Abstract
ATP-dependent chromatin-remodeling complexes (remodelers) are essential regulators of chromatin structure and gene transcription. How remodelers can act in a gene-selective manner has remained enigmatic. A yeast two-hybrid screen for proteins binding the Drosophila transcription factor Tramtrack69 (TTK69) identified MEP1. Proteomic characterization revealed that MEP1 is a tightly associated subunit of the NuRD remodeler, harboring the Mi2 enzymatic core ATPase. In addition, we identified the fly homolog of human Deleted in oral cancer 1 (DOC1), also known as CDK2-associated protein 1 (CDK2AP1), as a bona fide NuRD subunit. Biochemical and genetic assays supported the functional association between MEP1, Mi2, and TTK69. Genomewide expression analysis established that TTK69, MEP1, and Mi2 cooperate closely to control transcription. The TTK69 transcriptome profile correlates poorly with remodelers other than NuRD, emphasizing the selectivity of remodeler action. On the genes examined, TTK69 is able to bind chromatin in the absence of NuRD, but targeting of NuRD is dependent on TTK69. Thus, there appears to be a hierarchical relationship in which transcription factor binding precedes remodeler recruitment.
Chromatin is the natural template of the eukaryotic transcription machinery. Consequently, regulation of gene expression involves the interplay between sequence-specific transcription factors, the basal machinery, coregulators, and enzymes that modulate chromatin structure. ATP-dependent chromatin-remodeling factors (remodelers) constitute one class of enzymes that target chromatin. The basic biochemical activity of remodelers is to use the energy of ATP hydrolysis to move or eject nucleosomes (8). Although their in vitro activity might suggest that remodelers act in a generic way, it has become clear that different remodelers perform distinct, nonredundant functions. An early example of functional specialization was our finding that the Brahma (BRM) remodeling complexes, but not ISWI remodelers, act as chromatin-specific coactivators for the transcription factor ZESTE (15). Conversely, unlike ISWI, the BRM remodelers were unable to order a nucleosomal array. Moreover, several studies have demonstrated that different remodelers control distinct biological processes (4, 8, 24).
Currently, four major classes of remodelers are recognized, based on their ATPase and accessory subunits (8). These comprise the SWI/SNF, ISWI, CHD, and INO80 families. Mi2 (also known as CHD4) is the founding member of the CHD family of remodelers, characterized by the presence of a tandem chromodomain in the ATPase subunit (9). A unique aspect of NuRD is its coupling of remodeling and histone deacetylase (HDAC) activities in one complex. Although there is some variability between the various vertebrate NuRD complexes described so far, the key subunits of NuRD are the Mi2 ATPase, the protein deacetylases HDAC1 and HDAC2, metastasis-associated proteins MTA1, MTA2, and MTA3, the retinoblastoma-associated histone binding proteins RBP46 and RBP48, p66/68, and MBD2 and MBD3, harboring a methyl CpG binding domain (9). These vertebrate proteins have highly conserved Drosophila counterparts. However, to date, Drosophila NuRD has not been purified as a defined entity. Instead, a two-subunit complex comprising Mi2 and the fly homolog of the Caenorhabditis elegans Mog interacting, ectopic P granulocyte 1 (MEP1) protein has been isolated from Drosophila Kc cells (17). Thus, the identity of Drosophila NuRD has remained unclear.
Mi2 and NuRD play important roles in cell fate control during development. For example, the C. elegans homologs of Mi2 and MEP1 cooperate to maintain the distinction between germ line and soma in developing embryos by inhibiting the expression of germ line-specific genes in somatic cells (32). During lymphocyte development in mammals, NuRD acts as a corepressor for the BTB/POZ domain and the zinc finger transcription factor BCL6, repressing plasma cell-specific genes so as to promote differentiation toward B cells (11). In Drosophila, Mi2 has been implicated in the repression of homeotic genes by the GAP protein Hunchback (16). Moreover, the repression of proneural genes by the transcription factor Tramtrack69 (TTK69) so as to block a neuronal cell fate is dependent on Mi2 (27, 40). Indeed, Mi2 was identified as an interacting partner of TTK69 in a two-hybrid screen, and the two proteins colocalize on many loci on third-instar larval polytene chromosomes (27). Thus, Mi2 appears to act as transcriptional corepressor for TTK69.
TTK is an important Drosophila transcription factor, which is involved in various aspects of development and cell differentiation. As a result of alternative splicing, the ttk locus encodes two isoforms, TTK69 and TTK88. These proteins have different C-terminal zinc finger DNA-binding domains but share an N-terminal BTB/POZ protein-protein interaction domain (5, 14, 28, 39). TTK was first identified as a repressor of the pair-rule genes even skipped and fushi tarazu (5, 14) but is involved in numerous additional processes. For example, TTK controls selective cell fate decisions in the developing embryonic nervous system, in photoreceptor differentiation, and in sensory organ precursor differentiation (1, 3, 12, 13, 18, 21, 30, 37-39). During eye development, TTK blocks the differentiation of precursor cells to photoreceptors and promotes specific nonneuronal fates, such as cone cells. TTK is regulated posttranslationally through ubiquitin-mediated degradation triggered by the E3 ubiquitin ligase SINA, which is itself controlled by RAS-mitogen-activated protein (MAP) kinase signaling (21, 30). This ubiquitin-dependent developmental switch depends on the balance between the antagonistic activities of the deubiquitylating protease UBP64 and SINA (2).
Here we explored the interplay between the sequence-specific transcription factor TTK69 and the remodeler NuRD. A yeast two-hybrid screen identified MEP1 as a TTK69 partner. Our proteomic characterization of NuRD established MEP1 and fly CDK2AP1/DOC1 as bona fide subunits. TTK69 interacts genetically and functionally with both MEP1 and Mi2. We performed a genomewide expression analysis to determine the transcriptional circuitries controlled by TTK69 and NuRD. Their substantial overlap suggests that TTK is an important factor recruiting NuRD to its targets. Our analysis of chromatin association suggests a hierarchical relationship in which TTK69 recruits NuRD rather than the remodeler facilitating TTK69 binding.
MATERIALS AND METHODS
Protein-protein interaction assays.
Yeast two hybrid screens were performed as described previously (27) using a full-length TTK69 cDNA cloned into pLexA202 to screen a 0- to 24-h Drosophila embryonic cDNA library fused to the B42 activation domain in plasmid pJG4-5. Domain mapping was performed by using TTK69 deletion constructs cloned into pLexA202 and tested for their interaction with MEP1 residues 257 to 536 fused to the B42 activation domain in plasmid pJG4-5. The strength of interaction was tested by both colony growth and β-galactosidase assays (27). For glutathione S-transferase (GST)-tagged interaction assays, radiolabeled proteins were produced using the TnTQuick system (Promega) with the pLinkT7β vector. Glutathione beads containing 5 to 10 μg of the appropriate GST fusion protein made up to a 30-μl volume were mixed with 25 μl of the TnT reaction mixture and 50 μl of 2× pulldown buffer (40 mM Tris-HCl [pH 8], 200 mM NaCl, 800 mM KCl, 0.2% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitors), and the mixture was incubated at 4°C for 2 h. The beads were transferred to MobiCol columns (MoBiTec/VH Bioscience) and were washed 4 times with 1× pulldown buffer. Bound proteins were eluted by addition of 20 μl 3× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and boiling. A 10-μl volume of the resulting mixture, along with the input material, was analyzed by SDS-PAGE.
Antibodies, immunological procedures, protein purification, and mass spectrometry.
Polyclonal antibodies were generated by immunizing guinea pigs with GST fusion proteins expressed in Escherichia coli and were affinity purified as described previously (7). The following antigens were used: Mi2 amino acids (aa) 1290 to 1533, MTA1-like aa 132 to 476, MEP1 aa 681 to 800, TTK69 aa 333 to 546, and the full-length CG18292 (CDK2AP1/DOC1) protein. Immunization and affinity purification were carried out as described previously (7). Rabbit polyclonal antibodies against TTK69 (27) and ISWI (6) have been described previously. Coimmunoprecipitations (coIPs), immunoblotting, and immunolocalization on 3rd-instar larval salivary gland polytene chromosomes were performed as described previously (6). Embryo nuclear extracts were prepared from 0- to 12-h-old Drosophila embryos. Immunopurification procedures using affinity-purified antibodies directed against Mi2 or MEP1 and mass spectrometric analysis were all performed as described previously (6, 7). After affinity purification, beads were washed twice with HEMG buffer (25 mM HEPES-KOH [pH 7.6], 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 0.1% NP-40, and a cocktail of protease inhibitors) containing 200 mM KCl (HEMG/200), 5 times with HEMG/500, once with HEMG/200, and finally once with HEMG/200 lacking NP-40. Typical contaminants, also present in the products of immunopurification using beads coated with preimmune serum or antibodies directed against unrelated proteins, were omitted from Table 1. Immunodepletion was performed essentially as described previously (7), with the following adaptations. The nuclear extract was diluted with HEMG/100 to a final total protein concentration of 5 mg/ml and was then cleared by centrifugation. This extract was then incubated with protein A beads cross-linked with either anti-CDK2AP1/DOC1 (α-CDK2AP1/DOC1) antibodies or preimmune serum (mock control). After 2 h, the beads were removed by centrifugation. These steps were repeated 3 more times. Supernatants were then resolved by 8% SDS-PAGE and were analyzed by immunoblotting.
TABLE 1.
Mass spectrometric analysis of MEP1- and Mi2-associated proteins purified from Drosophila embryo nuclear extracts
| Protein identity | Mol mass (kDa) | MEP1 |
Mi2 |
Commentsa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mascot score | emPAI score | No. of unique peptides | % Sequence coverage | Mascot score | emPAI score | No. of unique peptides | % Sequence coverage | |||
| Mi2 (CG8103) | 225 | 8,311 | 11.87 | 98 | 54.1 | 9,323 | 14.83 | 112 | 62.1 | ATPase, NuRD enzymatic core |
| MEP1 (CG1244) | 175 | 3,817 | 9.97 | 40 | 53.2 | 3,315 | 6.02 | 37 | 46.3 | Zinc fingers, SUMO interaction motifs |
| MTA1-like (CG2244) | 97 | 2,610 | 3.94 | 39 | 55 | 3,047 | 6.77 | 43 | 59.1 | Metastasis-associated protein |
| p66/68-like (CG32067) | 95 | 2,229 | 2.33 | 22 | 48.1 | 2,480 | 2.24 | 24 | 48.8 | Transcriptional corepressor |
| RPD3 (CG7471) | 58 | 1,617 | 7.80 | 20 | 51.2 | 1,279 | 3.1 | 17 | 44.9 | Histone deacetylase |
| p55 CAF1 (CG4236) | 48 | 1,316 | 2.95 | 14 | 52.3 | 1,354 | 3.04 | 15 | 50.9 | Histone binding |
| MBD-like A (CG8208-PA) | 36 | 888 | 4.2 | 11 | 36.6 | 461 | 0.8 | 8 | 41.9 | Methyl-CpG binding domain-like isoform A |
| MBD-like B (CG8208-PB) | 25 | 481 | 2.69 | 7 | 46.5 | 300 | 0.92 | 5 | 31.4 | Methyl-CpG binding domain-like isoform B |
| CDK2AP1/DOC1 (CG18292) | 29 | 815 | 3.91 | 8 | 52 | 816 | 1.98 | 9 | 44.1 | CDK2-associated protein 1; deleted in oral cancer 1 |
| TTK69 (CG1856) | 69 | 478 | 0.28 | 7 | 21.8 | 70 | 0.08 | 1 | 5.4 | POZ domain, Zn finger DBD, sequence-specific transcriptional repressor |
| MAD (CG2662) | 50 | 681 | 1.09 | 9 | 34.5 | SMAD transcription factor | ||||
| dCtBP (CG7583) | 42 | 891 | 2.03 | 12 | 41.5 | Transcriptional corepressor | ||||
| Nejire/CBP (CG15319) | 343 | 1,044 | 0.16 | 17 | 9.8 | Histone acetyltransferase and coactivator | ||||
DBD, DNA binding domain.
Drosophila genetics.
All fly stocks were maintained under standard conditions, and crosses were performed using standard procedures. RNA interference (RNAi) lines (10) for MEP1 (strain 24533), Mi2 (strain 10766), and TTK69 (strain 10855) were obtained from the Vienna Drosophila RNAi Centre (http://stockcenter.vdrc.at). The GMR-Gal4 enhancer line was obtained from the Bloomington Drosophila Stock Center at Indiana University (http://flystocks.bio.indiana.edu/). All crosses were performed at 25°C and were repeated several times. Representative examples are shown in Fig. 3.
RNAi, genomewide expression analysis, and ChIP-qPCR.
RNAi in Drosophila S2 cells, RNA isolation, reverse transcription-quantitative PCR (RT-qPCR), microarray experiments, and data analysis were all performed as described previously (26). Details are available upon request. Chromatin immunoprecipitation (ChIP)-qPCR assays were performed, and results were quantified, as described previously (22, 25). Immunoprecipitations were performed with the following antibodies: α-Mi2, α-MEP1, α-TTK69, and α-BAP111 (6). Briefly, Drosophila S2 cells were cultured in Schneider's medium (catlog no. 21720-024; Invitrogen) and treated with double-stranded RNA (dsRNA) for 4 days. Double-stranded RNAs were synthesized using an Ambion Megascript T7 kit. RNA samples from three fully independent experiments were prepared and analyzed by RT-qPCR. Cross-linked chromatin was prepared from S2 cells and was sheared by sonication to an average length of 0.5 kbp. Chromatin was then incubated with the antibodies indicated in Fig. 6. Background ChIP levels, subtracted during data processing, were determined by using beads lacking specific antibodies. Following IP, the recovered DNA was analyzed by qPCR with SYBR green I, using the MyiQ single-color real-time PCR detection system (Bio-Rad). The data presented are the results of three independent biological replicate experiments.
ChIP primers were as follows: for ARK, 5′-ACCTGGCAGTGAATACCTTTGTTG-3′ and 5′-GTGTGACCATATTGAGCCGTATCG-3′; for Hairy, 5′-GAGCCGCAGATACACAGTACACAG-3′ and 5′-GCCGTTCGTGGTTTGCTGATTC-3′; for Engrailed, 5′-GAGCCACTGATTCTTCTG-3′ and 5′-TGTCGGAACAACAGTTGC-3′; for Sk1, 5′-AAAGCAAAGGCAAAAGCAACAG-3′ and 5′-GAGGGTGAACTAACCTTATTTTCC-3′; and for KCNQ, 5′-CGTTGTGGGCGGGTCAGG-3′ and 5′-TATTTGGGTTGTTGGGGTATGGC-3′. All other primer sequences will be made available upon request.
Microarray data accession number.
Raw expression data have been submitted to the ArrayExpress database (Microarray Informatics Team, EMBL) under accession no. E-TABM-1010 (http://www.ebi.ac.uk/microarray-as/ae/).
RESULTS
Transcription factor TTK69 binds the NuRD subunit MEP1.
Previously, we published a yeast two-hybrid screen that resulted in the identification of Mi2 as a TTK69-binding factor (27). Here we report an extension of this screen. Using full-length TTK69 as the bait, we isolated a partial cDNA encoding aa 257 to 536 of Drosophila MEP1, a 1,152-aa protein harboring multiple potential C2H2 zinc fingers and SUMO-interacting motifs (Fig. 1 A). To delineate the MEP1-binding domain of TTK69, we again utilized the two-hybrid assay. Surprisingly, we found that the same region that mediates Mi2 binding also suffices to recruit MEP1 (Fig. 1B) (27). A minimal binding region of about 100 amino acids (aa 392 to 500) is located directly upstream of the zinc fingers of TTK69 and is absent in TTK88. The other portions of TTK69 do not display robust binding to either MEP1 or Mi2. As an independent assay, we tested the ability of recombinant 35S-labeled TTK69 to bind GST-tagged MEP1 (Fig. 1C). TTK69 associates efficiently with immobilized MEP1 but does not bind GST. Together, our results suggest that TTK69 binds MEP1 directly.
FIG. 1.
TTK69 binds Drosophila MEP1. (A) A yeast two-hybrid screen identified Drosophila MEP1 (CG1244) as a TTK69-interacting protein. TTK69 interacts with MEP1 aa 257 to 536, encoded by a partial cDNA that was expressed fused to the activation domain (AD) in the original screen. The potential C2H2 zinc fingers are indicated. (B) A series of TTK69 truncation constructs fused to the LexA DNA-binding domain were used to map the MEP1-binding domain. The BTB/POZ domain, the double zinc finger DNA-binding domain, and the PEST domain are diagramed. Dark green indicates the TTK69 sequence; light green, the sequence shared with TTK88. A region spanning aa 317 to 500 retained full MEP1-binding activity. Amino acids 392 to 500 bound somewhat more weakly but still displayed robust binding to MEP1. The same constructs have been used previously to delineate the Mi2 binding domain (27), revealing an overlap between the MEP1 and Mi2 binding regions of TTK69. Representative streaks of yeast expressing the two-hybrid fusions are shown. The relative interaction strengths are indicated as follows: −, no detectable interaction; (+), very weak; +, weak; ++, strong; +++, very strong. (C) TTK69 binds MEP1 directly. [35S]methionine-labeled full-length TTK69 was incubated with either GST-MEP1 aa 257 to 536 or GST alone. Following washes, bound material was resolved by SDS-PAGE and visualized by autoradiography. Duplicate samples and 10% of the input were loaded.
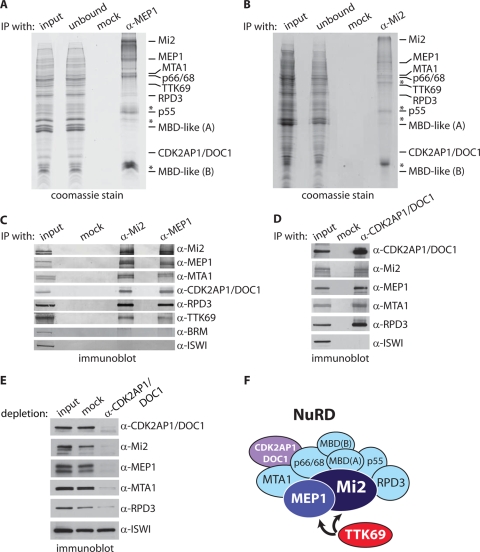
To determine the interaction network of MEP1, we took a proteomics approach to make an inventory of its associated proteins in Drosophila embryo nuclear extracts. Nuclear extracts were incubated with protein A Sepharose beads coated with affinity-purified antibodies directed against MEP1. Following extensive washes with a buffer containing 500 mM KCl and 0.1% NP-40, bound and unbound material was resolved by SDS-PAGE, followed by Coomassie staining (Fig. 2 A). Mass spectrometric analysis suggested that MEP1 is tightly associated with the full NuRD complex and with TTK69 (Fig. 2A and Table 1). No binding to the control beads occurred. Thus, in contrast to MEP1 isolated from Kc cells, which binds only Mi2 (17), our purification from embryo nuclear extracts suggests that MEP1 is part of NuRD. In parallel, we used affinity-purified antibodies directed against Mi2 to purify NuRD (Fig. 2B). Again, our mass spectrometric analysis revealed the presence of MEP1, all core NuRD subunits, and TTK69 (Fig. 2B and Table 1). No peptides unique for TTK88 were identified in the MEP1 or Mi2 purification. Our results suggest that Drosophila NuRD comprises Mi2, MEP1, MTA1, p66/68-like, RPD3, CAF1 p55 (the homolog of RbAp46/48), MBD-like protein isoforms A and B, and the fly homolog of CDK2-associated protein 1 (CDK2AP1). CDK2AP1 is a potential tumor suppressor also known as DOC1 (deleted in oral cancer 1) (31). Although not generally listed as such, CDK2AP1/DOC1 was identified previously as a protein associated with mammalian NuRD (19). Our independent identification of CDK2AP1 in Drosophila NuRD suggests that it might, in fact, be an overlooked subunit of NuRD. Finally, in our immunopurifications, we detected TTK69, confirming its binding to NuRD. However, based on its modest scores in the mass spectrometric analysis, we consider TTK69 a substoichiometric interacting factor and not a NuRD subunit.
FIG. 2.
MEP1 and CDK2AP1/DOC1 are Drosophila NuRD subunits. (A and B) Immunopurification (IP) of MEP1 (A) and Mi2 (B) from embryo nuclear extracts. Embryo nuclear extracts were incubated with protein A-Sepharose beads coated with either a control (anti-GST) antibody (mock) or an affinity-purified anti-MEP1 or anti-Mi2 antibody. Input, unbound material, and proteins retained on the beads after extensive washing were resolved by SDS-PAGE and visualized by Coomassie staining. Bands were excised, and proteins were identified by nanoflow liquid chromatography-tandem mass spectrometry. The mass spectrometry scores of the indicated proteins are listed in Table 1. (C) Coimmunoprecipitations of NuRD and TTK69 with MEP1. Embryo nuclear extracts were incubated either with preimmune serum (mock) or with an anti-Mi2 or anti-MEP1 antibody. Immunopurified proteins were resolved by SDS-PAGE and analyzed by immunoblotting using the indicated antibodies. Ten percent of the input material was loaded for reference. (D) CDK2AP1/DOC1 is a NuRD subunit. Results of coIPs using anti-CDK2AP1/DOC1 antibodies are shown. (E) The majority of NuRD is stably associated with CDK2AP1/DOC1. Nuclear extracts were immunodepleted with beads that were coated either with preimmune serum (mock) or with an antibody directed against CDK2AP1/DOC1. The supernatants were then resolved by SDS-PAGE and analyzed by Western immunoblotting with the indicated antibodies. Whereas NuRD subunits were strongly depleted, ISWI remained unaffected. Because RPD3 is part of multiple complexes, its depletion is expected to be less complete. (F) Cartoon summarizing our proteomic results. Drosophila NuRD comprises the ATPase Mi2, the HDAC RPD3, MTA1-like, CAF1 p55, p66/68-like, MBD-like isoforms A and B, MEP1, and CDK2AP1/DOC1. We note that mammalian CDK2AP1 was identified previously as a mammalian NuRD-associated protein by Le Guezennec et al. (19). We failed to detect additional proteins that have been reported incidentally as binding to NuRD. We do not consider TTK69 a NuRD subunit, although this cannot be formally concluded from the proteomics results. Rather, we view TTK69 as a transcription factor that interacts with NuRD by binding MEP1 and Mi2.
As an additional comparison of MEP1- and Mi2-associated factors, we performed coimmunoprecipitations (coIPs) from embryo nuclear extracts (Fig. 2C). Western immunoblotting confirmed the stable association of Mi2, MEP1, MTA1, CDK2AP1/DOC1, RPD3, and TTK69. BRM and ISW1 acted as negative controls, demonstrating the specificity of the coIPs. To obtain additional evidence that CDK2AP1/DOC1 is a core NuRD subunit, we performed coIPs using antibodies directed against this protein (Fig. 2D). Indeed, anti-CDK2AP1/DOC1 antibodies efficiently purified the NuRd complex, as illustrated by the copurification of Mi2, MEP1, MTA1, and RPD3. To investigate whether the majority of NuRD would be associated with CDK2AP1/DOC1, we immunodepleted an embryo nuclear extract using antibodies directed against this protein. Inspection of the CDK2AP1/DOC1-depleted extract revealed the concomitant loss of Mi2, MEP1, MTA1, and, to a somewhat lesser extent, RPD3 (Fig. 2E). In contrast, ISWI levels were not affected.
Collectively, our proteomic analysis, coIPs, and immune depletion experiments provide compelling evidence that MEP1 and CDK2AP1/DOC1 are tightly associated subunits of Drosophila NuRD. The composition of fly NuRD, based on our analysis, is diagramed in Fig. 2F. The results from our two-hybrid screen, in vitro interaction assay, proteomic survey, and coIPs all support the conclusion that TTK69 binds MEP1 directly. These findings suggest that MEP1 might act as a bridging factor between the DNA-binding transcription factor TTK69 and NuRD.
TTK69, MEP1, and Mi2 interact genetically.
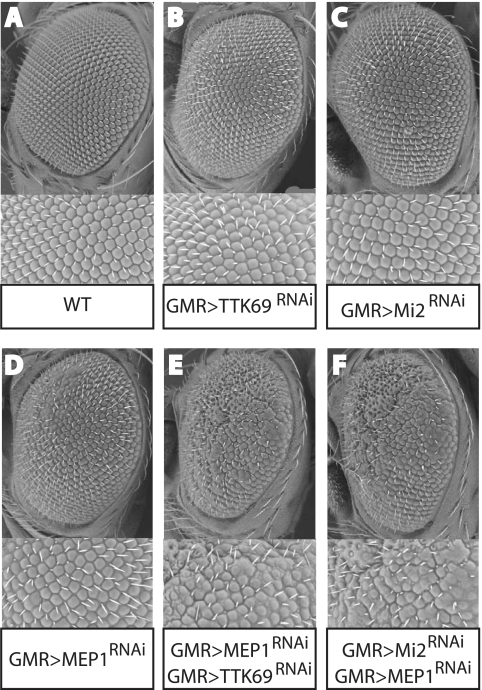
To complement our biochemical results and establish the in vivo significance of the interactions between TTK69, MEP1, and Mi2, we employed a genetic assay. We took advantage of the availability of fly lines expressing interfering RNA (RNAi) targeting TTK69, MEP1, or Mi2 (10). RNAi expression in these lines is under the control of the GAL4-upstream activation sequence (UAS) system, allowing the use of specific drivers to direct tissue-specific knockdowns. To lower TTK69, MEP1, or Mi2 levels in the developing eye, we used glass multiple reporter (GMR)-GAL4 to drive RNAi expression. Depletion of TTK69 (GMR>TTK69RNAi), MEP1 (GMR>MEP1RNAi) or Mi2 (GMR>Mi2RNAi) alone had only a slight effect on eye development and ommatidial arrangement (Fig. 3 A to D). However, the combined reduction of TTK69 and MEP1 (Fig. 3E) strongly affected ommatidial organization and caused a clear rough-eye phenotype. Likewise, the combined reduction of MEP1 and Mi2 synergistically affected eye development (Fig. 3F). These genetic interactions demonstrate that TTK69, MEP1, and Mi2 interact and cooperate in vivo.
FIG. 3.
TTK69, MEP1, and Mi2 interact genetically. Concomitant reduction of MEP1 and TTK69 or MEP1 and Mi2 causes an enhancement of the rough-eye phenotype. Representative scanning electron micrographs of adult eyes from flies with the indicated genotypes are shown. The GMR driver was used to direct the expression of specific interfering RNAs targeting TTK69 (B), Mi2 (C), or MEP1 (D), which had only a slight effect on eye development and the arrangement of ommatidia. However, a combined reduction in the levels of TTK69 and MEP1 (E) or Mi2 and MEP1 (F) strongly enhanced the rough-eye phenotype.
TTK69, MEP1 and Mi2 control overlapping transcriptomes.
To compare the genomewide binding pattern of TTK69 versus the NuRD subunits MEP1 and Mi2, we determined their distributions on larval salivary gland polytene chromosomes (Fig. 4). TTK69 colocalizes with MEP1 and Mi2 on a significant portion of their chromosomal binding sites. However, it is also clear that there are NuRD sites that lack TTK69 and, vice versa, that there are loci bound by TTK69 that are devoid of NuRD. We conclude that whereas TTK69 and NuRD also occupy unique loci, they colocalize on a substantial number of their binding sites.
FIG. 4.
TTK69 colocalizes with MEP1 and Mi2 on many loci. (A to E) The distributions of TTK69 (red) and MEP1 (green) on Drosophila salivary gland chromosomes were determined by immunostaining. (F to J) Likewise, the distributions of TTK69 (red) and Mi2 (green) were determined. DNA stained blue (with DAPI) in the merged panels. The colocalization of TTK69 and NuRD on many sites is demonstrated by the yellow staining in the merged panels and the similar patterns in split polytenes. However, TTK69 and NuRD also occupy unique loci.
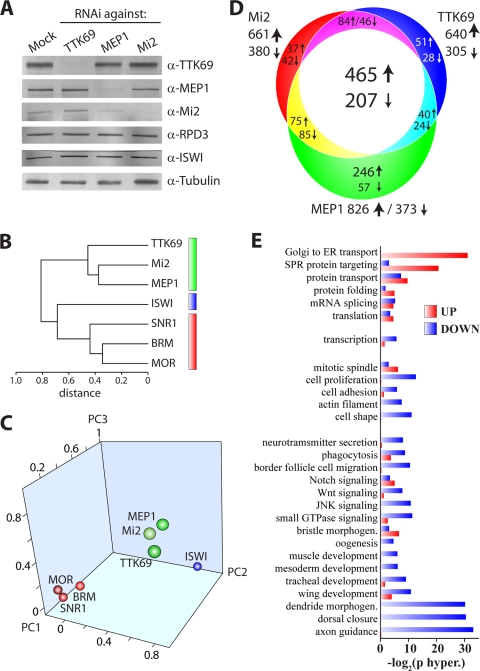
To investigate the level of transcriptional coregulation by TTK69, MEP1 and Mi2, we combined RNAi-mediated depletion with genomewide expression analysis. We treated S2 cells with dsRNA directed against TTK69, MEP1, or Mi2. Immunoblot experiments showed that loss of TTK69 did not affect the stability of the NuRD subunit Mi2, MEP1, or RPD3 (Fig. 5 A). Likewise, depletion of Mi2 or MEP1 left TTK69 levels unchanged. However, depletion of MEP1 did cause a reduction in Mi2 levels, whereas loss of Mi2 did not affect MEP1. RPD3 levels were unaffected by depletion of Mi2, MEP1, or TTK69. ISWI and tubulin acted as loading controls. Next, we extracted RNA from these cells or mock-treated cells. For each subunit, we performed three fully independent RNAi-mediated depletion experiments using distinct cell batches. For comparison, we used the expression analysis of cells depleted of ISWI, SNR1, Moira (MOR), or BRM (26). BRM, MOR, and SNR1 are three common core subunits of the BAP and PBAP remodelers, which represent the SWI/SNF class in Drosophila. The ISWI ATPase forms the enzymatic core of the ISWI class of remodelers, comprising NuRF and ACF/ChRAC.
FIG. 5.
Genomewide expression profiling reveals that TTK69, MEP1, and Mi2 control overlapping transcriptomes. (A) Selective depletion of TTK69, MEP1, and Mi2. S2 cells were either mock treated or treated with dsRNA against TTK69, MEP1, or Mi2. Whole-cell extracts were analyzed by Western immunoblotting using the indicated antibodies. ISWI and α-tubulin served as loading controls. (B) The TTK69 transcriptome correlates well with that of NuRD, but not with that of ISWI or (P)BAP. Agglomerative hierarchical cluster analysis of the expression profiles of TTK69, NuRD, ISWI, and the (P)BAP core subunits SNR1, BRM, and MOR was performed. Clustering is based on Spearman R values. TTK69 and NuRD (green), ISWI (blue), and (P)BAP (red) clusters are indicated. (C) Principal-component (PC) analysis of gene expression profiles reveals the close clustering of TTK69 with Mi2 and MEP1, but not with ISWI or the (P)BAP core subunits. (D) Venn diagram depicting the numbers of genes that are coordinately regulated by TTK69, Mi2, and MEP1. Arrows indicate either upregulation or downregulation following depletion. The numbers of genes affected are given. (E) GO analysis and biological pathway clustering of genes coordinately regulated by TTK69, Mi2, and MEP1. morphogen., morphogenesis.
The extracted RNA was labeled and hybridized with Affymetrics Drosophila Genome 2 arrays. Analysis of the expression data was performed as described previously (26). We used an unbiased statistical analysis of the whole data set to compare the impacts of the various proteins on gene expression. Hierarchical agglomerative clustering derived from Spearman's correlation analysis revealed a striking correlation between the effects of MEP1, Mi2, or TTK69 depletion. In contrast, neither ISWI depletion nor loss of (P)BAP subunits correlated well with the TTK69/NuRD cluster. Another way to uncover relationships between the gene expression profiles of different regulators is the application of principal-component analysis (PCA). PCA is a linear transformation that finds and projects original variables to the minimal principal components (PCs) that account for the maximal variance in the data set. About 76% of the variance in transcriptomes obtained after depletion of the 7 proteins analyzed here is explained by PC1 to PC3. Figure 5C shows the close clustering of the TTK69, Mi2, and MEP1 expression profiles, reflecting their high degree of correlation. The profiles of the (P)BAP core subunits and of ISWI were clearly separated from the TTK69/NuRD cluster, reflecting the fact that each regulates a specific set of genes.
Venn diagram analysis of genes that were affected significantly by the knockdowns demonstrates the substantial overlap between the TTK69-, MEP1-, and Mi2-dependent transcriptomes (Fig. 5D). Consistent with the well-established repressive functions of TTK69 and NuRD, roughly twice as many genes were upregulated as downregulated following depletion of TTK69, MEP1, or Mi2. Nonetheless, our results raise the possibility of a role for TTK69 and NuRD in gene activation as well as repression, although the latter function clearly appears to be more prevalent. Alternatively, the activating role of TTK69 and NuRD might be indirect, i.e., due to repression of anther repressor. Gene Ontology (GO) analysis revealed the strong overrepresentation of developmental signaling in processes regulated by TTK69 and NuRD (Fig. 5E). A striking feature of the GO category grouping is that whole classes of genes appear to be either mostly repressed or mostly activated, suggesting coordinate regulation. In conclusion, our genomewide analysis revealed substantial overlap between the TTK69, MEP1, and Mi2 transcriptomes, confirming that these factors collaborate closely.
TTK69 recruits NuRD to selective loci.
Does NuRD first open up chromatin to allow sequence-specific DNA-binding by TTK69? Or does TTK69 bind chromatin independently, followed by NuRD recruitment? To distinguish between these two scenarios, we combined RNAi-mediated knockdown in S2 cells with chromatin immunoprecipitations quantified by real-time PCR (ChIP-qPCR). All our ChIP data are the results of at least three fully independent biological replicates. Chromatin was extracted from S2 cells that were either mock treated or depleted of TTK69, MEP1, or Mi2 (Fig. 5A). For ChIP-qPCR analysis, we examined binding to the regulatory regions of three representative TTK69 target genes: Hairy, Engrailed (En), and Apaf1-related-killer (ARK; CG6829). The first two are classic TTK69-regulated genes, and ARK was identified as one of the potential targets of TTK69 and NuRD in our genomewide expression analysis.
First, we confirmed the derepression of Hairy, En, and ARK after TTK69, MEP1, or Mi2 knockdown by qPCR on mRNA isolated from S2 cells (Fig. 6 A). ChIPs using antibodies against TTK69 revealed that depletion of either MEP1 or Mi2 did not affect TTK69 binding (Fig. 6B). As expected, TTK69 knockdown led to a loss of the TTK69 ChIP signal. The (P)BAP target SK1 served as a negative control and was not bound by TTK69. These results show that the binding of TTK69 to the DNA loci tested is independent of chromatin remodeling by NuRD. ChIPs using antibodies against Mi2 revealed strongly reduced promoter binding due to a loss of TTK69, MEP1, or Mi2 itself (Fig. 6C). Because knockdown of MEP1 also caused a reduction in Mi2 protein levels (Fig. 5A), we cannot distinguish between loss of Mi2 and loss of recruitment. Like Mi2 binding, MEP1 binding was strictly dependent on TTK69 (Fig. 6D). However, loss of Mi2 only modestly affected MEP1 recruitment, suggesting that TTK69 can recruit MEP1 directly and independently of Mi2. As a control, ChIPs against BAP111 showed that BAP binding to its targets SK1 and KCNQ was unaffected by the knockdown of TTK69, MEP1, or Mi2 (Fig. 6E).
FIG. 6.
NuRD binding to selective promoters depends on TTK69, whereas TTK69 binding is independent of NuRD. (A) Upregulation of ARK, Hairy, and En after TTK69, MEP1, or Mi2 knockdown. RNA was extracted and quantified by RT-qPCR using appropriate primers. mRNA levels were normalized against those of CG11874, a gene whose expression did not change in our microarray experiments. Normalized mRNA levels were expressed relative to those in mock-treated cells. Graphs represent the results of three independent biological replicate experiments. Error bars represent the standard errors of the means. (B to E) ChIP-qPCR analysis of the binding of TTK69 (B), Mi2 (C), and MEP1 (D) to ARK, Hairy, and En. The BAP target SK1 is not bound by TTK69 or NuRD and serves as a negative control. ChIPs were performed on chromatin from mock-treated or RNAi-depleted cells for TTK69, MEP1, or Mi2. (E) The binding of BAP111 to its targets SK1 and KCNQ is not affected by RNAi against TTK69, MEP1, or Mi2. Cross-linked chromatin was prepared from mock-treated or RNAi-treated cells. All ChIP data are the results of at least 3 independent biological replicates. Error bars represent the standard errors of the means.
In summary, our ChIP results showed that NuRD recruitment required TTK69. In contrast, TTK69 binding was independent of NuRD. Loss of Mi2 caused only a modest reduction of MEP1 recruitment, suggesting that TTK69 binding to MEP1 suffices for promoter tethering. We conclude that, on the loci examined, TTK69 binds first and then recruits NuRD (Fig. 7).
FIG. 7.
TTK69 recruits NuRD to selective target loci. The models depict the hierarchical relationship between the sequence-specific transcription factor TTK69 and the ATP-dependent chromatin remodeling factor NuRD. (A) NuRD cannot bind TTK69 loci by itself, whereas TTK69 can bind its binding site independently of NuRD. (B) TTK69 can target NuRD through direct protein-protein interactions with its MEP1 and Mi2 (27) subunits. (C) TTK69 tethers the NuRD complex to specific loci, where it attenuates target gene transcription by modulating the local structure of chromatin. We note that the precise molecular nature of NuRD remodeling in vivo is still unclear. Our results support the notion that TTK69 recruits NuRD to selective loci, not the other way around. See the text for details.
DISCUSSION
Here we have studied the cooperation between the sequence-specific transcription factor TTK69 and the ATP-dependent chromatin-remodeling factor NuRD. One prevalent view of remodeler action is that remodelers act randomly to open up chromatin, creating a window of opportunity for sequence-specific transcription factors to bind their cognate DNA recognition sequences. A drawback of such a scenario is that it does not readily explain the functional specialization of remodelers and how they act in a gene-selective manner. In this study, we provide an example of transcription factor binding preceding remodeler recruitment through selective protein-protein interactions. In addition, we provide a detailed characterization of Drosophila NuRD.
As part of our efforts to understand the molecular mechanism underpinning transcription control by TTK69, we identified MEP1 as a TTK69-interacting protein. Subsequent proteomic and biochemical analyses established MEP1 and CDK2AP1/DOC1 as bona fide NuRD subunits. For example, the sequence coverage and emPAI scores (identification scores corrected for protein sequence length) of NuRD subunits in our Mi2 and MEP1 purifications were remarkably comparable (Table 1). Immunodepletion of CDK2AP1/DOC1 concomitantly removes key NuRD subunits, including Mi2, MEP1, and MTA1 (Fig. 2E), confirming that CDK2AP1/DOC1 is stably associated with the majority of NuRD complexes in the extract. Recently, the dMec complex, comprising solely MEP1 and Mi2, was isolated from Kc cells (17). Thus, as seen more commonly for chromatin-regulatory factors, MEP1 and Mi2 appear to be part of alternate assemblages. However, our results indicate that in embryo nuclear extracts, the majority of Mi2 and MEP1 exists as part of NuRD.
In addition, our biochemical analysis confirmed the association of MEP1 and TTK69. Genetic interaction assays provided independent functional evidence for cooperation between TTK69 and NuRD. TTK69 and NuRD colocalize on a substantial fraction of their binding sites but also occupy unique loci. Genomewide expression analysis revealed that the transcriptomes of TTK69, MEP1, and Mi2 overlap significantly. In contrast, the TTK69 gene expression profile correlated poorly with remodelers other than NuRD, reflecting their functional differentiation. Notably, some GO classes were largely repressed by TTK69 and NuRD, whereas others were mainly activated. Thus, it appears that some sets of functional gene classes are coordinately regulated by TTK69 and NuRD. Because both TTK69 and NuRD are commonly considered transcriptional repressors, their apparently positive role might be due to indirect effects. However, we note that a potential positive role for HDACs has been raised as well (36). Likewise, NuRD might directly activate certain target genes. Our ChIP analysis established that TTK69 could bind its targets independently of NuRD. Binding of NuRD, however, was critically dependent on TTK69. We conclude that TTK69 recruits NuRD to selective loci, not the other way around (Fig. 7). Of course, interactions of NuRD itself with DNA and histones are likely to contribute significantly to its targeting. Transcription factors other than TTK69 will also mediate NuRD recruitment to target loci. Conversely, there is no reason to assume that NuRD is the only transcriptional cofactor of TTK69.
Our work suggests that MEP1 is a genuine subunit of Drosophila NuRD. What is the relationship between MEP1 and NuRD in other organisms, including humans? Interestingly, previous studies have shown that in C. elegans, MEP1 associates with the homologs of Mi2 (LET-418) and RPD3 (HDAC-1) and functions in the repression of germ line genes in somatic cells (32). These observations are fully consistent with the notion that worm MEP1 is part of NuRD. Because MEP1 is critical for Mi2 function in flies (this study) and worms (32), we wondered whether there might also be a human ortholog. Although straightforward database inspection did not reveal a mammalian homolog, a search using the zinc finger domain of fly MEP1 revealed homology with human ZFHX1B/SIP1/ZEB2. A direct Clustal 2.0 alignment of ZFHX1B (35), Drosophila MEP1, and C. elegans MEP1 demonstrated a modest but suggestive homology between the 3 proteins (see Fig. S1 in the supplemental material). Importantly, it was recently reported that ZFHX1B interacts physically and functionally with NuRD (35). Thus, not only does ZFHX1B display structural similarity to MEP1; it also interacts with NuRD, making it an extremely attractive candidate mammalian ortholog. Additionally, ZFHX1B has been reported to interact with the SMAD transcription factors, the corepressor CtBP, and the acetyltransferase p300 (34). Suggestively, we identified the Drosophila homologs of these three proteins in our MEP1, but not Mi2, purifications (Table 1). These observations provide additional support for the notion that ZFHX1B and MEP1 might be orthologs; in addition, they indicate that MEP1 might participate in additional interactions, separate from NuRD.
Loss of heterozygosity of ZFHX1B/SIP1/ZEB2 has been implicated in the etiology of Mowat-Wilson syndrome (MWS), characterized by severe mental retardation and a range of additional defects. Mice lacking ZFHX1B display defects in early neurogenesis (33). Strikingly, a MWS-associated mutant form of ZFHX1B is unable to bind and recruit NuRD (35). In conclusion, we propose that MEP1 is an evolutionarily conserved NuRD subunit that is critical for targeting. In this sense, its function is reminiscent of that of selected signature subunits within the SWI/SNF class BAP and PBAP remodelers (6, 23, 24, 26).
Mi2 and MEP1 were identified in a screen for SUMO-dependent transcriptional repression (29). Both factors act downstream of SUMOylation of the transcription factor Sp3. MEP1 and Mi2 each interact with SUMO in vitro, and their recruitment to an integrated reporter gene is dependent on the SUMOylation of Sp3 (29). Extrapolating these observations made for Sp3, it is an attractive idea that TTK69 SUMOylation (20) will also modulate NuRD-dependent repression by TTK69. Indeed, we also observed an interaction between SUMO and Drosophila MEP1 and Mi2 (A. A. Travers and A. Bassett, unpublished results). However, thus far, we were unable to establish a direct function for SUMO in TTK69-directed silencing. We note that previous studies revealed a critical role for ubiquitylation/deubiquitylation of TTK in cell fate control (2, 21, 30). Likewise, it will be important to explore the role of SUMO signaling in TTK69/NuRD repression during development.
We identified the Drosophila homolog of the human CDK2-interacting protein CDK2AP1/DOC1 as a NuRD subunit. This small protein is a potential tumor suppressor (31), which thus far has received little attention. However, its identification in both mammalian (19) and fly NuRD strongly suggests that it is a conserved subunit, which may play a regulatory role. In our Mi2 and MEP1 purifications, we did not observe homologs of any of the other proteins incidentally reported as NuRD subunits in other organisms. Thus, based on the results of others and on this study, we surmise that the composition of NuRD as depicted in Fig. 2F represents its conserved core.
In conclusion, we present here a biochemical characterization of Drosophila NuRD. Our results showed that MEP1 and CDK2AP1 are two novel NuRD subunits. MEP1 plays a key role in linking NuRD to TTK69. Genomewide analysis of transcription revealed that TTK69 is critically required for a significant portion of NuRD-regulated genes. However, we note that TTK69 and NuRD also regulate genes independently of each other. Our results support a model in which TTK69 recruits NuRD to selective loci, rather than one in which NuRD-mediated chromatin remodeling is required for DNA binding by TTK69. We suggest that TTK69 belongs to a group of transcription factors, sometimes referred to as pioneer proteins, that bind chromatin independently of remodelers (41). We propose that, rather than acting in a generic fashion, specific remodelers cooperate with selective transcription factors in the control of gene expression.
Supplementary Material
Acknowledgments
We thank Alexander Brehm and Bastian Stielow for valuable comments on the manuscript and Zeliha Ozgur and Wilfred van Ijcken for microarray hybridizations.
This work was supported by a grant from the Dutch government (BSIK 03038, SCDD) to C.P.V.
Footnotes
Published ahead of print on 23 August 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Badenhorst, P. 2001. Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development 128:4093-4101. [DOI] [PubMed] [Google Scholar]
- 2.Bajpe, P. K., J. A. van der Knaap, J. A. Demmers, K. Bezstarosti, A. Bassett, H. M. van Beusekom, A. A. Travers, and C. P. Verrijzer. 2008. Deubiquitylating enzyme UBP64 controls cell fate through stabilization of the transcriptional repressor Tramtrack. Mol. Cell. Biol. 28:1606-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baonza, A., C. M. Murawsky, A. A. Travers, and M. Freeman. 2002. Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell Biol. 4:976-980. [DOI] [PubMed] [Google Scholar]
- 4.Bouazoune, K., and A. Brehm. 2006. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 14:433-449. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. L., S. Sonoda, H. Ueda, M. P. Scott, and C. Wu. 1991. Repression of the Drosophila fushi tarazu (ftz) segmentation gene. EMBO J. 10:665-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalkley, G. E., Y. M. Moshkin, K. Langenberg, K. Bezstarosti, A. Blastyak, H. Gyurkovics, J. A. Demmers, and C. P. Verrijzer. 2008. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol. Cell. Biol. 28:2920-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalkley, G. E., and C. P. Verrijzer. 2004. Immuno-depletion and purification strategies to study chromatin-remodeling factors in vitro. Methods Enzymol. 377:421-442. [DOI] [PubMed] [Google Scholar]
- 8.Clapier, C. R., and B. R. Cairns. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273-304. [DOI] [PubMed] [Google Scholar]
- 9.Denslow, S. A., and P. A. Wade. 2007. The human Mi-2/NuRD complex and gene regulation. Oncogene 26:5433-5438. [DOI] [PubMed] [Google Scholar]
- 10.Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova, M. Fellner, B. Gasser, K. Kinsey, S. Oppel, S. Scheiblauer, A. Couto, V. Marra, K. Keleman, and B. J. Dickson. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151-156. [DOI] [PubMed] [Google Scholar]
- 11.Fujita, N., D. L. Jaye, C. Geigerman, A. Akyildiz, M. R. Mooney, J. M. Boss, and P. A. Wade. 2004. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 119:75-86. [DOI] [PubMed] [Google Scholar]
- 12.Giesen, K., T. Hummel, A. Sollewerk, S. Harrison, A. Travers, and C. Klambt. 1997. Glial development in the Drosophila CNS requires concomitant activation of glial and repression of neuronal differentiation genes. Development 124:2307-2316. [DOI] [PubMed] [Google Scholar]
- 13.Guo, M., E. Bier, L. Y. Jan, and Y. N. Jan. 1995. tramtrack acts downstream of numb to specify distinct daughter cell fates during asymmetric cell divisions in the Drosophila PNS. Neuron 14:913-925. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, S. D., and A. A. Travers. 1990. The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J. 9:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kal, A. J., T. Mahmoudi, N. B. Zak, and C. P. Verrijzer. 2000. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 14:1058-1071. [PMC free article] [PubMed] [Google Scholar]
- 16.Kehle, J., D. Beuchle, S. Treuheit, B. Christen, J. A. Kennison, M. Bienz, and J. Muller. 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282:1897-1900. [DOI] [PubMed] [Google Scholar]
- 17.Kunert, N., E. Wagner, M. Murawska, H. Klinker, E. Kremmer, and A. Brehm. 2009. dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. EMBO J. 28:533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, Z. C., and Y. Li. 1999. Tramtrack69 is positively and autonomously required for Drosophila photoreceptor development. Genetics 152:299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Guezennec, X., M. Vermeulen, A. B. Brinkman, W. A. Hoeijmakers, A. Cohen, E. Lasonder, and H. G. Stunnenberg. 2006. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehembre, F., P. Badenhorst, S. Muller, A. Travers, F. Schweisguth, and A. Dejean. 2000. Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies. Mol. Cell. Biol. 20:1072-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, S., Y. Li, R. W. Carthew, and Z. C. Lai. 1997. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell 90:469-478. [DOI] [PubMed] [Google Scholar]
- 22.Mohd-Sarip, A., J. A. van der Knaap, C. Wyman, R. Kanaar, P. Schedl, and C. P. Verrijzer. 2006. Architecture of a polycomb nucleoprotein complex. Mol. Cell 24:91-100. [DOI] [PubMed] [Google Scholar]
- 23.Mohrmann, L., K. Langenberg, J. Krijgsveld, A. J. Kal, A. J. Heck, and C. P. Verrijzer. 2004. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24:3077-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohrmann, L., and C. P. Verrijzer. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681:59-73. [DOI] [PubMed] [Google Scholar]
- 25.Moshkin, Y. M., T. W. Kan, H. Goodfellow, K. Bezstarosti, R. K. Maeda, M. Pilyugin, F. Karch, S. J. Bray, J. A. Demmers, and C. P. Verrijzer. 2009. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol. Cell 35:782-793. [DOI] [PubMed] [Google Scholar]
- 26.Moshkin, Y. M., L. Mohrmann, W. F. van Ijcken, and C. P. Verrijzer. 2007. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol. Cell. Biol. 27:651-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murawsky, C. M., A. Brehm, P. Badenhorst, N. Lowe, P. B. Becker, and A. A. Travers. 2001. Tramtrack69 interacts with the dMi-2 subunit of the Drosophila NuRD chromatin remodelling complex. EMBO Rep. 2:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read, D., and J. L. Manley. 1992. Alternatively spliced transcripts of the Drosophila tramtrack gene encode zinc finger proteins with distinct DNA binding specificities. EMBO J. 11:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stielow, B., A. Sapetschnig, I. Kruger, N. Kunert, A. Brehm, M. Boutros, and G. Suske. 2008. Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol. Cell 29:742-754. [DOI] [PubMed] [Google Scholar]
- 30.Tang, A. H., T. P. Neufeld, E. Kwan, and G. M. Rubin. 1997. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell 90:459-467. [DOI] [PubMed] [Google Scholar]
- 31.Todd, R., J. McBride, T. Tsuji, R. B. Donoff, M. Nagai, M. Y. Chou, T. Chiang, and D. T. Wong. 1995. Deleted in oral cancer-1 (doc-1), a novel oral tumor suppressor gene. FASEB J. 9:1362-1370. [DOI] [PubMed] [Google Scholar]
- 32.Unhavaithaya, Y., T. H. Shin, N. Miliaras, J. Lee, T. Oyama, and C. C. Mello. 2002. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111:991-1002. [DOI] [PubMed] [Google Scholar]
- 33.Van de Putte, T., M. Maruhashi, A. Francis, L. Nelles, H. Kondoh, D. Huylebroeck, and Y. Higashi. 2003. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am. J. Hum. Genet. 72:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Grunsven, L. A., V. Taelman, C. Michiels, K. Opdecamp, D. Huylebroeck, and E. J. Bellefroid. 2006. deltaEF1 and SIP1 are differentially expressed and have overlapping activities during Xenopus embryogenesis. Dev. Dyn. 235:1491-1500. [DOI] [PubMed] [Google Scholar]
- 35.Verstappen, G., L. A. van Grunsven, C. Michiels, T. Van de Putte, J. Souopgui, J. Van Damme, E. Bellefroid, J. Vandekerckhove, and D. Huylebroeck. 2008. Atypical Mowat-Wilson patient confirms the importance of the novel association between ZFHX1B/SIP1 and NuRD corepressor complex. Hum. Mol. Genet. 17:1175-1183. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Z., C. Zang, K. Cui, D. E. Shones, A. Barski, W. Peng, and K. Zao. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen, Y., D. Nguyen, Y. Li, and Z. C. Lai. 2000. The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics 156:195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong, W. C., and C. Montell. 1993. tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev. 7:1085-1096. [DOI] [PubMed] [Google Scholar]
- 39.Xu, C., R. C. Kauffmann, J. Zhang, S. Kladny, and C. W. Carthew. 2000. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103:87-97. [DOI] [PubMed] [Google Scholar]
- 40.Yamasaki, Y., and Y. Nishida. 2006. Mi-2 chromatin remodeling factor functions in sensory organ development through proneural gene repression in Drosophila. Dev. Growth Differ. 48:411-418. [DOI] [PubMed] [Google Scholar]
- 41.Zaret, K. S., J. Watts, J. Xu, E. Wandzioch, S. T. Smale, and T. Sekiya. 2008. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harbor Symp. Quant Biol. 73:119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.