Abstract
Nucleosome-free regions (NFRs) at the 5′ and 3′ ends of genes are general sites of transcription initiation for mRNA and noncoding RNA (ncRNA). The presence of NFRs within transcriptional regulatory regions and the conserved location of transcription start sites at NFRs strongly suggest that the regulation of NFRs profoundly affects transcription initiation. To date, multiple factors are known to facilitate transcription initiation by positively regulating the formation and/or size of NFRs in vivo. However, mechanisms to repress transcription by negatively regulating the size of NFRs have not been identified. We identified four distinct classes of NFRs located at the 5′ and 3′ ends of genes, within open reading frames (ORFs), and far from ORFs. The ATP-dependent chromatin-remodeling enzyme Isw2 was found enriched at all classes of NFRs. Analysis of RNA levels also demonstrated Isw2 is required to repress ncRNA transcription from many of these NFRs. Thus, by the systematic annotation of NFRs across the yeast genome and analysis of ncRNA transcription, we established, for the first time, a mechanism by which NFR size is negatively regulated to repress ncRNA transcription from NFRs. Finally, we provide evidence suggesting that one biological consequence of repression of ncRNA, by Isw2 or by the exosome, is prevention of transcriptional interference of mRNA.
Eukaryotic cells compact their DNA into a nucleoprotein complex known as chromatin. The most basic repeating unit of chromatin is the nucleosome, consisting of ∼147 bp of DNA wrapped around an octamer of histone proteins (38). Nucleosomes are one of the most stable protein-DNA complexes known (38) and can effectively inhibit all DNA-dependent processes, including transcription, replication, repair, and recombination, by limiting the access of proteins to DNA (17). As a result, the mechanisms by which chromatin structure and nucleosome positions are specified and maintained in vivo are critical for the regulation of all DNA-dependent processes.
Genome-wide maps of nucleosome positions have recently been generated in a number of organisms, including Saccharomyces cerevisiae (1, 7, 19, 30, 33, 36, 37, 41, 50, 56, 63, 68, 69), Drosophila melanogaster (42), Caenorhabditis elegans (31, 60), Oryzias latipes (52), and humans (6, 54). Each of these organisms displays a characteristic chromatin structure spanning gene-coding regions and transcriptional regulatory regions. Gene-coding regions generally have high nucleosome occupancy with arrays of well-phased nucleosomes extending from the 5′ end of a gene. In contrast, transcriptional regulatory regions, such as promoters, enhancers, and terminators, have low nucleosome occupancy and often contain a nucleosome-free region (NFR). NFRs, also known as nucleosome-depleted regions (NDRs), typically represent regions with an increased accessibility to micrococcal nuclease (MNase) digestion. Thus, the term NFR refers to a deficiency in experimentally determined canonical nucleosomes and does not necessarily imply a complete lack of histones.
To date, predominately two major classes of NFRs, 5′-NFRs and 3′-NFRs, have been characterized. In S. cerevisiae, these NFRs are typically ∼80 to 300 bp in length and are flanked by two well-positioned nucleosomes that often contain the histone variant Htz1 (1, 50). 5′-NFRs, associated with the promoters of many genes, are highly enriched for sequence-specific transcription factor binding sites (7, 37, 63, 68) and demarcate the mRNA transcription start site (TSS) to their downstream edge (1, 37, 63). 3′-NFRs are located at the 3′ ends of genes and are enriched for transcription termination sites (TTSs) (41).
Recent genome-wide expression analyses have demonstrated that the majority of eukaryotic genomes are transcribed (11-13, 16, 26, 43-45, 49, 51, 55, 64, 66), resulting in the identification of numerous noncoding RNA (ncRNA) transcripts. In S. cerevisiae, many of these ncRNA transcripts were found to initiate at the upstream edge of 5′-NFRs between tandemly oriented genes or at 3′-NFRs (45, 66). However, whether these transcripts are subjected to active regulation is not known. The conserved locations of ncRNA TSSs around NFRs strongly suggest that NFRs are general locations of transcription initiation and that the mechanisms controlling NFR accessibility are critical to transcriptional regulation of ncRNA.
Multiple factors, including the physical properties of DNA (3, 29, 33, 41, 68, 69), transcription factors (5, 25), and chromatin regulators (5, 25), are known to positively regulate the formation and/or size of NFRs in vivo. The activities of these factors in establishing larger NFRs are thought to facilitate the initiation of transcription by allowing transcription factors greater access to DNA. Whether there are mechanisms to negatively regulate the size of NFRs in vivo is not known. However, we have recently shown in S. cerevisiae that the ATP-dependent chromatin-remodeling enzyme Isw2 functions at the 5′ and 3′ ends of genes to increase nucleosome occupancy within intergenic regions by sliding nucleosomes away from coding regions. Interestingly, Isw2 was also required to repress noncoding antisense transcripts from the 3′ end of three genes tested (63). Whether Isw2-dependent chromatin remodeling generally affects chromatin structure and ncRNA transcription around NFRs has not been established. We hypothesized that Isw2 may generally function to repress ncRNA transcription by negatively regulating the size of NFRs in vivo.
To test this model, we first analyzed data from multiple nucleosome mapping studies to systematically annotate a consensus set of NFRs across the S. cerevisiae genome. Our work identified two additional classes of NFRs apart from 5′- and 3′-NFRs that were located within open reading frames (ORF-NFRs) and far from ORFs (Other-NFRs). Isw2 targets were found to be significantly enriched at all classes of NFRs, thus identifying a previously unknown target of Isw2, ORF-NFRs. In addition, we employed custom strand-specific tiled microarrays to analyze ncRNA transcripts and found that Isw2 is globally required to repress initiation of cryptic RNA transcripts from NFRs by sliding nucleosomes toward NFRs to restrict their size. Finally, we provide evidence that a potential biological function for Isw2-dependent repression of some cryptic transcripts is to prevent transcriptional interference. To our knowledge, this is the first example in which the negative regulation of NFR size by a chromatin-remodeling enzyme is actively required to repress transcription of ncRNA from NFRs.
MATERIALS AND METHODS
Nucleosome positions.
Nucleosome positions used for the NFR annotation were derived from the following sources: (i) Whitehouse et al. (63), based on a Pearson correlation coefficient (r value) of ≥0.5; (ii) Lee et al. (37), using positions empirically determined as previously described (63) from the normalized log2 microarray signal and only nucleosomes with a Pearson correlation coefficient (r value) of ≥0.5; (iii) Mavrich et al. (41), with a read count of ≥3 from the combined Watson and Crick data; and (iv) Field et al. (19), using all uniquely mapped reads.
NFR annotations.
For the Whitehouse et al. (63), Lee et al. (37), and Mavrich et al. (41) data sets, a Gaussian distribution of linker lengths was fit to the frequency of linker lengths from each data set. Linkers with lengths less than or equal to the average plus 2 standard deviations were discarded. NFRs were then defined as those remaining linkers meeting at least one of the following criteria: (i) the average Pearson correlation coefficient (r value) (Whitehouse et al. [63] and Lee et al. [37]) or standard deviation (Mavrich et al. [41]) of the six surrounding nucleosomes (three immediately flanking nucleosomes on either side of the linker) is greater than or equal to the genome-wide average of all linkers from each data set; (ii) both of the immediately adjacent linkers (one on either side) have a length that is less than or equal to the average plus 2 standard deviations; or (iii) the midpoint of a Htz1-containing nucleosome (rank order of ≥3 from Albert et al. [1]) is present anywhere within one nucleosome distance (150 bp) upstream or downstream of the boundaries of the NFR. For the Field et al. (19) data set, a Gaussian distribution of linker lengths was fit to the frequency of region lengths sequenced fewer than two times. All linkers with lengths less than or equal to the average plus 2 standard deviations were discarded. All remaining linkers with at least one nucleosome distance (150 bp) flanking one side that was sequenced at least twice were annotated as NFRs. The regions identified as NFRs in at least three of the four data sets were annotated as core NFRs. NFRs separated by less than one nucleosome distance (150 bp) were combined into a single NFR. Annotated NFR locations are available for download at http://labs.fhcrc.org/tsukiyama.
Classification of NFRs.
Core NFRs were classified as 5′-NFRs, 3′-NFRs, ORF-NFRs, or Other-NFRs based on their location with respect to all nondubious ORFs. 5′- and 3′-NFRs were required to be within a single nucleosome distance (150 bp) of a nondubious ORF TSS or TTS, respectively, as experimentally determined by Nagalakshmi et al. (44). If no experimentally defined TSS or TTS was available for any particular ORF, the average length of all experimentally determined 5′ and 3′ untranslated regions (82 and 135 bp, respectively) was added to the translation start or termination site, respectively. NFRs contained completely within the coding region of a nondubious ORF were classified as ORF-NFRs. A single NFR could have met more than one criterion listed above and would thus have multiple classifications (5′-, 3′-, or ORF-NFR). NFRs not classified as 5′-, 3′-, or ORF-NFRs were classified as Other-NFRs. Tandem, divergent, and convergent NFRs were defined as individual NFRs classified as both a 5′- and 3′-NFR of tandemly oriented gene pairs, a 5′-NFR of two divergently oriented gene pairs, or a 3′-NFR of two convergently oriented gene pairs, respectively. Locations of annotated shared NFRs are available for download at http://labs.fhcrc.org/tsukiyama.
Genomic DNA isolation, fragmentation, and labeling.
Genomic DNA was isolated from wild-type (WT) cells by using Qiagen genomic DNA columns according to the manufacturer's protocols. Purified DNA was fragmented with DNase I to an average size of ∼25 to 75 bp. Fragmented DNA (30 ng) was heated at 70°C for 5 min. Then, 5 μl of buffer 4 (New England Biolabs [NEB]), 5 μl of 2.5 mM CoCl2 (NEB), 5 μl of 1 mM Cy3-dUTP (GE Healthcare), and 5 μl of 20,000 U ml−1 terminal deoxytransferase (NEB) were added to a total volume of 50 μl. The reaction mixture was incubated at 37°C for 3 h. Fifty microliters of H2O was added, and labeled DNA was purified with a gel filtration spin column and ethanol precipitated. The pellet was suspended in a final volume of 50 μl H2O.
RNA isolation, fragmentation, and cDNA labeling.
Yeast cells were grown to an optical density at 660 nm of 0.70 (±0.05), and standard hot acid-phenol extraction was used to isolate total RNA. Twenty-five micrograms of RNA and 12 μg of random hexamers were placed in 40 μl of H2O. The reaction mixture was incubated at 70°C for 5 min, 25°C for 5 min, and 4°C for 5 min. A 0.5-μl aliquot of 1-mg ml−1 actinomycin D, 20 μl of 5× SuperScript III buffer (Invitrogen), 8 μl of 0.1 M dithiothreitol, 1 μl of 40-U μl−1 RNase inhibitors (Roche), 4 μl of deoxynucleoside triphosphate mix (10 mM dATP, 10 mM dGTP, 10 mM dCTP, 8.5 mM dTTP, and 1.5 mM dUTP), 4 μl of 200-U μl−1 SuperScript III (Invitrogen), and 20 μl H2O were added, and the reaction mixture was incubated at 25°C for 10 min, 48°C for 90 min, and 70°C for 10 min. Two microliters of 0.1-mg ml−1 RNase A and 2 μl of 5,000-U μl−1 RNase H (NEB) were added and incubated at 37°C for 30 min. The reaction was stopped by phenol-chloroform extraction, and unincorporated random hexamers were removed with a gel filtration spin column. cDNA was ethanol precipitated, and the pellet was suspended in 80 μl H2O.
cDNA was fragmented with 5 μl of 10-U μl−1 APE1 (NEB), 5 μl of 2-U μl−1 uracil deglycosylase (NEB), and 10 μl buffer 4 (NEB) at 37°C for 60 min. The reaction was stopped by phenol-chloroform extraction and was purified with a gel filtration spin column. The fragmented cDNA was ethanol precipitated, and the pellet was suspended in 20 μl H2O.
Ten micrograms of fragmented cDNA was heated at 70°C for 5 min. Three microliters of buffer 4 (NEB), 3 μl of 2.5 mM CoCl2 (NEB), 5 μl of 1 mM Cy5-dUTP (GE Healthcare), and 3 μl of 20,000-U μl−1 terminal deoxytransferase (NEB) were added to the cDNA to a total volume of 30 μl. The reaction mixture was incubated at 37°C for 3 h. Fifty microliters H2O was added, and the reaction was purified with a gel filtration spin column and ethanol precipitated. The pellet was suspended in a final volume of 20 μl H2O.
Microarray hybridizations.
Five micrograms of labeled cDNA and 2.5 μg of labeled genomic DNA were competitively hybridized to custom strand-specific microarrays from NimbleGen. Microarrays tiled both strands of chromosomes III, VI, and XII with 50-mer probes overlapping, on average, by ∼42 bp with an ∼4-bp offset between strands. Biweight mean-adjusted log2 ratios were determined (Cy5/Cy3 ratios were generated using NimbleScan software). Raw and analyzed microarray hybridization files are available for download at http://labs.fhcrc.org/tsukiyama. The sequences of probes on microarrays are available upon request.
Identification of cryptic transcription units.
To identify probes with significantly different signals between strains, we used LIMMA (57), utilizing an object containing the log2 ratio data from all hybridizations as input, to generate P values (adjusted for multiple testing by using the method of Benjamini and Hochberg [6a] for each probe between strains. A 10-probe sliding window (∼80 bp) was used to identify regions that had at least 5 probes with a P value of ≤0.05. All probes within that window had to be less than or equal to 12 bp apart to account for missing or nontiled regions of the genome. Each region's start and end points were further trimmed so as to begin and end with a statistically significant probe. A Gaussian distribution was fit to a frequency plot of all the distances between adjacent regions. Regions that were closer to or equal in distance than the mean (∼60 bp) plus 2 standard deviations (∼30 bp) were combined into a single region. Finally, for each pair-wise comparison, a Gaussian distribution curve was fit to a frequency plot of the lengths of the regions, and means and standard deviations were calculated. Only regions that were greater than or equal to the mean plus 2 standard deviations were used, thus eliminating transcriptional units that were very small. Annotated cryptic transcriptional units are available for download at http://labs.fhcrc.org/tsukiyama.
Annotation of transcriptional interference loci.
Genes containing both a decreased sense coding transcript (mRNA) and an increased cryptic sense or antisense transcript directly overlapping the ORF (defined from the annotated TSS or TTS as described above for NFR annotation) were annotated as sense-sense or antisense-sense transcriptional interference loci. Annotated transcriptional interference loci are available for download at http://labs.fhcrc.org/tsukiyama.
Heat maps, graphs, and screen shots.
For all heat maps and average profiles, data were aligned as described below in the figure legends, and the signals were averaged into 20-bp nonoverlapping bins. The log2 signal, taken before averaging and binning, was required for profiles of Swr1 (61), Rpb3 (61), Rpo21 (61), Sua7 (61), TBP (61), Rsc9 (61), and Reb1 (61). The log2 signal, taken before averaging and binning, and an 11-bin moving average were required for profiles of H3K9Ac (47), H3K14Ac (47), H4Ac (47), H3K4Me1 (47), H3K4Me2 (47), H3K4Me3 (47), H3K36Me3 (47), H3K79Me3 (47), Esa1 (47), Gcn5 (47), and Rsc8 (5) (3-bin moving average). Additionally, chromosome X was omitted from the analysis due to a misalignment of data from Venters and Pugh (61). Each transcription factor binding site represents a binding P value of <0.05 and is conserved in at least one other yeast according to data reported by Harbison et al. (24).
All screen shots were taken using the Integrated Genome Browser program (46).
Microarray data accession number.
All raw and processed microarray data are available for download at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE23108.
RESULTS
Genome-wide annotation of NFRs.
It is generally accepted that mRNA transcription initiates at the edges of NFRs and can be facilitated by multiple factors that positively regulate the formation and/or size of NFRs. Recent studies have shown that ncRNA also initiates at the edges of NFRs (45, 66). However, the mechanisms for regulation of these transcripts are not well understood. Given the widespread prevalence of ncRNA detected throughout the S. cerevisiae genome (12, 13, 43-45, 66), it is critical to understand the mechanisms for regulation of ncRNAs. Isw2 is known to target the 3′ end of ∼250 genes and to repress noncoding transcription at three of the loci tested (63). These results suggest a possibility that Isw2 may be preferentially targeted to NFRs, where it functions as a unique chromatin regulator to repress ncRNA transcription by negatively restricting the size of NFRs. To test our model, we first systematically annotated NFRs genome-wide by using data from multiple independent nucleosome mapping data sets.
Global identification of NFRs was first done at the 5′ ends of genes by Yuan et al. (68). Subsequently, numerous studies recapitulated this group's findings and further showed that NFRs are also commonly present at the 3′ ends of genes (1, 30, 41, 45, 59, 66). Generally, these studies based their analyses on a single nucleosome mapping data set and identified NFRs as regions with a wider-than-average linker length or by fitting an idealized model to smoothed nucleosome signals. However, all nucleosome mapping data sets contain a large number of loci where nucleosomes are poorly defined, and while the overall agreement in nucleosome positions between studies is good, significant differences between all studies exist (30). Furthermore, some NFRs were identified only by their proximity to the ends of genes, thus precluding the annotation of NFRs located elsewhere in the genome. These reasons thus necessitated an unbiased and systematic annotation of NFRs.
Therefore, we developed an algorithm by using multiple criteria to systematically identify NFRs commonly found in multiple nucleosome mapping data sets (see Materials and Methods for details). These criteria were designed to eliminate NFRs from regions with poorly defined nucleosomes and to identify NFRs present in several nucleosome mapping data sets, thus mitigating the identification of spurious NFRs and enriching for NFRs with high confidence levels. This algorithm was applied to four independent nucleosome mapping data sets, two by using high-resolution microarrays and two by using high-throughput sequencing (19, 37, 41, 63). This algorithm identified a reference set of 6,589 core NFRs (Fig. 1 A). Because the annotated NFRs represent the overlapping regions identified from each data set and not necessarily the definitive edges of each NFR, the mean length and total genomic coverage (99 bp and ∼5.4%, respectively) are likely underestimates.
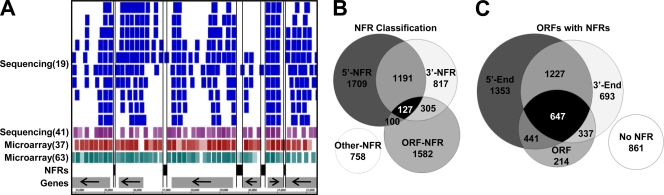
FIG. 1.
Systematic annotation of NFRs. (A) Representative screen shot (chromosome 12, 34,000 to 42,000 bp) displaying the annotated NFRs with respect to nucleosome positions. NFRs are shown as black boxes and nucleosomes are depicted as colored boxes, with lighter boxes representing delocalized nucleosomes (nucleosomes with a lower r value or higher standard deviation). Genes are shown as gray rectangles with arrows indicating the direction of coding transcription and black horizontal lines representing the boundaries of each core NFR. (B) A Venn diagram displaying the classification of the four NFRs, 5′-, 3′-, ORF-, and Other-NFRs (as described in Materials and Methods) of all 6,589 annotated NFRs. (C) Venn diagram of all 5,773 nondubious ORFs associated with at least one annotated NFR at the 5′ end, 3′ end, or within the middle of the ORF. Overlapping regions indicate ORFs associated with multiple NFRs. No NFR refers to nondubious ORFs not associated with any NFR.
It should be noted that during the course of this study, Jiang and Pugh (30) independently annotated NFRs genome-wide. In their study, NFRs were annotated based on the length of the linker regions from a compiled reference set of nucleosome positions, resulting in the identification of 14,467 NFRs. A direct comparison of both sets of NFRs revealed a statistically significant overlap (4,548 overlapping NFR regions; P < 10−300). However, many of the NFRs identified by Jiang and Pugh that were not present in our data set corresponded to regions of poorly defined nucleosomes or regions deficient of nucleosomes in only a single data set. Thus, for the purpose of this study, NFRs defined by our algorithm were more suitable.
Classification of NFRs.
NFRs were first identified around the TSSs at the 5′ ends of genes and, more recently, around TTSs at the 3′ ends of genes. Of the annotated NFRs, a total of 3,127 (averaging ∼111 bp in size and associated with ∼64% of nondubious ORFs) and 2,440 (averaging ∼109 bp in size and associated with ∼50% of nondubious ORFs) were classified as 5′-NFRs and 3′-NFRs, respectively (Fig. 1B). Because of the compact nature of the yeast genome, some 5′- and 3′-NFRs are shared between two neighboring genes in tandem, divergent, or convergent orientations. Therefore, we further classified 5′- and 3′-NFRs and identified 1,312, 555, and 484 shared tandem, divergent, and convergent NFRs, respectively (Fig. 1C). Thus, 2,438 (∼42%) nondubious genes contain a shared NFR at their 5′ end and 2,291 (∼40%) contain a shared NFR at their 3′ end.
Our unbiased genome-wide annotation of NFRs allowed us to further identify two additional classes of NFRs, ORF-NFRs and Other-NFRs (Fig. 1B). The ORF-NFR class comprises 2,114 long linkers (averaging ∼91 bp in size) that are located completely within the open reading frames of 1,639 genes. The remaining 758 NFRs that are not associated with any nondubious ORF were classified as Other-NFRs (averaging ∼88 bp in size) and represent NFRs located within long intergenic regions or upstream of tRNA genes and retrotransposons.
General properties of NFRs.
Previously, others have shown that nucleosomes immediately adjacent to 5′- and 3′-NFRs are well-positioned and that the phasing of nucleosomes progressively decreases with distance from NFRs (41). More recently, NFRs were also shown to have a sequence-intrinsic tendency to exclude nucleosomes (33, 69). Whether these properties are shared by ORF-NFRs or Other-NFRs is not known. We therefore compared the phasing and sequence-dependent exclusion of nucleosomes around each class of NFRs.
To this end, the nucleosome signals from two in vivo WT mapping data sets and one in vitro-reconstituted nucleosome mapping data set were analyzed around all NFRs (Fig. 2 A). As expected, the in vivo nucleosome maps displayed a prominent NFR that was flanked on both sides by an array of well-positioned nucleosomes whose phasing progressively decreased with distance from the NFR midpoint. The in vitro-reconstituted nucleosomes also exhibited a general depletion of nucleosomes at NFRs (Fig. 2A).
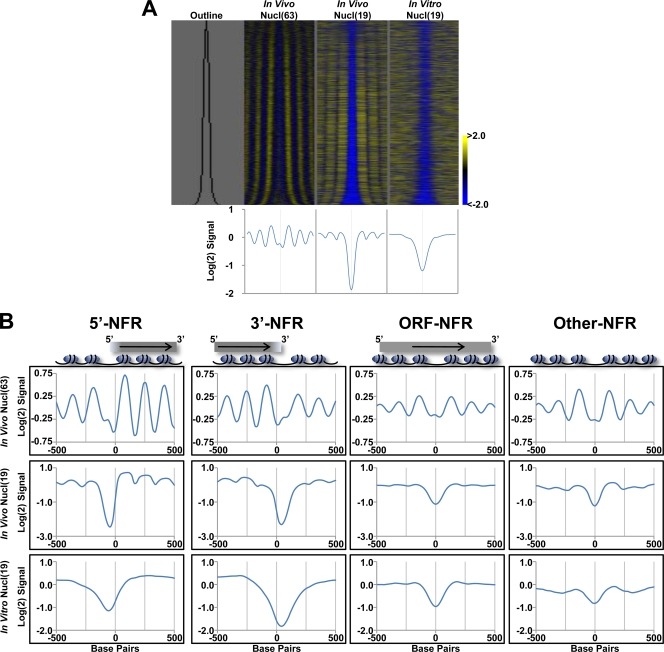
FIG. 2.
All classes of NFRs are surrounded by phased nucleosomes and have a sequence-intrinsic tendency to exclude nucleosomes. (A, Top) Nucleosome signals from two in vivo (19, 63) and one in vitro (19) nucleosome mapping data sets are shown around the annotated NFRs. NFRs for each data set were aligned based on their midpoint and then sorted by size, with the smallest shown at the top and the largest at the bottom. “Outline” marks the edges of each NFR in black. The nucleosome signals from 500 bp on either side of an NFR midpoint are shown. Yellow denotes high nucleosome occupancy, and cyan denotes lack of nucleosomes. (Bottom) Graph displaying the average nucleosome signals across each NFR from the data sets presented above. (B) NFRs were broken down into 5′-NFRs, 3′-NFRs, ORF-NFRs, and Other-NFRs, and the nucleosome signals were averaged for all NFRs within the class. 5′- and 3′-NFRs were realigned to the downstream or upstream edge of each NFR, respectively, such that transcription of the associated coding gene proceeds from left to right. ORF-NFRs are aligned by their midpoint such that transcription of the associated coding gene proceeds from left to right. Other-NFRs are aligned as for panel A.
Next, we individually compared the average in vivo and in vitro nucleosome profiles around each class of NFRs (Fig. 2B). This analysis revealed that all classes of NFRs are flanked by an array of highly positioned nucleosomes with phasing that progressively decreases with distance from the NFR in vivo. In vitro, all classes of NFRs exhibit a sequence-intrinsic property to exclude nucleosomes. However, the level of phasing or sequence-intrinsic exclusion of nucleosomes varies between classes. For example, 5′- and 3′-NFRs are generally bordered by the most highly phased nucleosomes and display the most prominent in vitro nucleosome exclusion, while ORF-NFRs and Other-NFRs have significantly reduced levels of phasing and sequence-intrinsic nucleosome exclusion.
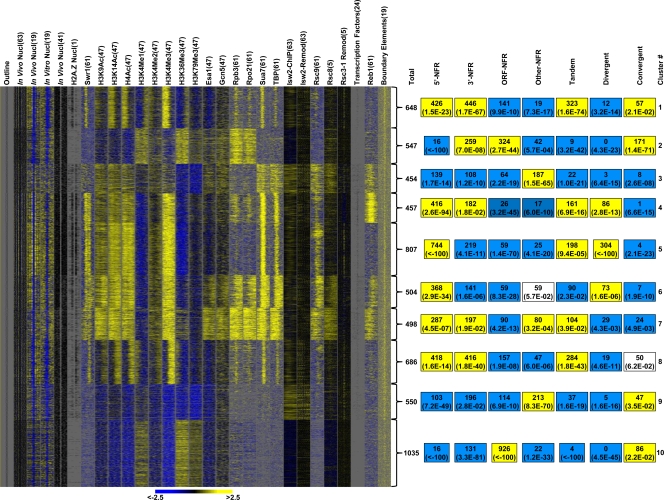
Because the in vivo and in vitro nucleosome profiles for each class of NFRs are different, we speculated that additional factors, beyond the sequence-dependent exclusion of nucleosomes, are required to establish the in vivo nucleosome architecture surrounding NFRs. As such, we analyzed the distribution of various histone modifications, transcriptional machineries, and chromatin-remodeling enzymes surrounding all NFRs. k-means clustering of this profile revealed distinct classes of NFRs that are enriched within different chromatin environments (Fig. 3). This result was consistent with our model that the chromatin architecture surrounding distinct classes of NFRs is differentially influenced by chromatin and transcription regulators in vivo. The observation that the annotated NFRs define boundaries for some histone modifications, such as H3K4me3, H4ac, and H3K14ac within clusters 1 and 8, as well as a histone variant, Htz1 within clusters 1, 4, 5, 6, and 8, provides additional support of the quality of the NFR annotation.
FIG. 3.
k-means clustering revealed distinct classes of NFRs that are enriched within different chromatin environments. (Left) The k-means clustering result (k = 10) for nucleosomes, the ChIP signals for histone modifications, the transcription factors, and the chromatin remodeling factors around the annotated NFRs. For each factor, NFRs were aligned by their midpoints, and the signals from 500 bp on either side of an NFR midpoint are shown. “Outline” marks the edges of each NFR in black. The in vivo (41) and H2A.Z (1) results display the locations of sequenced nucleosomes in black. Each transcription factor binding site, representing significant binding (P < 0.05) and conserved in at least one other yeast, (24) is displayed in black. Identified clusters are delineated by brackets. (Right) The total number of NFRs and the respective enrichment (P values, hypergeometric distribution) of NFR classes and subclasses, within each of the 10 identified clusters. Yellow indicates a positive enrichment, and cyan indicates negative enrichment.
Isw2 association with NFRs.
We next examined whether Isw2 is preferentially targeted to and functions around NFRs. To address these possibilities, we determined the total number of Isw2 targets, defined as regions where both enrichment of Isw2 chromatin immunoprecipitation (ChIP) signals and Isw2-dependent chromatin remodeling (63) take place, that are in close proximity to one of the annotated NFRs. Because we utilized a strict definition for Isw2 targets and restricted our analysis to the edges of the annotated core NFRs, our calculation for the association of Isw2 targets with NFRs is likely a significant underestimation. Nonetheless, our analysis revealed a striking association of Isw2 targets with NFRs. A total of 406 Isw2 targets (P = 9.5e−279), representing ∼48% of all Isw2 targets, were found within a single nucleosome distance (150 bp) of an annotated NFR. Classification of these NFRs targeted by Isw2 showed a statistical enrichment in all classes of NFRs: 5′-NFRs (217 Isw2 targets; P = 1.3e−148), 3′-NFRs (164 Isw2 targets; P = 2.1e−108), ORF-NFRs (50 Isw2 targets; P = 9.0e−14), and Other-NFRs (124 Isw2 targets; P = 4.1e−129). This analysis identified ORF-NFRs as a previously unknown class of Isw2 targets located within genes.
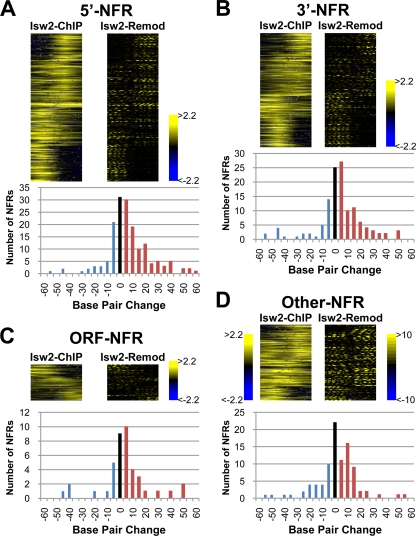
To further understand a role for Isw2 targeting NFRs, we first used self-organizing maps (SOMs) of Isw2 ChIP signals and Isw2-dependent chromatin remodeling around each class of Isw2 target NFRs (Fig. 4 A to D, top panels). This analysis revealed that with each class of NFRs, Isw2 tends to be enriched and remodels nucleosomes immediately adjacent to NFRs, consistent with the fact Isw2 preferentially targets NFRs. Second, plotting the distribution of the change in NFR size between WT and Δisw2 strains (63) revealed an increase in the size of many, but not all, NFRs in Δisw2 strains compared to WT (Fig. 4A to D, bottom panels). These results suggest that Isw2-dependent chromatin remodeling around target NFRs functions to restrict the size of many, but not all, target NFRs.
FIG. 4.
Isw2-dependent chromatin remodeling functions to restrict the size of NFRs. (A to D, top) NFRs targeted by Isw2 were grouped into 5′-NFRs, 3′-NFRs, ORF-NFRs, and Other-NFRs and aligned by their midpoint. Segments of 500 bp on either side of NFRs are shown. For Isw2-ChIP results, SOMs of Isw2-ChIP signals (63) were used to rearrange NFRs (top to bottom) with similar patterns adjacent to each other. Isw2-Remod, Isw2 remodeling signals (63) for NFRs aligned by SOMs of Isw2-ChIP signals as described above. (Bottom) Distribution of the change in NFR size between WT and Δisw2 strains (63). Red bars indicate an increase, black bars indicate no change, and cyan bars indicate a decrease in NFR size in Δisw2 strains compared to WT. (A) 5′-NFRs (203) that are Isw2 targets; (B) 3′-NFRs (153) that are Isw2 targets; (C) ORF-NFRs (51) that are Isw2 targets; (D) Other-NFRs (114) that are Isw2 targets.
Isw2-dependent repression of cryptic transcripts.
We next sought to determine a functional consequence for Isw2-dependent chromatin remodeling around target NFRs. We previously showed that Isw2 represses cryptic antisense transcripts from the 3′ ends of three genes (63). However, whether Isw2 generally functions to repress cryptic RNA transcription is currently unknown. Given that Isw2 is targeted and remodels chromatin around NFRs and that the TSSs of ncRNA are enriched around the edges of NFRs, we speculated that Isw2-dependent chromatin remodeling may result in repression of ncRNA transcription at NFRs.
To test our model, we hybridized total RNA from Isw2 deletion strains to high-resolution, strand-specific microarrays tiling chromosomes III, VI, and XII, covering ∼14% of the genome. Because the exosome complex efficiently degrades cryptic transcripts, deletion of components in the exosome pathway, either TRF4 or RRP6, in combination with ISW2 (Δisw2 trf4 and Δisw2 rrp6, respectively) is required to stabilize some cryptic transcripts (63). These mutations are not expected to alter the frequency of ncRNA transcription (28, 53) but are required for detection of cryptic RNA transcripts. Furthermore, because it is unclear how cryptic RNA transcripts are processed in vivo, especially in the absence of Trf4 or Rrp6, we avoided any selection or amplification of RNA (see Materials and Methods for details).
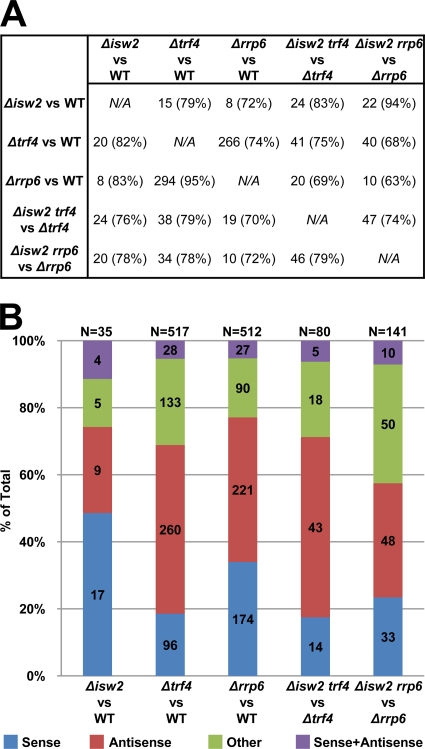
While the Δisw2 single mutant showed relatively few changes compared to WT (Δisw2 versus WT; 35 total Isw2-dependent cryptic transcripts), a total of 80 (mean length of 604 bases) and 141 (mean length of 411 bases) Isw2-dependent cryptic transcripts were identified in the Δtrf4 (Δisw2 trf4 versus Δtrf4) and Δrrp6 (Δisw2 rrp6-versus Δrrp6) backgrounds, respectively. Interestingly, while a larger number of Isw2-dependent cryptic transcripts were identified in Δisw2 rrp6 than in Δisw2 trf4, a more robust increase in cryptic RNA levels was observed in Δisw2 trf4 (data not shown). However, a comparison of cryptic transcripts identified in both double mutants revealed that 47 cryptic transcripts in Δisw2 trf4 cells directly overlapped with 46 cryptic transcripts in Δisw2 rrp6 cells (Fig. 5 A). These data suggest that Trf4 and Rrp6 have overlapping but distinct functions in cryptic RNA regulation.
FIG. 5.
Classification of cryptic transcripts. (A) Displayed are the total numbers of cryptic transcripts identified from a comparison of each strain pair that directly overlaps with a cryptic transcript identified from a different strain pair. The average percentage of overlapping base pairs of cryptic transcript is shown in parentheses. (B) Cryptic transcripts identified were classified as cryptic sense (sense), antisense, other, and sense plus antisense (representing cryptic transcripts that overlap two ORFs in both the sense and antisense direction) as described in Materials and Methods. The numbers within the columns denote the number of transcripts from each strain identified in a particular class. N denotes the total number of cryptic transcripts identified from the comparison of the pair of strains.
We next classified each identified cryptic transcript as either cryptic sense, antisense, or other (Fig. 5B) with respect to the transcriptional direction of an overlapping ORF (mean overlap with OFRs, 491 bp). The major class of Isw2-repressed cryptic transcripts in both Δtrf4 and Δrrp6 backgrounds was cryptic antisense, representing 54% (43 transcripts) and 34% (48 transcripts) of identified cryptic transcripts, respectively (Fig. 5B). A comparison of the cryptic RNA levels between each class of transcripts further revealed, as expected, that the exosome components have little contribution to Isw2-repressed cryptic sense transcripts, as shown by the same RNA levels in Δisw2 and Δisw2 trf4 or Δisw2 rrp6 strains (data not shown). In contrast, Isw2-repressed cryptic antisense transcripts generally require loss of both Isw2 and an exosome component for maximum derepression (data not shown), consistent with the known role of Trf4 and Rrp6 in the selective degradation of cryptic antisense transcripts.
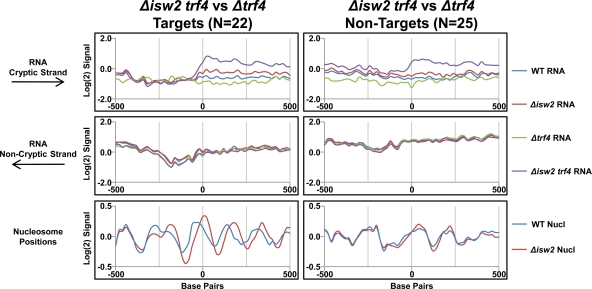
We next examined the relationships between Isw2-dependent chromatin remodeling and cryptic RNA transcription. We found that 22 (28%) and 41 (29%) Isw2-repressed cryptic RNA TSSs in Δisw2 trf4 and Δisw2 rrp6, respectively, are located within 300 bp of an Isw2 target. It should be noted that this is likely an underestimation, as many cryptic transcripts have multiple TSSs, resulting in blurring of microarray signals around the TSSs (see below). Strikingly, at these loci nucleosomes are preferentially shifted downstream of the cryptic TSSs in the absence of Isw2, compared to WT (Fig. 6 and data not shown). These results show that Isw2-dependent chromatin remodeling is often, but not always, associated with the repression of cryptic transcripts. For unknown reasons, we also found nucleosomes are more highly phased around cryptic TSSs in Δisw2 trf4 than in Δisw2 rrp6 (Fig. 6 and data not shown). Finally, an increased level of cryptic transcripts in Δisw2 trf4 was also observed in non-Isw2 targets (Fig. 6). However, the change in transcription, from the baseline to the peak of the cryptic transcript, was lower in nontargets than in targets. These data suggest that indirect effects of Isw2 on ncRNA transcription at nontargets do exist but that these effects tend to be smaller than the direct effects at Isw2 targets.
FIG. 6.
Isw2-dependent chromatin remodeling is associated with suppression of cryptic transcription. Cryptic transcripts identified in the Δisw2 trf4 versus Δtrf4 comparison were broken down into those detected as targets (defined as cryptic transcripts with TSSs within 300 bp of regions with both enriched Isw2-ChIP signals and Isw2-dependent chromatin remodeling [63]) and nontargets (defined as cryptic transcripts with TSSs farther than 300 bp from either enriched Isw2-ChIP signals or Isw2-dependent chromatin remodeling [63]). All cryptic transcripts were then aligned by their 5′ ends (0 bp denotes the 5′ end of each transcript) and oriented such that cryptic transcription occurs to the right. The average signals of RNA (top two panels) and nucleosomes (bottom panels, WT and Δisw2 [63]) around cryptic RNA TSSs are displayed.
5′ rapid amplification of cDNA ends (5′-RACE) was then performed at select loci to verify the cryptic transcripts identified by microarray analysis and to map TSS locations (Fig. 7). All three loci tested from a Δisw2 trf4 strain revealed capped transcripts with multiple TSSs that initiated from the edges of an NFR (within 150 bp) specifically targeted by Isw2. These data are consistent with previous studies that mapped the TSSs of cryptic transcripts (45, 66) and further confirm the role of Isw2-dependent chromatin remodeling in the repression of cryptic ncRNA transcription around NFRs.
FIG. 7.
5′-RACE confirmed Isw2 represses cryptic transcription from NFRs. (A to D) Results of 5′-RACE using Δtrf4 and Δisw2 trf4 strains. Capped, total (“All”), and noncapped transcripts were detected according to the manufacturer's (Ambion) protocol. Colored dots correspond to the quantified transcripts (graph, middle panel), which were normalized to the loading control ACT1. The bottom panel displays the mapped 5′-RACE TSS locations (denoted as the TSSs track) of each quantified transcript in relation to ORFs (designated by gray boxes in ORFs tracks, with arrows denoting the direction of transcription) and NFRs. The RNA signals (average of each replicate) from microarray data at corresponding loci are displayed as heat maps, with yellow denoting higher RNA signals and with cyan denoting lower RNA signals. The cryptic transcripts (Δisw2 trf4 versus trf4) identified on microarrays are shown by red boxes with the arrows designating the direction of the transcripts. (A) ACT1 - YFL039C (loading control); (B) YLL015W; (C) YCL030C; (D) YCR072C.
Transcriptional interference.
There are several potential biological reasons for the degradation or repression of cryptic transcripts by the exosome or Isw2. These include, but are not limited to, conservation of resources for transcription and translation, prevention of abnormal protein synthesis, and alleviation of transcriptional interference. In particular, recent reports in S. cerevisiae have demonstrated examples of transcriptional interference in the regulation of coding transcription (27, 39, 40). It is currently unknown how frequently transcriptional interference occurs on a global scale. Thus, to identify a potential biological role for the repression of ncRNA by Isw2 or the exosome, we identified genes whose mRNA levels were decreased when the levels of directly overlapping cryptic sense (sense-sense transcriptional interference) or antisense (antisense-sense transcriptional interference) transcripts were increased in each mutant strain analyzed.
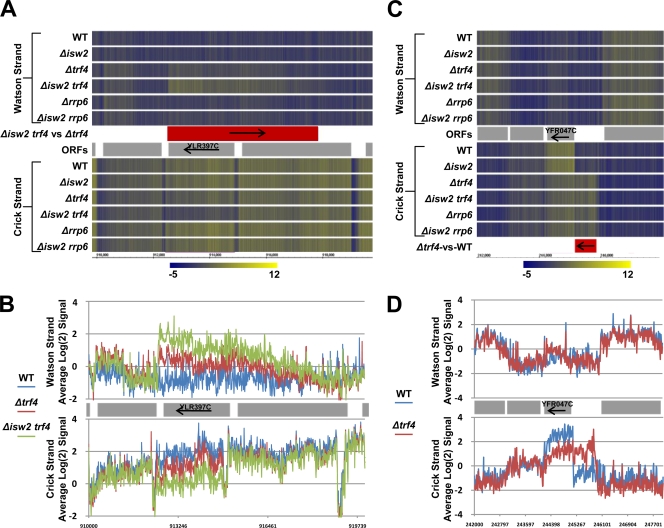
Potential antisense-sense transcriptional interference loci were identified in all strains analyzed, totaling 1 locus in the Δisw2 versus WT comparison, 36 loci in Δtrf4 versus WT, 19 loci in Δrrp6 versus WT, 5 loci in Δisw2 trf4 versus Δtrf4, and 3 loci in Δisw2 rrp6 versus Δrrp6 (one example locus is displayed in Fig. 8 A). In contrast, potential sense-sense transcriptional interference loci were less frequent, totaling nine loci in the Δtrf4 versus WT comparison and one locus in the Δisw2Δrrp6 versus Δrrp6 comparison (one example locus is displayed in Fig. 8B). Closer inspection revealed that many of the increased cryptic transcripts directly overlap the TSS of the repressed mRNA, totaling 1 locus in Δisw2 versus WT, 12 loci in Δtrf4 versusWT, 5 loci in Δrrp6 versus WT, 3 loci in Δisw2 trf4 versus Δtrf4, and 1 locus in Δisw2 rrp6 versus Δrrp6 for antisense-sense transcriptional interference and 9 loci in Δtrf4 versus WT and 1 locus in Δisw2 rrp6 versus Δrrp6 for sense-sense transcriptional interference. These data suggest the possibility that transcriptional interference may be caused by cryptic RNA transcription through the promoter of a gene, underscoring the importance of Isw2 and the exosome in the repression of ncRNA. Considering that our microarrays represent ∼14% of the genome, both sense-sense and antisense-sense transcriptional interference likely occurs at a high frequency throughout the genome. For example, assuming that chromosomes III, VI, and XII accurately represent the global picture, we estimate that ∼60 sense-sense and ∼250 antisense-sense transcriptional interference loci are present genome-wide in a Δtrf4 mutant alone.
FIG. 8.
Evidence for transcriptional interference. (A and C) The RNA signals (average of each replicate) for all the strains examined on high-density microarrays are displayed as heat maps. Arrows indicate the direction of transcription for either ORFs (denoted as gray rectangles) or annotated cryptic transcripts (denoted as red boxes). ORFs displaying transcriptional interference are labeled. Yellow indicates higher RNA signals, and cyan denotes lower RNA signals. (B and D) The RNA signals (average of each replicate) for strains relevant to the transcriptional interference locus are displayed as line graphs. Arrows indicate the direction of transcription for ORFs (denoted as gray rectangles). ORFs displaying transcriptional interference are labeled. (A and B) Example of antisense-sense transcription interference at YLR397C; (C and D) example of sense-sense transcription interference at YFR047C.
DISCUSSION
Genome-wide annotation of NFRs.
In this report we have annotated a consensus set of core NFRs across the yeast genome. This led to the identification of four distinct classes of NFRs, 5′-NFRs, 3′-NFRs, ORF-NFRs, and Other-NFRs. This annotation allowed the direct comparison of each class of NFRs. Examination of nucleosome positions surrounding NFRs showed that all classes of NFRs have a sequence-intrinsic tendency to exclude nucleosomes in vitro and are generally flanked by an array of highly positioned nucleosomes whose phasing progressively decreases with distance from the NFR in vivo. Additionally, k-means clustering of the distribution of chromatin and transcriptional regulators around all NFRs revealed that distinct classes of NFRs are surrounded by different chromatin environments in vivo (Fig. 3). Enrichment of TSSs around the edges of 5′-NFRs and TTSs toward the middle of 3′-NFRs has led to speculation that these NFRs play crucial roles in transcriptional regulation. In contrast, the functions of ORF-NFRs and Other-NFRs are not known. We speculate that, similar to 5′- and 3′-NFRs, ORF- and Other-NFRs may become sites of transcription initiation and/or termination under certain conditions. In fact, several recent studies have shown that deletion of a number of chromatin and transcription regulators, including Rpd3, Set2, Spt6, and Spt16, or a nutritional shift of WT cells, leads to initiation of cryptic RNA transcription within ORFs (8, 9, 32). While the TSSs of these transcripts have not been mapped on a global scale, it is possible that some of these cryptic transcripts initiate at the edges of ORF-NFRs. Thus, our annotation of NFRs lays the foundation for a better understanding of the relationship between chromatin architecture and any DNA-dependent processes that are affected by NFRs.
The yeast genome is very compact, with intergenic regions averaging ∼500 bp. As a result, promoters and terminators are generally within close proximity. Consistently, we found that a significant fraction of tandem, divergent, and convergent gene pairs contain a shared NFR within their transcriptional regulatory regions at the 5′ or 3′ end of the gene. How cells prevent RNA polymerases from colliding or interfering with each other at shared NFRs is unknown. We speculate that if the genes associated with shared NFRs were transcribed at different times, this would mitigate the likelihood of colliding or interfering RNA polymerases. Alternatively, it is possible that the collision of RNA polymerases is not a frequent event and does not pose significant problems for cells.
In vivo functions of Isw2.
In this study, we showed that Isw2 targets are significantly enriched in all classes of NFRs, and we identified a novel class of Isw2 targets within ORFs (ORF-NFRs). Strikingly, for all classes of NFRs targeted by Isw2, Isw2-dependent chromatin remodeling was found to restrict the size of many, but not all, NFRs by sliding nucleosomes toward the middle of NFRs. To our knowledge, this is the first example in which a functional role for a chromatin-remodeling enzyme has been shown to decrease the size of NFRs in vivo. For example, previous reports demonstrated that the RSC complex is required at a subset of gene promoters, or 5′-NFRs, to exclude nucleosomes and increase the size of NFRs (5, 25). These results demonstrate that the accessibility of NFRs is under dynamic control by multiple chromatin regulators, which is consistent with the idea that NFRs play highly important roles in vivo.
It was recently found that ncRNA is transcribed throughout the S. cerevisiae genome (12, 13, 43-45, 66). However, how these transcripts are regulated is not well understood. Several studies have shown that a number of other chromatin regulators and transcription factors appear to function cotranscriptionally with the elongating RNA polymerase to repress cryptic transcripts from initiating within ORFs (8, 9, 32). In contrast, we found a large fraction of cryptic transcripts repressed by Isw2 initiate around NFRs and are antisense to known ORFs. These results revealed a mechanism by which transcription of cryptic ncRNA can be repressed by restricting the size of NFRs. In addition, our results and those of others (8, 9, 32) collectively establish that a large number of chromatin regulators are used to control cryptic RNA transcription in vivo. This suggests the possibility that there may be a significant number of unidentified mechanisms used to control ncRNA transcription in vivo.
Considering the widespread cryptic RNA transcription that occurs in all eukaryotes (11-13, 16, 26, 43-45, 49, 51, 55, 64, 66), it is likely that many of the mechanisms for repressing cryptic RNA transcription are conserved in eukaryotes. ISWI homologues are necessary for normal development in D. melanogaster (14), C. elegans (2), Xenopus laevis (15, 65), and mice (58). In fact, the ISWI family is known to play key roles in a variety of essential biological processes, including transcription (2, 4, 14, 18, 22, 34, 35, 63, 67), global chromatin structure (14), DNA replication (10, 48, 62), cell cycle progression (21), ribosomal DNA silencing (70, 71), and cohesin loading (23). However, the underlying mechanisms by which these ISWI homologues are required for these processes are not fully understood. Because the subunits of the ISWI complex are highly conserved (20), we speculate that ISWI may be required to repress cryptic RNA transcription in other eukaryotes, similar to Isw2 in S. cerevisiae. If this is the case, misregulation of cryptic RNA transcripts caused by loss of ISWI could trigger widespread RNA interference (RNAi), which might lead to the observed developmental defects. While S. cerevisiae has no functional RNAi pathway, our work revealed a large number of loci potentially exhibiting transcriptional interference upon loss of Isw2. These results underscore the importance for future investigations into Isw2 function and establish this remodeling enzyme as a model for elucidating the functions and regulatory mechanisms of cryptic RNA transcription and the modulation of chromatin structure.
Acknowledgments
We thank J. Rodriguez, M. Hogan, N. Bogenschutz, and T. Au for helpful discussions and critical reading of the manuscript.
This research was supported by grant R01GM058564 to T.T. A.N.Y. was supported by Developmental Biology Predoctoral Training Grant T32HD007183 from the National Institute of Child Health and Human Development.
Footnotes
Published ahead of print on 30 August 2010.
REFERENCES
- 1.Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton, S. C. Schuster, and B. F. Pugh. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446:572-576. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, E. C., X. Lu, and H. R. Horvitz. 2006. C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development 133:2695-2704. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. D., and J. Widom. 2001. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell. Biol. 21:3830-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badenhorst, P., M. Voas, I. Rebay, and C. Wu. 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16:3186-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badis, G., E. T. Chan, H. van Bakel, L. Pena-Castillo, D. Tillo, K. Tsui, C. D. Carlson, A. J. Gossett, M. J. Hasinoff, C. L. Warren, M. Gebbia, S. Talukder, A. Yang, S. Mnaimneh, D. Terterov, D. Coburn, A. Li Yeo, Z. X. Yeo, N. D. Clarke, J. D. Lieb, A. Z. Ansari, C. Nislow, and T. R. Hughes. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32:878-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 6a.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57:289-300. [Google Scholar]
- 7.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, V., G. Chua, N. N. Batada, C. R. Landry, S. W. Michnick, T. R. Hughes, and F. Winston. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, N., R. A. Poot, I. Kukimoto, C. Garcia-Jimenez, G. Dellaire, and P. D. Varga-Weisz. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32:627-632. [DOI] [PubMed] [Google Scholar]
- 11.Core, L. J., J. J. Waterfall, and J. T. Lis. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David, L., W. Huber, M. Granovskaia, J. Toedling, C. J. Palm, L. Bofkin, T. Jones, R. W. Davis, and L. M. Steinmetz. 2006. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U. S. A. 103:5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, C. A., and M. Ares, Jr. 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 103:3262-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deuring, R., L. Fanti, J. A. Armstrong, M. Sarte, O. Papoulas, M. Prestel, G. Daubresse, M. Verardo, S. L. Moseley, M. Berloco, T. Tsukiyama, C. Wu, S. Pimpinelli, and J. W. Tamkun. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5:355-365. [DOI] [PubMed] [Google Scholar]
- 15.Dirscherl, S. S., J. J. Henry, and J. E. Krebs. 2005. Neural and eye-specific defects associated with loss of the imitation switch (ISWI) chromatin remodeler in Xenopus laevis. Mech. Dev. 122:1157-1170. [DOI] [PubMed] [Google Scholar]
- 16.Dutrow, N., D. A. Nix, D. Holt, B. Milash, B. Dalley, E. Westbroek, T. J. Parnell, and B. R. Cairns. 2008. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat. Genet. 40:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrenhofer-Murray, A. E. 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 271:2335-2349. [DOI] [PubMed] [Google Scholar]
- 18.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Field, Y., N. Kaplan, Y. Fondufe-Mittendorf, I. K. Moore, E. Sharon, Y. Lubling, J. Widom, and E. Segal. 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 4:e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flaus, A., D. M. Martin, G. J. Barton, and T. Owen-Hughes. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34:2887-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fyodorov, D. V., M. D. Blower, G. H. Karpen, and J. T. Kadonaga. 2004. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 18:170-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 23.Hakimi, M. A., D. A. Bochar, J. A. Schmiesing, Y. Dong, O. G. Barak, D. W. Speicher, K. Yokomori, and R. Shiekhattar. 2002. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418:994-998. [DOI] [PubMed] [Google Scholar]
- 24.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartley, P. D., and H. D. Madhani. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, Y., B. Vogelstein, V. E. Velculescu, N. Papadopoulos, and K. W. Kinzler. 2008. The antisense transcriptomes of human cells. Science 322:1855-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hongay, C. F., P. L. Grisafi, T. Galitski, and G. R. Fink. 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127:735-745. [DOI] [PubMed] [Google Scholar]
- 28.Houseley, J., and D. Tollervey. 2008. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim. Biophys. Acta 1779:239-246. [DOI] [PubMed] [Google Scholar]
- 29.Iyer, V., and K. Struhl. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14:2570-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, C., and B. F. Pugh. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 10:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, S. M., F. J. Tan, H. L. McCullough, D. P. Riordan, and A. Z. Fire. 2006. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 16:1505-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan, N., I. K. Moore, Y. Fondufe-Mittendorf, A. J. Gossett, D. Tillo, Y. Field, E. M. Leproust, T. R. Hughes, J. D. Lieb, J. Widom, and E. Segal. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazzaro, M. A., D. Pepin, N. Pescador, B. D. Murphy, B. C. Vanderhyden, and D. J. Picketts. 2006. The imitation switch protein SNF2L regulates steroidogenic acute regulatory protein expression during terminal differentiation of ovarian granulosa cells. Mol. Endocrinol. 20:2406-2417. [DOI] [PubMed] [Google Scholar]
- 36.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 37.Lee, W., D. Tillo, N. Bray, R. H. Morse, R. W. Davis, T. R. Hughes, and C. Nislow. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39:1235-1244. [DOI] [PubMed] [Google Scholar]
- 38.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 39.Martens, J. A., L. Laprade, and F. Winston. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429:571-574. [DOI] [PubMed] [Google Scholar]
- 40.Martens, J. A., P. Y. Wu, and F. Winston. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19:2695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mavrich, T. N., I. P. Ioshikhes, B. J. Venters, C. Jiang, L. P. Tomsho, J. Qi, S. C. Schuster, I. Albert, and B. F. Pugh. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mavrich, T. N., C. Jiang, I. P. Ioshikhes, X. Li, B. J. Venters, S. J. Zanton, L. P. Tomsho, J. Qi, R. L. Glaser, S. C. Schuster, D. S. Gilmour, I. Albert, and B. F. Pugh. 2008. Nucleosome organization in the Drosophila genome. Nature 453:358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miura, F., N. Kawaguchi, J. Sese, A. Toyoda, M. Hattori, S. Morishita, and T. Ito. 2006. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. U. S. A. 103:17846-17851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagalakshmi, U., Z. Wang, K. Waern, C. Shou, D. Raha, M. Gerstein, and M. Snyder. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neil, H., C. Malabat, Y. d'Aubenton-Carafa, Z. Xu, L. M. Steinmetz, and A. Jacquier. 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457:1038-1042. [DOI] [PubMed] [Google Scholar]
- 46.Nicol, J. W., G. A. Helt, S. G. Blanchard, Jr., A. Raja, and A. E. Loraine. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale data sets. Bioinformatics 25:2730-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 48.Poot, R. A., L. Bozhenok, D. L. van den Berg, S. Steffensen, F. Ferreira, M. Grimaldi, N. Gilbert, J. Ferreira, and P. D. Varga-Weisz. 2004. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat. Cell Biol. 6:1236-1244. [DOI] [PubMed] [Google Scholar]
- 49.Preker, P., J. Nielsen, S. Kammler, S. Lykke-Andersen, M. S. Christensen, C. K. Mapendano, M. H. Schierup, and T. H. Jensen. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322:1851-1854. [DOI] [PubMed] [Google Scholar]
- 50.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samanta, M. P., W. Tongprasit, H. Sethi, C. S. Chin, and V. Stolc. 2006. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc. Natl. Acad. Sci. U. S. A. 103:4192-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasaki, S., C. C. Mello, A. Shimada, Y. Nakatani, S. Hashimoto, M. Ogawa, K. Matsushima, S. G. Gu, M. Kasahara, B. Ahsan, A. Sasaki, T. Saito, Y. Suzuki, S. Sugano, Y. Kohara, H. Takeda, A. Fire, and S. Morishita. 2009. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science 323:401-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmid, M., and T. H. Jensen. 2008. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 33:501-510. [DOI] [PubMed] [Google Scholar]
- 54.Schones, D. E., K. Cui, S. Cuddapah, T. Y. Roh, A. Barski, Z. Wang, G. Wei, and K. Zhao. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seila, A. C., J. M. Calabrese, S. S. Levine, G. W. Yeo, P. B. Rahl, R. A. Flynn, R. A. Young, and P. A. Sharp. 2008. Divergent transcription from active promoters. Science 322:1849-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shivaswamy, S., A. Bhinge, Y. Zhao, S. Jones, M. Hirst, and V. R. Iyer. 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 6:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- 58.Stopka, T., and A. I. Skoultchi. 2003. The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. U. S. A. 100:14097-14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun, W., W. Xie, F. Xu, M. Grunstein, and K. C. Li. 2009. Dissecting nucleosome free regions by a segmental semi-Markov model. PLoS One 4:e4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valouev, A., J. Ichikawa, T. Tonthat, J. Stuart, S. Ranade, H. Peckham, K. Zeng, J. A. Malek, G. Costa, K. McKernan, A. Sidow, A. Fire, and S. M. Johnson. 2008. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 18:1051-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venters, B. J., and B. F. Pugh. 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19:360-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vincent, J. A., T. J. Kwong, and T. Tsukiyama. 2008. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 15:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitehouse, I., O. J. Rando, J. Delrow, and T. Tsukiyama. 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450:1031-1035. [DOI] [PubMed] [Google Scholar]
- 64.Wilhelm, B. T., S. Marguerat, S. Watt, F. Schubert, V. Wood, I. Goodhead, C. J. Penkett, J. Rogers, and J. Bahler. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453:1239-1243. [DOI] [PubMed] [Google Scholar]
- 65.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86-90. [DOI] [PubMed] [Google Scholar]
- 66.Xu, Z., W. Wei, J. Gagneur, F. Perocchi, S. Clauder-Munster, J. Camblong, E. Guffanti, F. Stutz, W. Huber, and L. M. Steinmetz. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yasui, D., M. Miyano, S. Cai, P. Varga-Weisz, and T. Kohwi-Shigematsu. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419:641-645. [DOI] [PubMed] [Google Scholar]
- 68.Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626-630. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Y., Z. Moqtaderi, B. P. Rattner, G. Euskirchen, M. Snyder, J. T. Kadonaga, X. S. Liu, and K. Struhl. 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 16:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou, Y., and I. Grummt. 2005. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 15:1434-1438. [DOI] [PubMed] [Google Scholar]
- 71.Zhou, Y., R. Santoro, and I. Grummt. 2002. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21:4632-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]