroenteritis showed higher homogeneity of phenotypes and higher levels of fitness than isolates from any other sources.
In previous work (5), we reported high genetic homogeneity among the same set of isolates, as evaluated by molecular genotyping and CGH, with the two preepidemic isolates being the most divergent. However, an initial cell invasion assay also suggested that differences between isolates might be higher than predicted from the genomic data. The results presented here confirm this observation. We found a much higher heterogeneity of phenotypes in all the assays than that estimated by the previous genotyping analysis, with isolates belonging to identical genetic profiles showing very dissimilar behaviors, e.g., isolates 251/01 and 8/02, both previously indistinguishable from the reference strain by all the genotyping methods assayed (5), here nevertheless showed great disparity of phenotype. Thus, isolates that apparently belong to the same clone seem to have markedly different virulence capacities. These findings should be considered when drawing conclusions about potential virulence from genomic comparisons, since usually a single isolate of each lineage is analyzed. Our results are in line with those previously reported by Lu et al., who found high heterogeneity of in vivo and in vitro phenotypes among S. Enteritidis isolates belonging to the same phage type (PT4 or PT8) (32).
It has been reported previously that among strains of S. Enteritidis belonging to a single multilocus genotype, there were subpopulations with distinct capacities to attach and invade avian ovary granulosa cells (39). Isolates were divided into those recovered from eggs or human infections and those isolated from chicken cecal samples. Most S. Enteritidis isolates obtained from eggs or clinical sources were positive for both the attachment and invasive phenotypes, whereas among strains recovered from the chicken ceca, there was a significant excess of attachment-negative phenotypes and more attachment-negative, invasion-negative phenotypes than expected (39). In contrast, we report here significant differences in invasive characteristics between isolates recovered from human gastroenteritis cases and those derived from nonhuman or human systemic infections, using a human intestinal epithelial cell line instead of avian cells. The vast majority of strains (13 out of 14) isolated from human gastroenteritis during the epidemic and postepidemic periods showed proficient invasion into Caco-2 cells, in contrast to the more variable behavior displayed by isolates recovered from other sources, including systemic infections in humans (Fig. 1C). These findings suggest a correlation between the capacity of internalization into human enterocytes and the ability to cause intestinal disease in humans. Overall, these results suggest the existence of a predominant subpopulation of strains with a PT4-like genetic profile, which is fully invasive for human epithelial cells, that was potentially responsible for the epidemics of S. Enteritidis gastroenteritis in Uruguay.
Motility is postulated to enable the bacterium to penetrate the thick mucus that covers the intestinal mucosa and to promote contact with epithelial surfaces, which is often required for effective infection (40). In a recent report, it was demonstrated that motility allows S. Typhimurium to benefit from the nutrients released in the context of an inflamed gut in the mouse model of colitis (43). Here we show that a high proportion of natural isolates (10 out of 29) displayed impaired motility, but only 2 of those were recovered from human gastroenteritis cases during the epidemic (Fig. 1A), reinforcing the view that this feature is important for developing gastroenteritis in humans. Interestingly, three out of four isolates recovered from invasive disease in humans showed defects in motility (Fig. 1A). Several studies have reported the importance of motility for in vitro cell invasion, but no clear explanation has been suggested for this phenomenon (24, 46, 47). We observed that all the hypoinvasive isolates showed adhesion levels similar to or even higher than that of the PT4 reference strain, indicating that the defect in invasion is not due to a lower capacity of the nonmotile isolates to attach to the cells. Of note, all isolates tested possess all known Salmonella pathogenicity island 1 (SPI1), regulatory, and fliC genes required for invasion of cultured epithelial cells, as deduced from our previous microarray analysis (5, 15, 36, 49). However, altered expression of any of these genes or DNA sequence heterogeneity could be involved in the observed invasion defects.
As the inflammatory response to Salmonella infection by the intestinal epithelia is considered to be a determinant for eliciting diarrhea (50), we evaluated a set of isolates, looking for differences in their capacities to elicit a proinflammatory response by Caco-2 cells in vitro. No significant differences were observed in the transcriptional responses elicited by each isolate and PT4, except for TNF-α upregulation elicited by isolate 31/88. No correlation was found between invasiveness and transcriptional responses, i.e., hypoinvasive and fully invasive isolates elicit similar proinflammatory responses. In this respect, it was previously reported that altering the invasive capacity into epithelial cell lines by mutations of the SPI1 type three secretion system does not affect the ability of Salmonella to trigger a proinflammatory response in this system (44, 52).
Several reports have shown that genes involved in bacterial cell wall structure and function, amino acid and nucleic acid metabolism, motility, and stress responses are important for survival in egg albumen or are induced in eggs (8, 19, 31). We have found three isolates markedly affected in survival in egg albumen. One of them, isolate 47/03, lacks rpoS that codes for the stress-induced sigma factor and SEN4286, a gene involved in a type I restriction modification system (5), two features that may explain its deficiency in survival under these conditions of stress. Isolate 31/88 lacks genes coding for cytochrome C synthesis, ferredoxin, alcohol dehydrogenase, aldehyde dehydrogenase, and ethanolamine utilization (5), which could explain its survival-deficient phenotype (25). Isolate 254/01 was indistinguishable from PT4 genomically (5), suggesting that other underlying factors not detected in our CGH analysis may cause the difference in survival.
It is of note that all three egg isolates assayed (251/01, 254/01, and 32/02) showed diminished survival in albumen. A possible explanation is that they may have been originally deposited in the egg yolk, an environment more permissive for bacterial persistence than albumen.
Overall, the analysis of survival in egg albumen discriminated between isolates originating from humans and isolates originating from nonhuman sources, with the former having a significantly higher frequency of isolates more resistant to egg albumen (Fig. 3B). This is consistent with the finding that consumption of contaminated eggs is a major source of human infection with S. Enteritidis.
The chicken virulence assays also revealed high heterogeneity of behaviors, but no significant differences related to the origin of the isolates was found. However, this analysis may be affected by the low number of isolates studied.
The two preepidemic isolates performed deficiently in most of the assays conducted, suggesting that differences in their genomic content correlated with phenotype and giving a plausible explanation for their inability to become epidemic. Strain 53/94, which was isolated immediately before the beginning of the epidemic, showed good fitness in cell invasion and albumen survival tests and moderate fitness in chicken invasion tests, indicating a shift in virulence compared with that of previous isolates. Interestingly, the genetic profile of this isolate is more similar to that seen for the epidemic isolates (Table 1).
Therefore, a correlation between genetic makeup and phenotype can be suggested for the old preepidemic isolates. Some common genetic features between these isolates are the lack of ΦSE20 and the presence of several phage-associated genes (regions 10 to 12 and gogB [5]), which are absent in all other S. Enteritidis isolates analyzed here. However, absence of ΦSE20 was also observed in isolates 53/94 and 206/99, both of which displayed good fitness in almost all assays tested. Acquisition of DNA sequences such as pathogenicity islands is regarded as a potentially important contributor to the evolution of virulence in pathogens. However, loss of function may also be important, as seen in Shigella and Escherichia coli, where a large deletion (around 190 kb) of the genome is involved in enhanced virulence (33). In this regard, we might speculate that the loss of regions 10 to 12 and/or gogB improved the virulence of S. Enteritidis, rendering strains more prone to cause an epidemic. Alternatively, other features contained in the sequences of the old preepidemic isolates and not discriminated by our genotyping analyses may be involved in their virulence-associated phenotypes. Solving the complete genomic sequences of these isolates will shed light on this issue, and this is our future direction.
Acknowledgments
This work was supported by a project grant from the Wellcome Trust (grant 078168/Z/05/Z). L.B. was supported by a fellowship from the Fundacion Manuel Perez, Facultad de Medicina, Uruguay.
We thank Marcelo López and Heather Brooks for excellent technical assistance in the chicken and cell culture experiments.
Footnotes
Published ahead of print on 27 August 2010.
REFERENCES
- 1.Anderle, P., M. Rumbo, F. Sierro, R. Mansourian, P. Michetti, M. A. Roberts, and J. P. Kraehenbuhl. 2005. Novel markers of the human follicle-associated epithelium identified by genomic profiling and microdissection. Gastroenterology 129:321-327. [DOI] [PubMed] [Google Scholar]
- 2.Barrow, P. A. 1991. Experimental infection of chickens with Salmonella enteritidis. Avian Pathol. 20:145-153. [DOI] [PubMed] [Google Scholar]
- 3.Bäumler, A. J., B. M. Hargis, and R. M. Tsolis. 2000. Tracing the origins of Salmonella outbreaks. Science 287:50-52. [DOI] [PubMed] [Google Scholar]
- 4.Betancor, L., F. Schelotto, A. Martínez, M. Pereira, G. Algorta, M. A. Rodríguez, R. Vignoli, and J. A. Chabalgoity. 2004. Random amplified polymorphic DNA and phenotyping analysis of Salmonella enterica serovar Enteritidis isolates collected from humans and poultry in Uruguay from 1995 to 2002. J. Clin. Microbiol. 42:1155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betancor, L., L. Yim, M. Fookes, A. Martinez, N. R. Thomson, A. Ivens, S. Peters, C. Bryant, G. Algorta, S. Kariuki, F. Schelotto, D. Maskell, G. Dougan, and J. A. Chabalgoity. 2009. Genomic and phenotypic variation in epidemic-spanning Salmonella enterica serovar Enteritidis isolates. BMC Microbiol. 9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betancor, L., M. Pereira, A. Martinez, G. Giossa, M. Fookes, K. Flores, P. Barrios, V. Repiso, R. Vignoli, N. Cordeiro, G. Algorta, N. Thomson, D. Maskell, F. Schelotto, and J. A. Chabalgoity. 2010. Prevalence of Salmonella enterica in poultry and eggs in Uruguay during an epidemic due to Salmonella enterica serovar Enteritidis. J. Clin. Microbiol. 48:2413-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolton, A. J., M. P. Osborne, and J. Stephen. 2000. Comparative study of the invasiveness of Salmonella serotypes Typhimurium, Choleraesuis and Dublin for Caco-2 cells, HEp-2 cells and rabbit ileal epithelia. J. Med. Microbiol. 49:503-511. [DOI] [PubMed] [Google Scholar]
- 8.Clavijo, R. I., C. Loui, G. L. Andersen, L. W. Riley, and S. Lu. 2006. Identification of genes associated with survival of Salmonella enterica serovar Enteritidis in chicken egg albumen. Appl. Environ. Microbiol. 72:1055-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogan, T. A., and T. J. Humphrey. 2003. The rise and fall of Salmonella Enteritidis in the UK. J. Appl. Microbiol. 94:114S-119S. [DOI] [PubMed] [Google Scholar]
- 10.Danyluk, M. D., M. Nozawa-Inoue, K. R. Hristova, K. M. Scow, B. Lampinen, and L. J. Harris. 2008. Survival and growth of Salmonella Enteritidis PT 30 in almond orchard soils. J. Appl. Microbiol. 104:1391-1399. [DOI] [PubMed] [Google Scholar]
- 11.Das, E., G. C. Gurakan, and A. Bayindirli. 2006. Effect of controlled atmosphere storage, modified atmosphere packaging and gaseous ozone treatment on the survival of Salmonella Enteritidis on cherry tomatoes. Food Microbiol. 23:430-438. [DOI] [PubMed] [Google Scholar]
- 12.Dibb-Fuller, M. P., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145(Pt. 5):1023-1031. [DOI] [PubMed] [Google Scholar]
- 13.Eckmann, L., M. F. Kagnoff, and J. Fierer. 1993. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect. Immun. 61:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichelberg, K., C. C. Ginocchio, and J. E. Galan. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176:4501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24-29. [DOI] [PubMed] [Google Scholar]
- 16.Finlay, B. B., and J. H. Brumell. 2000. Salmonella interactions with host cells: in vitro to in vivo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella Typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galanis, E., D. M. Lo Fo Wong, M. E. Patrick, N. Binsztein, A. Cieslik, T. Chalermchikit, A. Aidara-Kane, A. Ellis, F. J. Angulo, H. C. Wegener, and World Health Organization Global Salm-Surv. 2006. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg. Infect. Dis. 12:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gantois, I., R. Ducatelle, F. Pasmans, F. Haesebrouck, and F. Van Immerseel. 2008. Salmonella enterica serovar Enteritidis genes induced during oviduct colonization and egg contamination in laying hens. Appl. Environ. Microbiol. 74:6616-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gewirtz, A. T., P. O. Simon, Jr., C. K. Schmitt, L. J. Taylor, C. H. Hagedorn, A. D. O'Brien, A. S. Neish, and J. L. Madara. 2001. Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J. Clin. Invest. 107:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guard-Petter, J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421-430. [DOI] [PubMed] [Google Scholar]
- 22.Hald, T., D. Vose, H. C. Wegener, and T. Koupeev. 2004. A Bayesian approach to quantify the contribution of animal-food sources to human salmonelosis. Risk Anal. 24:255-269. [DOI] [PubMed] [Google Scholar]
- 23.Humphrey, T. J. 1994. Contamination of egg shell and contents with Salmonella enteritidis: a review. Int. J. Food Microbiol. 21:31-40. [DOI] [PubMed] [Google Scholar]
- 24.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang, H., C. Loui, R. I. Clavijo, L. W. Riley, and S. Lu. 2006. Survival characteristics of Salmonella enterica serovar Enteritidis in chicken egg albumen. Epidemiol. Infect. 134:967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller, L. H., C. E. Benson, K. Krotec, and R. J. Eckroade. 1995. Salmonella enteritidis colonization of the reproductive tract and forming and freshly laid eggs of chickens. Infect. Immun. 63:2443-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller, L. H., D. M. Schifferli, C. E. Benson, S. Aslam, and R. J. Eckroade. 1997. Invasion of chicken reproductive tissues and forming eggs is not unique to Salmonella enteritidis. Avian Dis. 41:535-539. [PubMed] [Google Scholar]
- 28.Kim, S. H., and C. I. Wei. 2007. Invasiveness and intracellular growth of multidrug-resistant Salmonella and other pathogens in Caco-2 cells. J. Food Sci. 72:M72-M78. [DOI] [PubMed] [Google Scholar]
- 29.Langridge, G. C., J. Wain, and S. Nair. 18 August 2008, posting date. Chapter 8.6.2.2, Invasive salmonellosis in humans. In A. Bock et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. doi: 10.1128/ecosal.8.6.2.2. [DOI]
- 30.La Ragione, R. M., W. A. Cooley, P. Velge, M. A. Jepson, and M. J. Woodward. 2003. Membrane ruffling and invasion of human and avian cell lines is reduced for aflagellate mutants of Salmonella enterica serotype Enteritidis. Int. J. Med. Microbiol. 293:261-272. [DOI] [PubMed] [Google Scholar]
- 31.Lu, S., P. B. Killoran, and L. W. Riley. 2003. Association of Salmonella enterica serovar enteritidis yafD with resistance to chicken egg albumen. Infect. Immun. 71:6734-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, S., A. R. Manges, Y. Xu, F. C. Fang, and L. W. Riley. 1999. Analysis of virulence of clinical isolates of Salmonella enteritidis in vivo and in vitro. Infect. Immun. 67:5651-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormick, B. A., S. I. Miller, D. Carnes, and J. L. Madara. 1995. Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect. Immun. 63:2302-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohle-Boetani, J. C., J. Farrar, P. Bradley, J. D. Barak, M. Miller, R. Mandrell, P. Mead, W. E. Keene, K. Cummings, S. Abbott, and S. B. Werner. 2009. Salmonella infections associated with mung bean sprouts: epidemiological and environmental investigations. Epidemiol. Infect. 137:357-366. [DOI] [PubMed] [Google Scholar]
- 36.Penheiter, K. L., N. Mathur, D. Giles, T. Fahlen, and B. D. Jones. 1997. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol. Microbiol. 24:697-709. [DOI] [PubMed] [Google Scholar]
- 37.Rabsch, W., H. Tschäpe, and A. J. Bäumler. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3:237-247. [DOI] [PubMed] [Google Scholar]
- 38.Raffatellu, M., R. P. Wilson, D. Chessa, H. Andrews-Polymenis, Q. T. Tran, S. Lawhon, S. Khare, L. G. Adams, and A. J. Bäumler. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73:146-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saeed, A. M., S. T. Walk, M. Arshad, and T. S. Whittam. 2006. Clonal structure and variation in virulence of Salmonella Enteritidis isolated from mice, chickens, and humans. J. AOAC Int. 89:504-511. [PubMed] [Google Scholar]
- 40.Schmitt, C. K., J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sierro, F., B. Dubois, A. Coste, D. Kaiserlian, J. P. Kraehenbuhl, and J. C. Sirard. 2001. Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 98:13722-13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stecher, B., A. J. Macpherson, S. Hapfelmeier, M. Kremer, T. Stallmach, and W. D. Hardt. 2005. Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect. Immun. 73:3228-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stecher, B., M. Barthel, M. C. Schlumberger, L. Haberli, W. Rabsch, M. Kremer, and W. D. Hardt. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell. Microbiol. 10:1166-1180. [DOI] [PubMed] [Google Scholar]
- 44.Tallant, T., A. Deb, N. Kar, J. Lupica, M. J. de Veer, and J. A. DiDonato. 2004. Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-kappa B and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol. 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorns, C. 2000. Bacterial food-borne zoonoses. Rev. Sci. Tech. 19:226-239. [DOI] [PubMed] [Google Scholar]
- 46.van Asten, F. J., H. G. Hendriks, J. F. Koninkx, and J. E. van Dijk. 2004. Flagella-mediated bacterial motility accelerates but is not required for Salmonella serotype Enteritidis invasion of differentiated Caco-2 cells. Int. J. Med. Microbiol. 294:395-399. [DOI] [PubMed] [Google Scholar]
- 47.Van Asten, F. J., H. G. Hendriks, J. F. Koninkx, B. A. Van der Zeijst, and W. Gaastra. 2000. Inactivation of the flagellin gene of Salmonella enterica serotype Enteritidis strongly reduces invasion into differentiated Caco-2 cells. FEMS Microbiol. Lett. 185:175-179. [DOI] [PubMed] [Google Scholar]
- 48.Velge, P., A. Cloeckaert, and P. Barrow. 2005. Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet. Res. 36:267-288. [DOI] [PubMed] [Google Scholar]
- 49.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 50.Winter, S. E., P. Thiennimitr, S. P. Nuccio, T. Haneda, M. G. Winter, R. P. Wilson, J. M. Russell, T. Henry, Q. T. Tran, S. D. Lawhon, G. Gomez, C. L. Bevins, H. Russmann, D. M. Monack, L. G. Adams, and A. J. Baumler. 2009. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype Typhimurium infection. Infect. Immun. 77:1904-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, S. K., L. Eckmann, A. Panja, and M. F. Kagnoff. 1997. Differential and regulated expression of C-X-C, C-C, and C-chemokines by human colon epithelial cells. Gastroenterology 113:1214-1223. [DOI] [PubMed] [Google Scholar]
- 52.Zeng, H., A. Q. Carlson, Y. Guo, Y. Yu, L. S. Collier-Hyams, J. L. Madara, A. T. Gewirtz, and A. S. Neish. 2003. Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J. Immunol. 171:3668-3674. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, S., R. L. Santos, R. M. Tsolis, S. Stender, W. D. Hardt, A. J. Bäumler, and L. G. Adams. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]