Abstract
The candidate tumor suppressor BAP1 is a deubiquitinating enzyme (DUB) involved in the regulation of cell proliferation, although the molecular mechanisms governing its function remain poorly defined. BAP1 was recently shown to interact with and deubiquitinate the transcriptional regulator host cell factor 1 (HCF-1). Here we show that BAP1 assembles multiprotein complexes containing numerous transcription factors and cofactors, including HCF-1 and the transcription factor Yin Yang 1 (YY1). Through its coiled-coil motif, BAP1 directly interacts with the zinc fingers of YY1. Moreover, HCF-1 interacts with the middle region of YY1 encompassing the glycine-lysine-rich domain and is essential for the formation of a ternary complex with YY1 and BAP1 in vivo. BAP1 activates transcription in an enzymatic-activity-dependent manner and regulates the expression of a variety of genes involved in numerous cellular processes. We further show that BAP1 and HCF-1 are recruited by YY1 to the promoter of the cox7c gene, which encodes a mitochondrial protein used here as a model of BAP1-activated gene expression. Our findings (i) establish a direct link between BAP1 and the transcriptional control of genes regulating cell growth and proliferation and (ii) shed light on a novel mechanism of transcription regulation involving ubiquitin signaling.
Posttranslational modification of proteins with ubiquitin plays a central role in a wide variety of biological processes in eukaryotic cells (44, 64). Depending on the nature of the modification (e.g., poly- versus monoubiquitination), modified substrates can be either degraded by the proteasome or regulated at the level of their activity and function (4, 45). Ubiquitination is reversible, and a significant repertoire of proteases, termed deubiquitinating enzymes (DUBs), are emerging as critical regulators of ubiquitin signaling (40, 46).
BAP1 (BRCA1-associated protein 1) was originally isolated as a nuclear DUB that interacts with, and enhances the growth-suppressive effect of, the tumor suppressor BRCA1 (19). BAP1 also acts in a BRCA1-independent manner; its overexpression in cells lacking BRCA1 has been shown to inhibit cell proliferation and tumor growth (60). Interestingly, recent studies indicate that RNA interference (RNAi)-mediated depletion of BAP1 can also exert an inhibitory effect on cell proliferation (31, 36, 41). Although the exact molecular mechanisms are largely unknown, these data suggest that BAP1 controls cell cycle progression. In further support of this notion, homozygous inactivating mutations in BAP1 have been found in subsets of lung carcinoma and breast cancer cell lines, suggesting that this DUB is a tumor suppressor (19, 67).
BAP1 is a member of the ubiquitin carboxyl hydrolase (UCH) family, including UCH-L1, UCH-L3, and UCH-L5 (UCH37), all of which possess a conserved catalytic domain containing an invariant histidine, cysteine, and aspartic acid catalytic triad (20). Although UCH family members were initially associated with the maturation and turnover of ubiquitin, these enzymes possess isopeptidase activity and thus might selectively regulate protein stability or activity (32, 35, 41). Remarkably, BAP1 possesses a large C-terminal domain, not present in other UCH members, which is predicted to play an important role in regulating and coordinating its DUB activity through selective association with potential substrates or regulatory components.
Host cell factor 1 (HCF-1) is a chromatin-associated protein initially identified as part of a multiprotein complex comprising the viral coactivator VP16 and the POU domain transcription factor Oct-1 (23). During herpes simplex virus infection, this complex is recruited to the enhancer/promoter of the immediate-early gene to activate viral gene expression (23). HCF-1 was further shown to interact, often through a tetrapeptide sequence termed the HCF-1 binding motif (HBM), with specific members of diverse classes of transcription factors, including E2F1, Krox20, Sp1, and GA binding protein (GABP). This suggests a crucial role for HCF-1 in regulating the expression of a plethora of genes involved in diverse cellular processes (7, 10, 16, 22, 28-30, 34, 58, 62). HCF-1 also associates with chromatin-modifying enzymes, most notably methyltransferases (Set1, MLL1, MLL5), acetyltransferases (hMOF), and deacetylases (histone deacetylase 1 [HDAC1], HDAC2) (8, 11, 39, 58, 68, 72). Most recently, HCF-1 was shown to recruit LSD1 to demethylate the repressive mark histone H3 lysine 9 and to promote the trimethylation of histone H3 lysine 4 by Set1, a mark associated with active genes (26). Although HCF-1 has been associated mostly with transcription activation, this regulator is also involved in transcription repression (6, 58, 68). It is thought that sequence-specific DNA-binding transcription factors are responsible for the differential recruitment of distinct HCF-1 complexes to either positively or negatively regulate target gene expression. For instance, HCF-1 has been shown to regulate the G1/S transition of the cell cycle through specific interaction with either E2F4 or E2F1, which repress or activate E2F target genes, respectively (58). Despite these findings, the manner in which HCF-1 is selectively recruited to coordinate the assembly of diverse chromatin-modifying complexes that tightly regulate gene expression remains an area of active investigation.
BAP1 was recently shown to interact, through a NHNY sequence (HBM) located in its middle region, with the kelch motif of HCF-1; moreover, this interaction appears to be required for cell proliferation (31, 36). Ectopic expression studies indicate that BAP1 can deubiquitinate HCF-1 (31, 36), although the significance of this event remains to be elucidated. Additional proteins identified by virtue of their copurification with BAP1 have also been reported recently; most of them are involved in the regulation of chromatin-associated processes, particularly transcription (31, 54). These include the forkhead transcription factors FOXK1 and FOXK2, the histone acetyltransferase HAT1, the human homologs of additional sex combs ASXL1 and ASXL2, the histone lysine demethylase KDM1B (LSD2), and the ubiquitin-conjugating E2 enzyme UBE20. Interestingly, very recently, the drosophila polycomb group protein Calypso was found to be the ortholog of BAP1. Calypso associates with ASX to form the transcription complex PR-DUB, which, in turn, deubiquitinates histone H2A and regulates hox gene expression (47). However, it should be noted that the association of human BAP1 with several additional partners, as described above, suggests a substantially more complex network of functional interactions.
Here we establish that mammalian BAP1 is assembled into high-molecular-weight multiprotein complexes containing transcription factors and cofactors, including HCF-1. We reveal novel BAP1-interacting partners, including the transcription factor Yin Yang 1 (YY1), a zinc finger protein that possesses dual functionality by either activating or repressing gene expression depending on its association with specific transcription coactivators or corepressors at specific target gene promoters (for reviews, see references 13 and 51). We show that BAP1 interacts directly with YY1 and that HCF-1 is required for this interaction in vivo. Finally, in providing a model for BAP1-mediated control of gene expression, we demonstrate that this DUB is a direct coactivator of cox7c, a nuclear gene encoding a component of the mitochondrial respiratory chain. Our data provide novel molecular insight into the involvement of deubiquitination in the control of gene expression.
MATERIALS AND METHODS
Plasmids and antibodies.
Retroviral constructs that express N-terminal Flag-hemagglutinin (HA)-tagged wild-type (WT) or mutant forms of human BAP1 were generated by subcloning the cDNA into the POZ-N plasmid provided by Y. Nakatani (38). The catalytically inactive BAP1 construct, POZ-BAP1 (C91S), was generated by site-directed mutagenesis. The BAP1 mutant deleted in the NHNY sequence corresponding to the HCF-1 binding domain (ΔHBM) was generated by PCR-based subcloning of 2 fragments ligated in frame into POZ-N. The Gal4-BAP1 and Gal4-BAP1 catalytically inactive (C91S) constructs were generated by PCR amplification of the Gal4 DNA binding sequence and ligation in frame into pCDNA.3 BAP1. The Gal4-BAP1 ΔHBM construct was generated by subcloning BAP1 ΔHBM in frame into pCDNA.3 containing the Gal4 DNA binding sequence. Short hairpin RNAs (shRNAs) for human BAP1 (hBAP1) (shBAP1 #1 and shBAP1 #2) and for hHCF-1 were generated as described previously (57); the targeted sequences are GGCTGAGATTGCAAACTATGAG, GGTTTCAGCCCTGAGAGCAAAG, and GGCAGTGCTCTGATTTCCAATC, respectively. The constructs used to produce recombinant full-length glutathione S-transferase (GST)-YY1 and various deletion fragments have been described previously (25). Constructs to produce recombinant full-length GST-BAP1 and various deleted forms were obtained by PCR amplification of various fragments, which were cloned into pGEX4T1. The construct for producing recombinant human His-tagged YY1 has been described previously (56). A construct to produce recombinant human His-tagged BAP1 was generated by subcloning BAP1 cDNA into pET30a+. The shRNA constructs for YY1 and the nontarget sequence have been described previously (56). The pCGN-HCF-1 vector (65) was used for subcloning HCF-1 into the pcDNA.3/HA vector.
Monoclonal anti-BAP1 (C4) and anti-YY1 (H10), polyclonal anti-BAP1 (H300), and anti-TFIID (N12) were from Santa Cruz. Monoclonal anti-HCF-1 (M2) (66) and polyclonal anti-HCF-1 (N18) (14) were described previously. Polyclonal anti-HCF-1 (A301-400A) was from Bethyl Laboratories. Monoclonal anti-RNA polymerase II (H14) was from Covance. Polyclonal anti-histone H3 (06-755), polyclonal anti-histone H3 trimethylated at lysine 27 (H3 K27 me3) (17-622), and monoclonal anti-β-actin (MAB1501) were from Millipore. As controls for immunoprecipitation (IP) and chromatin immunoprecipitation (ChIP), the polyclonal anti-green fluorescent protein (anti-GFP) (FL), anti-HA (Y-11), and rabbit IgG (sc-2027) antibodies from Santa Cruz were used.
Cell culture, RNAi, and immunoblotting.
HeLa cervical cancer cells, U2OS osteosarcoma cells, and Phoenix virus-producing cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Cells were transfected with either a nontargeting control or a BAP1 RNA interference (RNAi) plasmid using Lipofectamine 2000 (Invitrogen). For transient RNAi experiments, shRNA vectors were mixed with the pBABE puromycin resistance-encoding vector, and transfected cells were selected by addition of 2 μg/ml of puromycin for 2 days as described previously (1). The small interfering RNA (siRNA) smart pools for human HCF-1, BAP1, and a nontarget control were from Dharmacon and were transfected into HeLa or U2OS cells using Lipofectamine 2000.
Total-cell extracts were prepared in lysis buffer (50 mM Tris-HCl [pH 7.3], 5 mM EDTA, 50 mM KCl, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], and a protease inhibitor cocktail [Sigma]), and protein concentrations were determined by Bradford assays. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were conducted according to standard procedures.
Purification of BAP1-associated proteins and coimmunoprecipitation.
HeLa or U2OS cell lines stably expressing Flag-HA-BAP1 (WT, C91S, or ΔHBM) were generated following retroviral transduction and 4 rounds of selection using magnetic beads as previously described (38). HeLa (∼9 × 109) or U2OS (∼0.5 × 109) cells were used for the purification of BAP1-associated proteins, essentially as previously described (38). Mass spectrometry analysis was provided by the Proteomics Platform of the Quebec Genomics Center (CHUQ Research Center, Laval University, Quebec, Canada). Standard coimmunoprecipitations using appropriate antibodies were conducted as previously described (56).
Immunodepletion was conducted on HeLa nuclear extracts (∼100 μg of proteins) by overnight incubation at 4°C with 2 μg of an anti-HCF-1 or anti-BAP1 polyclonal antibody in IP buffer (50 mM Tris [pH 7.3], 150 mM NaCl, 5 mM EDTA, 10 mM NaF, 1% Triton X-100, 1 mM PMSF, and a protease inhibitor cocktail [Sigma]). The anti-HA (Y-11) polyclonal antibody was used as a control. The immunocomplexes were incubated for 7 h at 4°C with protein G agarose beads (Sigma) that were saturated with 1% bovine serum albumin (BSA) in IP buffer. After centrifugation, the flowthrough and bead fractions were collected. The immune complexes were washed once with the IP buffer supplemented with 1% BSA. Bound proteins were eluted from the beads with Laemmli buffer and were subjected, along with the flowthrough fractions, to Western blotting.
Chromatin fractions were prepared and digested with micrococcal nuclease (MNase) as previously described (15). Briefly, the nuclear pellet was resuspended in 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 0.3 M sucrose, 0.1% Triton X-100, and protease inhibitor cocktail. Following MNase treatment (3 U/ml for 10 min), the reaction was ended with 5 mM (each) EGTA and EDTA. The samples were then centrifuged at 13,000 × g for 10 min at 4°C to obtain the soluble chromatin fraction.
Glycerol gradient and gel filtration analysis.
Molecular mass separation of native BAP1 complexes from nuclear extracts was conduced using a 10 to 40% glycerol gradient prepared in 20 mM Tris-HCl (pH 7.9), 100 mM KCl, 5 mM MgCl2, 1 mM PMSF, 0.1% NP-40, and 10 mM 2-mercaptoethanol. The samples were centrifuged for 12 h at 50,000 rpm (SW55 Ti rotor; Beckman) at 4°C. Individual fractions were then collected from top to bottom and were analyzed by Western blotting. The C-terminal binding protein (CtBP) corepressor complex, estimated to have a molecular mass of 1.3 to 1.5 MDa, was used as a reference (52).
Gel filtration analysis of purified BAP1 complexes was conducted using a Superose 6 HR gel exclusion chromatography column. Eluted fractions were analyzed by silver staining and Western blotting. The native molecular size markers used for column calibration were thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and albumin (66 kDa) (obtained from GE Healthcare).
Deubiquitination assay on Ub-AMC.
Deubiquitination assays on ubiquitin-C-terminal 7-amino-4-methylcoumarin (Ub-AMC) were conducted as previously described (32) with the following modifications. Purified BAP1 complexes (WT, C91S, and ΔHBM) and recombinant His-BAP1 were adjusted to the same amount of BAP1 protein (125 ng; 1.5 pmol) and were incubated individually with 37.5 pmol of Ub-AMC (Boston Biochem) in 100 μl of assay buffer (50 mM Tris [pH 7.3], 0.25 mM EDTA, 10% dimethyl sulfoxide [DMSO], and 1 mM DTT) for 1,200 s. Fluorescence was measured using a fluorimeter (Cytofluor; PerSeptive Biosystems) at excitation and emission wavelengths of 380 nm and 460 nm, respectively.
In vitro interaction assays.
Recombinant GST fusion proteins were purified using glutathione agarose beads (Sigma), and 2 to 3 μg of beads containing bound proteins was incubated with 10 μl of in vitro-translated [35S]methionine-labeled HCF-1 (TNT T7 Quick Coupled Transcription/Translation system; Promega), 1 μg His-YY1, or 1 μg His-BAP1 for 6 to 8 h at 4°C in pulldown buffer (50 mM Tris [pH 7.5], 50 mM NaCl, 0.02% Tween 20, 1 mM PMSF, and 500 μM dithiothreitol). The beads were washed extensively with the same buffer, and bound proteins were eluted in Laemmli buffer and subjected to autoradiography or Western blotting.
Immunofluorescence.
Cells were fixed for 20 min using 3% paraformaldehyde prepared in phosphate-buffered saline (PBS). Cells were then permeabilized with 0.5% NP-40 in PBS for 20 min and were washed with PBS containing 0.1% NP-40. Cells were further incubated in blocking solution (PBS containing 0.1% NP-40 and 10% FBS) and were stained with a monoclonal anti-BAP1 antibody. An anti-mouse antibody conjugated to Alexa Fluor 594 dye (Invitrogen) was used as a secondary antibody. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Z-stacks were acquired using a Leica DMRE microscope with an HCX PL APO 63× (numerical aperture, 1.32 to 0.6) oil CS objective and a Retiga EX (Qimaging) camera and were deconvoluted with the Openlab 3.1.1 program. Red-green-blue (RGB) profiles were generated by the WCIF ImageJ program (NIH).
Cell synchronization and cell cycle analysis.
U2OS cells were synchronized at the G1/S border using a thymidine double-block protocol (17). The DNA content of cells was analyzed essentially as described previously (1). Briefly, cells were harvested by trypsinization and were fixed with 70% ethanol. After one wash with PBS, cells were treated with 100 μg/ml RNase A (Sigma-Aldrich) for 30 min at 37°C, stained with 50 μg/ml propidium iodide (Sigma-Aldrich), and analyzed using a FACScan flow cytometer equipped with CellQuest software (Becton Dickinson).
Genome-wide gene expression analysis and quantitative reverse transcription-PCR (qRT-PCR) analysis of individual mRNAs.
U2OS cells, transfected with a nontarget control shRNA (shControl) or shRNAs targeting BAP1, were selected with a puromycin-containing medium and were then synchronized at the G1/S border to allow comparative analysis of gene expression. RNA was prepared using the Trizol reagent (Invitrogen) and the RNeasy kit (Qiagen). The generation of cDNA and biotinylated cRNA, as well as hybridization to Human genome Hu133 plus 2.0 arrays (containing 47,000 transcripts and transcript variants; Affymetrix), was conducted according to the One-Cycle target labeling protocol of the GeneChip expression analysis technical manual from Affymetrix (Genome Québec Innovation Centre, Montréal, Canada). Gene expression levels from shControl and shRNAs were subjected to comparative analysis using FlexArray expression analysis software (version 1.1; Michal Blazejczyk, Mathieu Miron, and Robert Nadon, Genome Quebec, Montreal, Canada, 2007 [http://genomequebec.mcgill.ca/FlexArray]). A functional analysis of genes deregulated following BAP1 depletion was conducted using Ingenuity pathways analysis, version 8.5 (3).
Levels of individual mRNAs in BAP1-depleted cells were determined by RT-PCR. Total mRNA (prepared as described above) was used for reverse transcription with SuperScript III reverse transcriptase and oligo(dT)12-18 primers (Invitrogen). The cDNAs obtained were subjected to PCR amplification for bap1 (forward primer, ATGAATAAGGGCTGGCTGGAGCTG; reverse primer, GGGTATCAGCTGGTGGGCAAAGAA), cox7c (forward, GGTCCGTAGGAGCCACTATG; reverse, CAGTTGGTGTCTTACTACAAGG), p107 (forward, ACGACTTGGCGAATCAGGACCATA; reverse, GGCTGCCATTGAACTTGTACAGCA), ccne2 (forward, GATGCTCCTAAAGTTCTTCTACC; reverse, CACATTCTGAAATACTGTCCCAC), cdc6 (forward, GGAAGCCTTTACCTTTCTGGTG; reverse, CAGCTGGCCTGGATACCTCTTC), skp2 (forward, GCTGAAGAGCAAAGGGAGTGAC; reverse, GGCGATACCACCTCTTACAAAC), and β-actin (forward, GGACTTCGAGCAAGAGATGGCCA; reverse, CTCCTTCTGCATCCTGTCGGCAATG). The mRNA levels were normalized to gapdh expression (forward, CATGTTCGTCATGGGTGTGAACC; reverse, GGTCATGAGTCCTTCCACGATAC).
Chromatin immunoprecipitation analysis.
ChIP experiments were conducted essentially as described previously (2) with the following modifications. U2OS cells (5 × 106) were cross-linked with 1% formaldehyde in PBS at room temperature for 10 min with prior incubation in 1.5 mM ethylene glycol bis(succinimidylsuccinate) (EGS; Sigma-Aldrich) in PBS for 30 min at room temperature as described previously (42, 73). Following quenching with glycine (125 mM) for 5 min, cells were scraped in cold PBS. The cells were first washed with buffer A (50 mM Tris-HCl [pH 8.0], 0.1% NP-40, 2 mM EDTA, 10% glycerol, 1 mM PMSF, and a protease inhibitor cocktail [Sigma]) and were then sonicated in buffer B (50 mM Tris-HCl [pH 8.0], 1% SDS, 10 mM EDTA, 1 mM PMSF, and a protease inhibitor cocktail) to generate 300- to 600-bp fragments. After centrifugation and preclearing for 1 h, the suspension was incubated overnight with polyclonal anti-HCF-1, anti-BAP1, anti-YY1, or a nonrelevant antibody used as a control. Immunocomplexes were recovered with protein A-agarose beads (Millipore), and the DNA was purified after de-cross-linking with phenol-chloroform. Real-time PCR was conducted using a SYBR green detection kit (Invitrogen) on an iCycler iQ apparatus (Bio-Rad). Quantification was conducted using the 2−ΔΔCT method, where ΔΔCT is calculated as follows: (ChIP threshold cycle [CT] − input CT of the control antibody) − (ChIP CT − input CT of the target antibody). The results are shown as a ratio of the target gene promoter versus the reference gene promoter. The promoter of the target gene cox7c (forward primer, CAGGAATCCTAGACCTAAGC; reverse primer, ACGCGACAAAGCGGAAATCG) and the promoter of the reference gene β-globin (forward, GGCTGTCATCACTTAGACCTC; reverse, GGTTGCTAGTGAACACAGTTG) were used. The amplification efficiencies of all primer sets were verified before qPCR analysis. All experiments were conducted at least 3 times, and the data shown are the results of a representative experiment.
Luciferase reporter assays.
HeLa cells were transfected with various amounts of a Gal4-BAP1, Gal4-BAP (C91S), Gal4-BAP1 ΔHBM, BAP1, or Gal4 expression plasmid along with 500 ng of the Gal4-thymidine kinase (TK)-luciferase or 500 ng of the TK-luciferase reporter plasmid. The pEGFP-N2 construct (10 ng) was also included to ensure equal transfection efficiencies under the different conditions. Luciferase activity was measured 2 days posttransfection using a luciferase assay kit (Promega).
Microarray data accession number.
The gene expression data for both BAP1 shRNAs have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE23035.
RESULTS
BAP1 is assembled into high-molecular-weight multiprotein complexes and interacts with the transcription factor YY1.
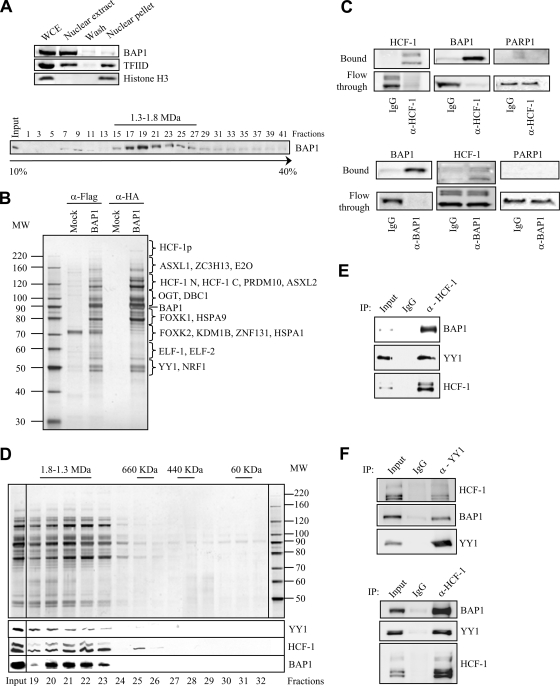
HeLa nuclear extracts in which nearly all the nuclear BAP1 protein was recovered were prepared (Fig. 1 A, top). Glycerol density gradient fractionation of these extracts showed that most of the endogenous protein was detected as a peak in the high-molecular-weight fractions (∼1.3 to 1.8 MDa), suggesting that BAP1 is assembled into multiprotein complexes (Fig. 1A, bottom). To identify these potential complexes, we generated a stable HeLa cell line expressing Flag-HA-BAP1 and conducted a large-scale double immunopurification of the protein using anti-Flag and anti-HA columns. Silver staining of the eluted material revealed that several polypeptides copurify with BAP1 (Fig. 1B). These proteins are specific, since no apparent protein bands were detected in the HA elution from the mock purification. Of note, most of the proteins copurifying with BAP1 were readily detectable following the anti-Flag purification step. Nonetheless, to ensure high specificity, the HA-eluted material was used for mass spectrometry analysis to identify BAP1-interacting partners. Several recently reported as well as novel BAP1-interacting proteins were recovered (Fig. 1B). As reflected by the protein sequence coverage and the number of peptides identified for each protein, the most abundant polypeptides identified include the transcriptional regulator HCF-1, the forkhead transcription factors FOXK1 and FOXK2, the O-linked N-acetylglucosamine transferase (OGT), the human homologs of additional sex combs ASXL1 and ASXL2, the ETS-related transcription factors ELF-1 and ELF-2, and the E2 enzyme UBE20. Less-abundant BAP1-interacting proteins comprise specific transcription factors and cofactors such as YY1, ZNF131, PRDM10, and the histone H3 K4 demethylase KDM1B. To validate these results, we also established a U2OS osteosarcoma cell line stably expressing Flag-HA-BAP1. Using this model cell type, we found that although a small-scale cell preparation was used, most of the major BAP1-associated proteins were recovered following purification. These include HCF-1, OGT, ASXL1/ASXL2, FOXK1/FOXK2, and KDM1B.
FIG. 1.
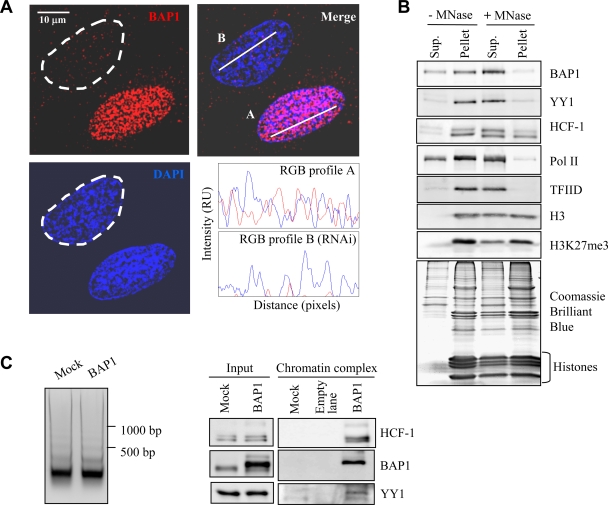
BAP1 assembles high-molecular-weight multiprotein complexes containing the YY1 transcription factor. (A) (Top) Extraction of cellular BAP1 protein. HeLa nuclei isolated with hypotonic buffer were extracted with 300 mM KCl for 30 min in order to obtain the nuclear extract and the chromatin/nuclear matrix pellet fractions. The nuclear pellet was washed once. All fractions were resuspended in the same volume and were used for the immunodetection of BAP1. TFIID was detected as a marker for the transcriptional machinery and histone H3 as a marker for chromatin. WCE, whole-cell extract. (Bottom) Endogenous BAP1 migrates in high-molecular-weight fractions. A HeLa nuclear extract was fractionated using glycerol density gradient ultracentrifugation. Fractions collected from the top to the bottom were subsequently used for the immunodetection of BAP1. The gradient was calibrated with the previously purified CtBP complex, whose estimated molecular size is ∼1.3 to 1.5 MDa. (B) Purification of BAP1-associated proteins. A HeLa cell line stably expressing Flag-HA-BAP1 was used for sequential double immunopurification using anti-Flag antibody and anti-HA antibody columns. The Flag- or HA-eluted proteins were separated by SDS-PAGE and detected by silver staining. The mock purification was conducted using a stable cell line generated with the empty vector. Several regions were cut from the gel, and the polypeptides were identified by mass spectrometry. MW, molecular weight (in thousands). (C) Immunodepletion of HCF-1 (top) or BAP1 (bottom) from nuclear extracts using an excess of an anti-HCF-1 or anti-BAP1 polyclonal antibody. A nonrelevant anti-HA polyclonal antibody was used as a control IgG. BAP1 and HCF-1 were immunodetected in the beads and the flowthrough fractions. The nuclear protein PARP1 was detected as a negative control. (D) BAP1 forms high-molecular-weight multiprotein complexes. Fractionation of the BAP1-purified material was performed using a Superose 6 HR gel filtration column. The eluted complexes were detected by silver staining. BAP1, HCF-1, and YY1 were detected by immunoblotting. MW, molecular weight (in thousands). (E) Reciprocal immunoprecipitation. The Flag-purified BAP1 material was used as input for additional immunoprecipitations with a polyclonal antibody against HCF-1 or a nonrelevant anti-GFP antibody (IgG control). The immunocomplexes were extensively washed, and YY1, HCF-1, and BAP1 were detected by immunoblotting. (F) Interaction of endogenous HCF-1, BAP1, and YY1. A HeLa nuclear extract was used for immunoprecipitation with a polyclonal antibody against YY1 (top), a polyclonal antibody against HCF-1 (bottom), or a nonrelevant anti-GFP antibody (IgG control). The immunocomplexes were washed, and YY1, HCF-1, and BAP1 were detected by immunoblotting.
Stoichiometric amounts of HCF-1 copurify with BAP1, as evidenced by the facts that a large number of peptides were obtained following mass spectrometry analysis (data not shown) and the intensities of the silver-stained bands were similar for the two proteins (Fig. 1B). Since the majority of endogenous BAP1 protein migrates within a high-molecular-weight fraction (Fig. 1A), we reasoned that all of the cellular BAP1 might be complexed with HCF-1. In fact, nearly all BAP1 protein could be immunodepleted from nuclear extracts by using an excess of an anti-HCF-1 antibody (Fig. 1C, top panels). As expected, virtually all HCF-1 protein was recovered in the bead fraction. As a negative control, the nuclear protein PARP1 was shown to remain in the extracts. Next, we immunodepleted BAP1 using a specific antibody and observed that although nearly all the BAP1 was recovered, only a minor fraction of HCF-1 was depleted (Fig. 1C, bottom panels). This indicates that (i) HCF-1 is highly abundant relative to BAP1 and (ii) essentially all cellular BAP1 is complexed with HCF-1. Thus, HCF-1 could be a major scaffold protein for BAP1 and might play a critical role in coordinating the association of this DUB with other partners to form specific transcription regulatory complexes.
To provide insight into the potential role of BAP1 as a gene-specific transcription regulator, we focused in this study on characterizing the interaction of BAP1/HCF-1 with YY1. The anti-HA-eluted material was fractionated using size exclusion chromatography, which revealed that BAP1 is assembled into ∼1.3- to 1.8-MDa multiprotein complexes (Fig. 1D). These complexes contain the transcription factor YY1, HCF-1, and very likely additional components. Next, the anti-Flag-purified BAP1 material was used as input for immunoprecipitation using an anti-HCF-1 antibody, and both YY1 and BAP1 were coimmunoprecipitated (Fig. 1E). These results strongly suggest the existence of at least one complex simultaneously containing BAP1, HCF-1, and YY1. Moreover, the interactions of endogenous YY1 with HCF-1 and BAP1 were also confirmed (Fig. 1F).
The DUB activity is not required for the formation of BAP1 complexes.
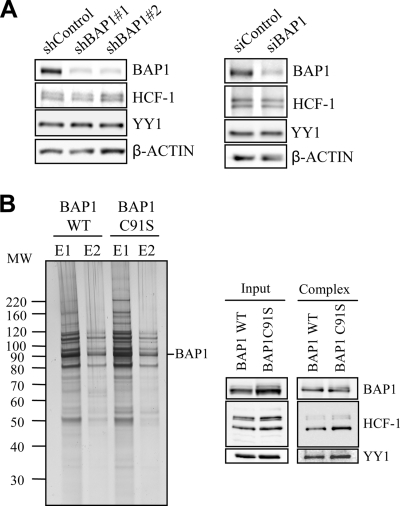
It was recently shown that BAP1 can disassemble K48 ubiquitin chains on HCF-1, suggesting that this DUB might regulate the stability of HCF-1 and possibly other substrates (31, 36). In addition, ubiquitin peptides were detected following mass spectrometry analysis of BAP1-associated proteins, suggesting that some polypeptides were ubiquitinated (data not shown). Thus, we first tested whether loss of BAP1 function affects the stability of YY1. Knockdown of BAP1 using two shRNAs resulted in its substantial depletion, whereas no significant changes were observed in steady-state levels of YY1 or HCF-1 (Fig. 2A, left). These results were confirmed using a pool of 4 different siRNAs targeting BAP1 (Fig. 2A, right). Next, we sought to determine whether DUB activity is required for the assembly of BAP1 complexes. For this purpose, a stable cell line expressing BAP1 in which the catalytic cysteine was mutated (C91S) was generated. Since wild-type BAP1 and BAP1 C91S are not highly expressed, we did not observe a significant difference in cell proliferation between these two conditions (data not shown). Importantly, the purified complexes containing either BAP1 or its catalytically inactive form are essentially indistinguishable (Fig. 2B, left). These results were confirmed by immunoblotting for some of the associated components, i.e., YY1 and HCF-1 (Fig. 2B, right).
FIG. 2.
DUB activity is not required for the assembly of BAP1 complexes or for YY1 stability. (A) Depletion of BAP1 does not affect the steady-state levels of YY1 and HCF-1. (Left) HeLa cells were transfected with either a nontargeting control plasmid (shControl) or a BAP1 shRNA plasmid along with the pBABE puromycin resistance-encoding vector, and transfected cells were selected by addition of puromycin 2 days prior to harvesting for Western blotting using the indicated antibodies. (Right) The siRNA smart pools for human BAP1, or a nontarget control, were transfected into U2OS cells and expressed for 3 days prior to harvesting for Western blotting using the indicated antibodies. (B) BAP1 catalytic activity is not required for the formation of BAP1 complexes. A HeLa cell line stably expressing a Flag-HA-BAP1 catalytically inactive mutant (C91S) was used along with the wild-type control cells for double immunopurification of BAP1 complexes. (Left) Silver staining was conducted on fractions from two elutions (E1 and E2) with an HA peptide. (Right) Immunoblotting for YY1, HCF-1, and BAP1 was conducted.
BAP1 interacts directly with YY1 in vitro, and HCF-1 is required for complex formation in vivo.
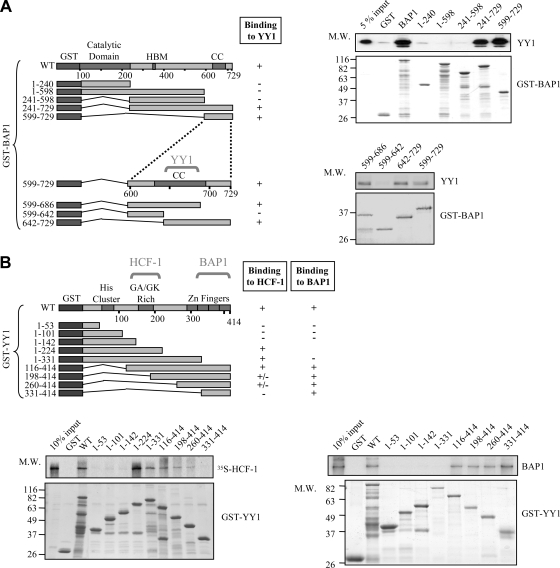
To provide further insight into the interaction of YY1 with HCF-1/BAP1, we generated recombinant proteins, including various deletion mutants, and conducted in vitro GST pulldown assays. We found that BAP1 interacts directly with YY1. The C-terminal region of BAP1 (amino acids [aa] 599 to 729), encompassing the coiled-coil domain, is necessary and sufficient for this interaction (Fig. 3A, top right). We used smaller GST-BAP1 deletion fragments within the aa 599-to-729 region and identified the coiled-coil domain as the interacting motif (Fig. 3A, bottom right). Thus, BAP1 could simultaneously bind YY1 and HCF-1. Next, we demonstrated that in vitro-translated full-length [35S]Met-labeled HCF-1 interacts directly with YY1 and that the central region of the latter (aa 142 to 260), which contains the GA/GK-rich domain, is required for this interaction (Fig. 3B, bottom left). Finally, we determined that the zinc finger region of YY1 (aa 331 to 414) is necessary and sufficient for interaction with BAP1 (Fig. 3B, bottom right). Taken together, these data indicate that BAP1, HCF-1, and YY1 interact using nonoverlapping domains and thus can form a ternary complex involving binary binding for each protein.
FIG. 3.
YY1 interacts with HCF-1 and BAP1 in vitro. (A) Interaction between YY1 and BAP1 mutants in vitro. Various GST deletion fragments of BAP1 bound to glutathione beads were incubated with His-YY1 for 8 h, and following extensive washes, the bead-associated complexes were analyzed by Coomassie blue staining for GST-BAP1 fragments and by Western blotting for YY1. HBM, HCF-1 binding motif; CC, coiled-coil domain; M.W., molecular weight (in thousands). (B) Interaction between HCF-1 or BAP1 and various YY1 mutants in vitro. (Bottom left) Interaction between YY1 and HCF-1 in vitro. Various GST deletion fragments of YY1 bound to glutathione beads were incubated with in vitro-translated 35S-labeled HCF-1 for 8 h, and following purification, HCF-1 was analyzed by autoradiography. (Bottom right) Identification of the YY1 domain required for interaction with BAP1. Various GST deletion fragments of YY1 were incubated with His-BAP1 for 8 h, and the bead-associated complexes were analyzed by Coomassie blue staining and Western blotting for BAP1.
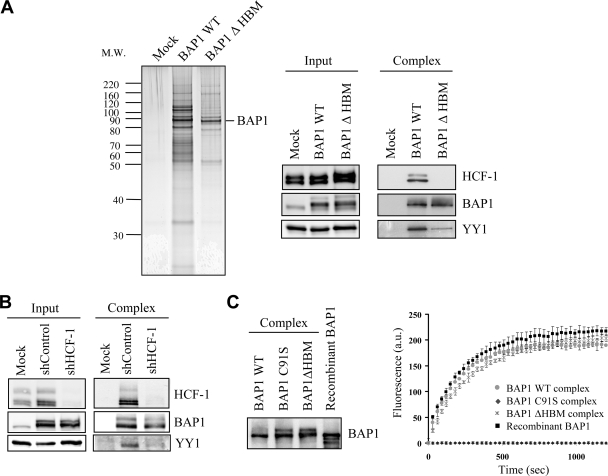
To further characterize these interactions in vivo, a stable cell line expressing BAP1 lacking the HBM was generated and used for the double immunopurification of BAP1-associated proteins. Silver staining of the eluted proteins reveals that while some polypeptide bands appeared similar for wild-type and mutant BAP1, several other bands were absent or substantially reduced in the elution of the mutant BAP1 (Fig. 4A, left). As expected, HCF-1 was not detected in the elution of the mutant BAP1 (31, 36) (Fig. 4A, right). Significantly, the interaction of BAP1 with YY1 was dramatically weaker for the mutant lacking HBM than for wild-type BAP1, suggesting that HCF-1 is required for optimal interaction between YY1 and BAP1 in vivo. Of note, neither YY1 nor HCF-1 levels were changed upon expression of BAP1 lacking HBM. Next, we depleted HCF-1 using shRNA and immunopurified BAP1. As expected, substantially reduced levels of HCF-1 were observed following BAP1 purification (Fig. 4B), and the interaction of BAP1 with YY1 was again reduced. Taken together, these data indicate that BAP1, HCF-1, and YY1 form a ternary complex in vivo, strongly suggesting a functional link between these proteins. We then sought to determine whether the DUB activity of BAP1 is modulated by its interacting partners by similar amounts of BAP1, either recombinant or assembled into complexes, i.e., WT, ΔHBM, and C91S (Fig. 4C, left). Deubiquitination assays with the substrate ubiquitin-AMC (Fig. 4C, right) were conducted. As expected, no activity could be detected for the catalytically inactive form of BAP1 used as a control. However, recombinant BAP1 and BAP1 complexes (wild type or ΔHBM) exhibited similar DUB activities.
FIG. 4.
HCF-1 is required for the formation of a ternary complex with BAP1 and YY1 in vivo. (A) HCF-1 is required for the proper assembly of BAP1 complexes. A HeLa cell line stably expressing Flag-HA-BAP1 lacking the HBM was used for immunopurification with anti-Flag and anti-HA antibodies. (Left) The eluted material was used for SDS-PAGE and silver staining. (Right) Detection of BAP1, HCF-1, and YY1 by immunoblotting. WT BAP1 was used as a control. (B) Depletion of HCF-1 destabilizes the BAP1 interaction with YY1. A HeLa cell line stably expressing Flag-HA-BAP1 was transfected with either a nontargeting control plasmid (shControl) or an HCF-1 shRNA plasmid along with the pBABE puromycin resistance-encoding vector, and transfected cells were selected by addition of puromycin 2 days prior to harvesting for double immunopurification of BAP1. The eluted proteins were detected by Western blotting using the indicated antibodies. (C) Cleavage of Ub-AMC by various BAP1 complexes (WT, C91S, and ΔHBM) and recombinant BAP1. (Left) Equal quantities of BAP1 were used for deubiquitination reactions with 37.5 pmol of Ub-AMC. (Right) The release of AMC was monitored by fluorescence spectroscopy (excitation wavelength, 380 nm; emission wavelength, 460 nm). All experiments were repeated at least 3 times, and the data presented are means ± standard deviations. a.u., arbitrary units.
BAP1 is associated with transcriptionally active chromatin.
Most of the BAP1-interacting proteins are known to be involved in chromatin-associated processes, suggesting a role for BAP1 in regulating gene expression. BAP1 has been shown to associate with chromatin (31). In our study, we found that this protein is mostly excluded from heterochromatic regions, as indicated by the nearly mutually exclusive staining between BAP1 and the highly packed chromatin, i.e., regions strongly stained with DAPI (Fig. 5A). Thus, we set out to determine whether BAP1 is associated with transcriptionally active regions by isolating the chromatin fraction and conducting short-term incubations with micrococcal nuclease (MNase) to release accessible nucleosomes. Nearly all the BAP1 was recovered in the soluble fraction (Fig. 5B). As expected, the basal transcription factor TFIID and RNA Pol II were also recovered predominantly in the soluble fraction. HCF-1 and YY1 were found in this fraction as well, but to a lesser extent than BAP1 or RNA Pol II. Histone H3 was only partially recovered, indicating that a fraction less accessible to MNase, the heterochromatin, remained in the pellet. Consistent with this, histone H3 trimethylated at lysine 27, which is associated with transcriptional repression and compacted chromatin (49), was found predominantly in the pellet. These results suggest that BAP1 is associated with actively transcribed regions, where it might form complexes with HCF-1, YY1, and other regulators to control gene expression. Although BAP1/HCF-1 and YY1 were found on chromatin, the possibility remained that these proteins coexist in different complexes. To determine whether BAP1/HCF-1/YY1 indeed form a complex on chromatin, we immunopurified BAP1 from the chromatin fraction following digestion with MNase. We found that BAP1 immunoprecipitated both HCF-1 and YY1 from this fraction (Fig. 5C, right). Of note, MNase digestion was nearly complete, as indicated by the release of mononucleosomes (Fig. 5C, left).
FIG. 5.
A BAP1/HCF-1/YY1 complex is associated with euchromatin regions. (A) Immunolocalization of BAP1 in U2OS cells, indicating that this DUB is mostly excluded from heterochromatic regions. To ensure the specificity of immunostaining, U2OS cells were transiently transfected with a siRNA against BAP1, and at 3 days posttransfection, cells were used for immunostaining with an anti-BAP1 monoclonal antibody. Following the acquisition of Z-stack images, RGB profiles were generated by the WCIF ImageJ program (NIH). Although most of the cells are depleted of BAP1, some were not transfected and show normal BAP1 expression. In the top left image, the cell delimited by the dashed line has been depleted of BAP1 by RNAi. The other cell shown presumably did not receive the siRNA and expresses normal levels of BAP1. The intensities of fluorescence signals for BAP1 (red) and DNA (blue) at the white bars in the top right image are shown in relative units (RU) at the bottom right. (B) BAP1 and other components of the BAP1 complexes are associated with euchromatin. The chromatin/nuclear matrix fraction was treated with micrococcal nuclease (MNase) to release nucleosomes. Proteins were detected in the soluble (Sup.) and pellet fractions by immunoblotting or Coomassie blue staining. (C) Purification of BAP1/HCF-1/YY1 from the chromatin fraction. The chromatin fraction of HeLa cells stably expressing Flag-HA-BAP1 was digested with MNase (3 U/ml) for 10 min. Following centrifugation at 13,000 × g for 10 min, an aliquot was used for phenol-chloroform extraction of DNA and agarose gel analysis (left). Immunopurification of BAP1 was conducted with the prepared chromatin fraction. The eluted proteins were detected using antibodies against BAP1, YY1, and HCF-1 (right).
BAP1 is a transcriptional coactivator and regulates the expression of genes involved in numerous cellular processes.
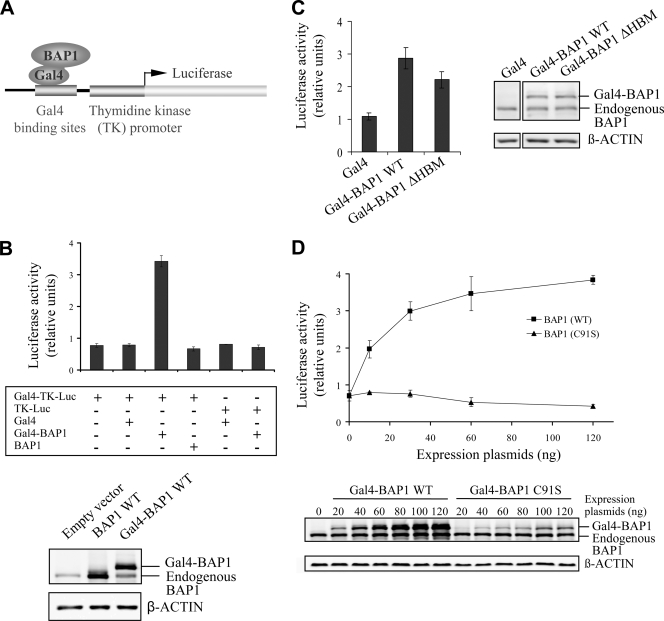
To elucidate the role of BAP1 in transcription regulation, a well-established transcription reporter assay was used (24). This consists of targeting a protein of interest, fused in frame with the GAL4 DNA binding domain, to the luciferase reporter driven by a promoter containing GAL4 binding sites and the thymidine kinase promoter (Fig. 6A). A fusion between the DNA binding domain of Gal4 (aa 1 to 147) and BAP1 was generated and expressed in HeLa cells by transient transfection (Fig. 6B, bottom). Gal4-BAP1 activated transcription of the reporter gene by 3- to 4-fold (Fig. 6B, top). This effect was not observed following expression of BAP1 alone, the Gal4 domain alone, or Gal4-BAP1 along with a thymidine kinase reporter lacking Gal4 binding sites. Taken together, these results suggest that transcription activation by Gal4-BAP1 requires DNA binding and is not an indirect effect. A Gal4-BAP1 mutant lacking the HBM, expressed at the same levels as the wild type (Fig. 6C, right), also activated transcription, although less efficiently than the wild-type form (Fig. 6C, left). Importantly, a Gal4-BAP1 catalytically inactive mutant (C91S) was unable to activate transcription, suggesting that BAP1 regulates gene expression in a DUB activity-dependent manner (Fig. 6D, top). We note that although BAP1 C91S was expressed at lower levels than the wild type for the same quantity of transfected DNA (Fig. 6D, bottom), no reporter activation was observed with BAP1 C91S over a wide range of plasmid concentrations.
FIG. 6.
BAP1 activates transcription in a DUB activity-dependent manner. (A) Schematic representation of the Gal4 transcription system. A transcription reporter assay was conducted by targeting BAP1 to the Gal4-TK-luciferase construct by using the Gal4-BAP1 fusion protein. In this process, a protein of interest, fused in frame to the GAL4 DNA binding domain, is targeted to the luciferase reporter driven by a promoter containing GAL4 binding sites and the thymidine kinase proximal promoter. (B) Gal4-BAP1 activates transcription. HeLa cells were transfected with 100 ng of a Gal4-BAP1, BAP1, or Gal4 expression plasmid along with 500 ng of the Gal4-TK-luciferase or TK-luciferase reporter plasmid. Equal expression of various BAP1 constructs was confirmed by Western blotting using anti-BAP1 (bottom), and luciferase activity was measured (top) at 2 days posttransfection. (C) HCF-1 is essentially dispensable for Gal4-BAP1 transcriptional activity. The Gal4 reporter assay was conducted using 500 ng Gal4-TK-luciferase and an equal amount of WT Gal4-BAP1 or Gal4-BAP1 ΔHBM. The expression of BAP1 constructs was monitored by Western blotting (right), and luciferase activity was measured (left) at 2 days posttransfection. (D) The catalytic activity of BAP1 is required for transcription activation. The Gal4 reporter assay was conducted using 500 ng Gal4-TK-luciferase and various amounts of WT Gal4-BAP1 or the catalytically inactive mutant (C91S). The expression of BAP1 constructs was monitored by Western blotting (bottom), and luciferase activity was measured (top) at 2 days posttransfection. All experiments were repeated at least 3 times, and results from a representative experiment are shown. Data are presented as means ± standard deviations.
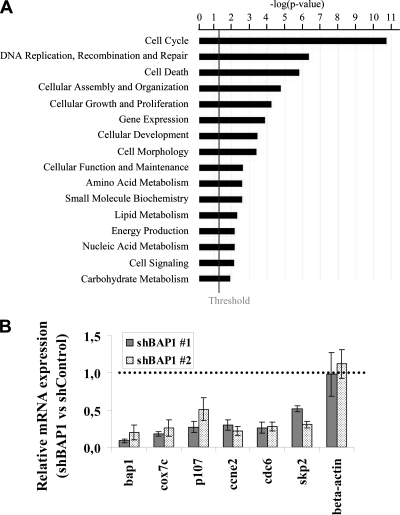
In order to identify potential BAP1 target genes, global mRNA expression profiling using microarrays was conducted following BAP1 depletion in U2OS cells using two shRNA constructs and a nontargeting shRNA as a control. The gene expression data for both shRNAs are deposited in the Gene Expression Omnibus (GEO) NCBI database (accession number GSE23035). Using the cutoff 2-fold difference relative to the control, we found that BAP1 depletion resulted in significantly elevated or decreased expression of about 249 genes (137 upregulated and 112 downregulated). Among these genes, several are associated with cell cycle progression, DNA damage signaling/repair, or survival and metabolism, suggesting that BAP1 participates in diverse cellular processes (Fig. 7A and Table 1). Interestingly, several E2F target genes, including skp2, p107, cdc2, and cdc25a, were downregulated. The effect of BAP1 knockdown on the expression of some of these genes and others was further validated by RT-PCR (Fig. 7B).
FIG. 7.
BAP1 regulates the expression of genes involved in numerous cellular processes. (A) Functional analysis of genes deregulated following BAP1 depletion. The bar chart was generated by Ingenuity pathways analysis, version 8.5, using 1,244 genes deregulated by both shBAP1s (fold change, less than 0.7 or more than 1.5). P values were calculated using the Fisher exact test. The smaller the P value, the less likely that the association is random. The vertical line marks the cutoff for significance (P, 0.05). (B) RT-PCR analysis of selected genes. U2OS cells were transfected with either a nontargeting control plasmid or a BAP1 shRNA plasmid along with the pBABE puromycin resistance-encoding vector, and transfected cells were selected by addition of puromycin 24 h prior to synchronization at the G1/S border by the double-thymidine-block method. mRNA quantification was conducted by real-time RT-PCR analysis. All experiments were repeated at least 3 times, and the data are presented as means ± standard deviations.
TABLE 1.
Genes deregulated following BAP1 depletion are associated with different cellular functions
| Gene function and namea | Gene designation | shBAP1 #1 |
shBAP1 #2 |
||
|---|---|---|---|---|---|
| Fold changeb | P | Fold changeb | P | ||
| Cell cycle | |||||
| Cyclin E2 | CCNE2 | 0.18 | 5.15E−07 | 0.18 | 6.05E−06 |
| CDC5 cell division cycle 5-like (Schizosaccharomyces pombe) | CDC5L | 0.30 | 1.23E−06 | 0.65 | 1.02E−05 |
| S-phase kinase-associated protein 2 (p45) | SKP2 | 0.39 | 2.07E−05 | 0.28 | 1.84E−05 |
| Cell division cycle 2 | CDC2 | 0.39 | 1.38E−05 | 0.33 | 3.55E−05 |
| Cell division cycle 25A | CDC25A | 0.42 | 2.41E−04 | 0.40 | 1.85E−07 |
| Retinoblastoma-like 1 (p107) | p107 | 0.44 | 2.11E−05 | 0.53 | 6.88E−04 |
| E2F transcription factor 5 | E2F5 | 2.10 | 9.17E−05 | 4.00 | 3.74E−07 |
| Cyclin D2 | CCND2 | 2.32 | 8.10E−07 | 3.01 | 6.18E−08 |
| DNA replication | |||||
| Replication factor C (activator 1) 3 | RFC3 | 0.42 | 7.01E−06 | 0.29 | 5.79E−05 |
| CDC6 cell division cycle 6 homolog (Saccharomyces cerevisiae) | CDC6 | 0.46 | 1.58E−05 | 0.34 | 5.78E−06 |
| CDC45 cell division cycle 45-like (S. cerevisiae) | CDC45L | 0.47 | 1.91E−04 | 0.31 | 6.95E−05 |
| Geminin | GMNN | 0.48 | 2.41E−05 | 0.56 | 1.97E−05 |
| DNA repair | |||||
| CHK1 checkpoint homolog (S. pombe) | CHEK1 | 0.38 | 9.19E−07 | 0.51 | 5.31E−07 |
| RAD51-like 1 (S. cerevisiae) | RAD51L1 | 0.44 | 2.24E−04 | 0.47 | 5.41E−06 |
| Ubiquitin-specific peptidase 1 | USP1 | 0.48 | 1.06E−05 | 0.40 | 3.67E−05 |
| Breast cancer 1, early onset | BRCA1 | 0.48 | 5.43E−05 | 0.44 | 2.19E−04 |
| BRCA1-interacting protein C-terminal helicase 1 | BRIP1 | 0.49 | 1.19E−04 | 0.37 | 5.13E−05 |
| Survival/apoptosis | |||||
| Platelet-derived growth factor receptor, alpha polypeptide | PDGFRA | 0.20 | 6.26E−05 | 0.18 | 1.05E−05 |
| Baculoviral IAP repeat-containing 5 (survivin) | BIRC5 | 0.41 | 4.92E−06 | 0.44 | 1.73E−04 |
| Transcriptional adaptor 3 (NGG1 homolog, yeast)-like | TADA3L | 0.43 | 2.33E−06 | 0.51 | 5.14E−04 |
| Tumor necrosis factor receptor superfamily, member 10b | TNFRSF10B | 2.13 | 1.25E−07 | 2.93 | 4.56E−08 |
| BCL2-related protein A1 | BCL2A1 | 3.72 | 1.14E−05 | 5.81 | 4.48E−07 |
| Metabolism | |||||
| Carbonic anhydrase II | CA2 | 0.22 | 8.99E−06 | 0.45 | 4.26E−06 |
| Cytochrome c oxidase subunit VIIc | COX7C | 0.23 | 7.85E−07 | 0.57 | 1.48E−05 |
| UDP-glucuronate decarboxylase 1 | UXS1 | 0.30 | 8.45E−06 | 0.27 | 7.60E−07 |
| ELOVL family member 6 | ELOVL6 | 0.32 | 2.79E−06 | 0.52 | 1.32E−05 |
| N-Acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) | NPL | 0.41 | 3.38E−06 | 0.65 | 9.79E−05 |
| Cytochrome b5 reductase 1 | CYB5R1 | 0.45 | 5.21E−07 | 0.63 | 3.34E−05 |
| BRCA1-associated protein 1 | BAP1 | 0.33 | 4.59E−05 | 0.33 | 7.46E−07 |
Genes are selected as representative examples of different cellular functions.
Values are shBAP1/shControl ratios.
BAP1 is recruited by YY1 to regulate cox7c gene expression.
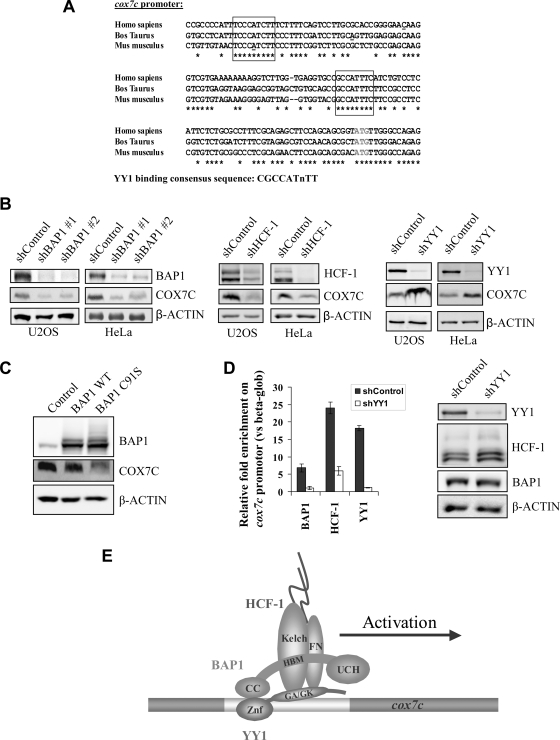
It is not known whether BAP1 assembles complexes that can be recruited to specific promoters to activate transcription. In light of our data, we reasoned that BAP1 might be recruited by YY1 to regulate gene expression. To investigate this possibility, we selected cox7c, one of the most downregulated genes based on our microarray data. cox7c encodes a subunit of the holoenzyme that mediates the terminal step of the mitochondrial electron transport chain. The bovine cox7c promoter has been shown to contain two YY1 binding sites, mutations in which abrogate most of the promoter activity (50). These sites are highly conserved in mouse and human sequences (Fig. 8 A). First, we confirmed that COX7C protein levels were also downregulated following BAP1 depletion in U2OS and HeLa cells (Fig. 8B). Moreover, similar results were obtained following knockdown of HCF-1 (Fig. 8B). Interestingly, depletion of YY1 induced a significant increase in COX7C expression in both HeLa and U2OS cells (Fig. 8B). To determine whether BAP1 regulates cox7c expression in a DUB activity-dependent manner, we transduced U2OS cells with retroviral particles to overexpress either wild-type BAP1 or the BAP1 C91S mutant (Fig. 8C). BAP1 C91S significantly inhibited the expression of COX7C protein, an effect not observed with the wild-type form. Of note, BAP1 C91S is a bona fide dominant negative mutant, since it competes with wild-type BAP1 for assembly of the same multiprotein complexes (Fig. 2B). To further characterize the role of the BAP1/HCF-1/YY1 complex in regulating gene expression, we conducted ChIP assays and found that these proteins are all enriched on the promoter region of cox7c but not on the β-globin promoter (Fig. 8D, left). Moreover, YY1 depletion by shRNA significantly decreased the enrichment of BAP1 and HCF-1 on the cox7c promoter, indicating an essential role of YY1 in targeting BAP1/HCF-1 to specific gene-regulatory regions (Fig. 8D, left, and E). Of note, shRNA-mediated depletion of YY1 did not affect either HCF-1 or BAP1 levels (Fig. 8D, right).
FIG. 8.
YY1 recruits BAP1 to coactivate cox7c expression. (A) Alignment of cox7c promoter sequences from various mammalian species: Homo sapiens (NC000005.9 [reference assembly GRCH37]), Bos taurus (NC007305.4 [reference assembly Btau_4.2]), and Mus musculus (NC000079.5 [reference assembly C57BL/6J]). The YY1 binding sites are boxed. The transcription start site is underlined. (B) Expression of COX7C following depletion of BAP1, HCF-1, or YY1. COX7C protein levels following transfection with BAP1 (left), HCF-1 (center), YY1 (right), or nontarget control (shControl) shRNAs in U2OS or HeLa cells are shown. Following transfection and selection with puromycin for 2 days, cells were harvested for immunoblotting. (C) Downregulation of COX7C following the expression of catalytically inactive BAP1. U2OS cells were transduced with retroviral particles to overexpress either BAP1 or its catalytically inactive form (C91S). After 3 days, cells were harvested for Western blotting using the indicated antibodies. (D) cox7c promoter occupancy by YY1, BAP1, and HCF-1. A YY1 shRNA was expressed in U2OS cells by transfection and selection with puromycin for 2 days before harvesting for ChIP (left) or Western blotting (right). ChIP was conducted by using polyclonal antibodies against BAP1, HCF-1, or YY1. An IgG was used as a control. The enrichment of factors was calculated versus the β-globin promoter, used as a control. All experiments were repeated at least 3 times, and the results of a representative experiment are shown. Data are presented as means ± standard deviations. (E) Model representing the recruitment of BAP1 and HCF-1 to the cox7c promoter by the transcription factor YY1.
DISCUSSION
In this study, we identified novel BAP1-interacting proteins and showed that nearly all cellular BAP1 forms high-molecular-weight multiprotein complexes with several transcription factors and cofactors. The associated partners are likely to play critical roles in targeting BAP1 to potential substrates, thereby regulating its function. Based on the relative abundance of BAP1-associated proteins purified from HeLa or U2OS cells, and on data from other studies (31, 36, 54), it appears that HCF-1, ASXL1 and/or ASXL2, OGT, and FOXK1 and/or FOXK2 might form a BAP1 core complex. This minimal complex may selectively associate with additional regulators or transcription factors to form specific functional complexes in a cell type- and/or promoter-dependent manner. Indeed, substoichiometric levels of several transcription factors copurified with BAP1. These factors are involved in a wide range of cellular processes, suggesting that BAP1 might exert a much broader role in regulating cell function than previously appreciated. Consistent with this notion, BAP1 depletion by RNAi induced profound changes in the expression of genes mediating and/or controlling numerous cellular pathways. Further studies will be needed to investigate how BAP1, via selective interactions with specific transcription factors and cofactors, regulates specific biological responses.
We provided strong evidence that BAP1 is a transcriptional coactivator, as follows. (i) BAP1 associates with transcriptionally active chromatin. (ii) BAP1 acts as an activator, in a DUB activity-dependent manner when targeted to a promoter by using the Gal4 system. (iii) Genomewide expression analysis reveals a considerable number of genes downregulated following BAP1 depletion. (iv) BAP1 directly occupies the cox7c promoter, and depletion of BAP1 results in the downregulation of this gene. It is also possible that BAP1 possesses dual coactivator/corepressor functions, depending on its association with specific transcription factors and cofactors on the regulatory elements of target genes. In agreement with the latter hypothesis, some BAP1-interacting proteins, including HCF-1, YY1, OGT, and ASXL, are known to interact with both coactivators and corepressors (5, 9, 12, 43, 53, 68, 70, 71). In addition, a significant number of genes were upregulated following the depletion of BAP1. This suggests that BAP1 might exert a repressive effect on their promoters, although these genes could constitute indirect targets, i.e., their upregulation could result from secondary changes induced by BAP1 depletion.
Using YY1 as a model for sequence-specific transcription factors that interact with BAP1/HCF-1, we demonstrated that these three proteins form a ternary complex in vivo that can associate with chromatin. Moreover, we found that BAP1 and HCF-1 are recruited by YY1 to coactivate cox7c, a gene previously reported to depend on YY1 binding sites for transcriptional activation (50). While depletion of BAP1 or HCF-1 reduces expression of cox7c, in contrast, depletion of YY1 induces an increase in the expression of this gene. These results suggest that YY1 possesses a dual function as both a repressor and an activator of cox7c, depending on its association with the HCF-1/BAP1 coactivator complex. A similar repression/activation mechanism by YY1 has been previously shown for the murine beta interferon promoter (37, 63). Consistent with this model, YY1 interacts with HCF-1 through the central region containing a GA/GK-rich domain, previously shown to be involved in interactions with HDACs (70). This suggests that the association of YY1 with HCF-1/BAP1 is mutually exclusive with respect to its interaction with HDAC corepressive complexes. With respect to cox7c expression, it is well-known that nucleus-encoded mitochondrial genes, including components of the cytochrome c oxidase complex, are not constitutively expressed but rather are subject to tight regulation by several transcription factors and cofactors, depending on the state of cell growth, energy balance, and other tissue-specific needs (18). Therefore, such genes are expected to oscillate between activation and repression states.
It is not clear at this time whether BAP1 regulates all YY1 target genes. It is possible that it might regulate only a subset of these targets, perhaps those on which YY1 acts as an activator only or on which YY1 might exert a dual activator/repressor function. Other transcription factors might dictate the specificity via interaction with YY1. Indeed, YY1 is well known to interact with numerous transcription factors, such as SP1, C-myc, and E2Fs (25, 48, 74). HCF-1, via additional interactions, might also contribute to the selectivity of BAP1 recruitment to specific YY1 target genes. In this respect, it is not surprising that Gal4-BAP1 lacking HBM is only slightly impaired in transcription activation, suggesting that the interaction between HCF-1 and BAP1 might be involved mostly in the recruitment of the latter to specific promoters.
Precisely how the assembled BAP1/HCF-1/YY1 complex acts to induce the activation of cox7c or other target genes remains to be established. Nonetheless, the data suggest that the molecular mechanism involves ubiquitin signaling and the deubiquitination of specific substrates on target promoters. BAP1 might be continuously needed to prevent the degradation of HCF-1 (31, 36). Although the stability of the total cellular pool of HCF-1 is not significantly affected by BAP1 depletion, it is nonetheless possible that BAP1 stabilizes HCF-1 only on specific promoters following recruitment by YY1 or other transcription factors. Consistent with this, a BAP1 catalytically inactive mutant exerts a dominant negative effect on cox7c expression. It is also plausible that HCF-1 association with BAP1 and YY1 targets the DUB activity to deubiquitinate histones, specific transcription factors, or components of the general transcription machinery. Consistent with this, the drosophila BAP1, Calypso, deubiquitinates monoubiquitinated H2A, a histone mark associated with gene repression (47). However, Calypso does not possess HBM, and thus, the mammalian BAP1 appears to associate selectively with HCF-1 and numerous other proteins not found with Calypso. In addition, in contrast to Calypso, whose activity on ubiquitin-AMC is very low when not associated with ASX, the recombinant mammalian BAP1 appears to have the same activity as complexed BAP1. We note that although BAP1 partners do not affect its DUB activity on ubiquitin-AMC, this does not exclude the possibility of their effect in the context of a physiological substrate in vivo.
Our results also shed light on the biological function of BAP1. This DUB was previously shown to be required for proper cell cycle progression, particularly the G1/S transition (31, 41), Moreover, we observed similar effects in U2OS cells (data not shown), the cell type used here for global gene expression analysis. We also provided molecular insight linking BAP1 to the control of cell cycle genes, including subsets of E2F targets. In addition, HCF-1 is known to be required for a normal G1/S transition and was recently shown to play a major role in regulating the expression of E2F target genes by promoting histone H3 K4 trimethylation (21, 58). Thus, BAP1 might play a pivotal role in regulating the G1/S transition under normal and possibly stress conditions. Supporting this view, BAP1 is phosphorylated on an ATM/ATR consensus motif in response to DNA damage (33, 55), suggesting that these critical DNA damage-responsive checkpoint kinases might regulate BAP1 DUB activity and thus its function in controlling the expression of cell cycle genes.
BAP1 might also participate in transcriptional regulatory programs that coordinate cell growth with the cell cycle. For instance, in addition to cox7c, the expression of several mitochondrial and general metabolism genes are shown here to be deregulated upon BAP1 knockdown. Interestingly, recent bioinformatics and genome-wide promoter occupancy studies indicated that YY1 binding sites are enriched in the promoter regions of nuclear genes that encode mitochondrial proteins (59, 69). Moreover, NRF1, a major regulator of mitochondrial respiration, copurifies with BAP1 (Fig. 1B), and both YY1 and NRF1 binding sites are frequently found in close proximity in a large number of promoters of genes encoding mitochondrial proteins (59, 69). Furthermore, HCF-1 has been found to interact with, and increase the transcriptional activity of, peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1), a major transcriptional regulator of mitochondrial biogenesis (27, 61). Thus, BAP1 might play an important role in dynamically controlling transcriptional responses that coordinate mitochondrial function. Such responses, in turn, could constitute targets of stress signaling pathways (e.g., induced by DNA damage) that orchestrate adaptive metabolic responses.
In summary, our work indicates that BAP1 associates with several transcription factors and cofactors and is a gene-specific transcription regulator. As such, our findings establish a framework for further studies to (i) delineate the exact role of BAP1 in regulating the expression of genes involved in cell cycle progression and (ii) define how deregulation of BAP1 function contributes to tumorigenesis.
Acknowledgments
This work was supported by grants to E.B.A. from the Terry Fox Foundation (grant 018144) and to Y.S. from the National Institutes of Health (grant GM053874). E.B.A. is a scholar of Le Fonds de la Recherche en Santé du Québec. H.Y. is a scholar of the Cole Foundation and the Canadian Institutes of Health Research. N.M. is supported by a scholarship from Le Fonds Québécois de la Recherche sur la Nature et les Technologies. S.D. is supported by a scholarship from the Islamic Development Bank.
We are grateful to Winship Herr for the generous gift of the HCF-1 construct and antibodies. We thank Stephania Bottardi for advice and help.
Footnotes
Published ahead of print on 30 August 2010.
REFERENCES
- 1.Affar, E. B., F. Gay, Y. Shi, H. Liu, M. Huarte, S. Wu, C. Tucker, E. Li, and Y. Shi. 2006. Essential dosage-dependent functions of the transcription factor Yin Yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 26:3565-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottardi, S., J. Ross, V. Bourgoin, N. Fotouhi-Ardakani, E. B. Affar, M. Trudel, and E. Milot. 2009. Ikaros and GATA-1 combinatorial effect is required for silencing of human gamma-globin genes. Mol. Cell. Biol. 29:1526-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvano, S. E., W. Xiao, D. R. Richards, R. M. Felciano, H. V. Baker, R. J. Cho, R. O. Chen, B. H. Brownstein, J. P. Cobb, S. K. Tschoeke, C. Miller-Graziano, L. L. Moldawer, M. N. Mindrinos, R. W. Davis, R. G. Tompkins, and S. F. Lowry. 2005. A network-based analysis of systemic inflammation in humans. Nature 437:1032-1037. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z. J., and L. J. Sun. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33:275-286. [DOI] [PubMed] [Google Scholar]
- 5.Cho, Y. S., E. J. Kim, U. H. Park, H. S. Sin, and S. J. Um. 2006. Additional sex comb-like 1 (ASXL1), in cooperation with SRC-1, acts as a ligand-dependent coactivator for retinoic acid receptor. J. Biol. Chem. 281:17588-17598. [DOI] [PubMed] [Google Scholar]
- 6.Dejosez, M., J. S. Krumenacker, L. J. Zitur, M. Passeri, L. F. Chu, Z. Songyang, J. A. Thomson, and T. P. Zwaka. 2008. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 133:1162-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delehouzee, S., T. Yoshikawa, C. Sawa, J. Sawada, T. Ito, M. Omori, T. Wada, Y. Yamaguchi, Y. Kabe, and H. Handa. 2005. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells 10:717-731. [DOI] [PubMed] [Google Scholar]
- 8.Dou, Y., T. A. Milne, A. J. Tackett, E. R. Smith, A. Fukuda, J. Wysocka, C. D. Allis, B. T. Chait, J. L. Hess, and R. G. Roeder. 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121:873-885. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, C. L., I. Lee, S. Bloyer, S. Bozza, J. Chevalier, A. Dahl, C. Bodner, C. D. Helgason, J. L. Hess, R. K. Humphries, and H. W. Brock. 2010. Additional sex combs-like 1 belongs to the enhancer of trithorax and polycomb group and genetically interacts with Cbx2 in mice. Dev. Biol. 337:9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freiman, R. N., and W. Herr. 1997. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 11:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiki, R., T. Chikanishi, W. Hashiba, H. Ito, I. Takada, R. G. Roeder, H. Kitagawa, and S. Kato. 2009. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 459:455-459. [DOI] [PubMed] [Google Scholar]
- 12.Gambetta, M. C., K. Oktaba, and J. Muller. 2009. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325:93-96. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, S., G. Akopyan, H. Garban, and B. Bonavida. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25:1125-1142. [DOI] [PubMed] [Google Scholar]
- 14.Goto, H., S. Motomura, A. C. Wilson, R. N. Freiman, Y. Nakabeppu, K. Fukushima, M. Fujishima, W. Herr, and T. Nishimoto. 1997. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 11:726-737. [DOI] [PubMed] [Google Scholar]
- 15.Groisman, R., J. Polanowska, I. Kuraoka, J. Sawada, M. Saijo, R. Drapkin, A. F. Kisselev, K. Tanaka, and Y. Nakatani. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357-367. [DOI] [PubMed] [Google Scholar]
- 16.Gunther, M., M. Laithier, and O. Brison. 2000. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol. Cell. Biochem. 210:131-142. [DOI] [PubMed] [Google Scholar]
- 17.Harper, J. V. 2005. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol. Biol. 296:157-166. [DOI] [PubMed] [Google Scholar]
- 18.Hock, M. B., and A. Kralli. 2009. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 71:177-203. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, D. E., M. Proctor, S. T. Marquis, H. P. Gardner, S. I. Ha, L. A. Chodosh, A. M. Ishov, N. Tommerup, H. Vissing, Y. Sekido, J. Minna, A. Borodovsky, D. C. Schultz, K. D. Wilkinson, G. G. Maul, N. Barlev, S. L. Berger, G. C. Prendergast, and F. J. Rauscher III. 1998. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16:1097-1112. [DOI] [PubMed] [Google Scholar]
- 20.Johnston, S. C., C. N. Larsen, W. J. Cook, K. D. Wilkinson, and C. P. Hill. 1997. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 Å resolution. EMBO J. 16:3787-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Julien, E., and W. Herr. 2003. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 22:2360-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knez, J., D. Piluso, P. Bilan, and J. P. Capone. 2006. Host cell factor-1 and E2F4 interact via multiple determinants in each protein. Mol. Cell. Biochem. 288:79-90. [DOI] [PubMed] [Google Scholar]
- 23.Kristie, T. M., Y. Liang, and J. L. Vogel. 2009. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim. Biophys. Acta 1799:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J. S., K. M. Galvin, R. H. See, R. Eckner, D. Livingston, E. Moran, and Y. Shi. 1995. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 9:1188-1198. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. S., K. M. Galvin, and Y. Shi. 1993. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc. Natl. Acad. Sci. U. S. A. 90:6145-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, Y., J. L. Vogel, A. Narayanan, H. Peng, and T. M. Kristie. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 15:1312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, J., P. Puigserver, J. Donovan, P. Tarr, and B. M. Spiegelman. 2002. Peroxisome proliferator-activated receptor gamma coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 277:1645-1648. [DOI] [PubMed] [Google Scholar]
- 28.Lu, R., P. Yang, S. Padmakumar, and V. Misra. 1998. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J. Virol. 72:6291-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luciano, R. L., and A. C. Wilson. 2003. HCF-1 functions as a coactivator for the zinc finger protein Krox20. J. Biol. Chem. 278:51116-51124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luciano, R. L., and A. C. Wilson. 2000. N-terminal transcriptional activation domain of LZIP comprises two LxxLL motifs and the host cell factor-1 binding motif. Proc. Natl. Acad. Sci. U. S. A. 97:10757-10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machida, Y. J., Y. Machida, A. A. Vashisht, J. A. Wohlschlegel, and A. Dutta. 2009. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 284:34179-34188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason, D. E., J. Ek, E. C. Peters, and J. L. Harris. 2004. Substrate profiling of deubiquitin hydrolases with a positional scanning library and mass spectrometry. Biochemistry 43:6535-6544. [DOI] [PubMed] [Google Scholar]
- 33.Matsuoka, S., B. A. Ballif, A. Smogorzewska, E. R. McDonald III, K. E. Hurov, J. Luo, C. E. Bakalarski, Z. Zhao, N. Solimini, Y. Lerenthal, Y. Shiloh, S. P. Gygi, and S. J. Elledge. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160-1166. [DOI] [PubMed] [Google Scholar]
- 34.Mazars, R., A. Gonzalez-de-Peredo, C. Cayrol, A. C. Lavigne, J. L. Vogel, N. Ortega, C. Lacroix, V. Gautier, G. Huet, A. Ray, B. Monsarrat, T. M. Kristie, and J. P. Girard. 2010. The thap-zinc finger protein thap1 associates with coactivator HCF-1 and O-GLcNAc transferase: a link between DYT6 and DYT3 dystonias. J. Biol. Chem. 285:13364-13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misaghi, S., P. J. Galardy, W. J. Meester, H. Ovaa, H. L. Ploegh, and R. Gaudet. 2005. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 280:1512-1520. [DOI] [PubMed] [Google Scholar]
- 36.Misaghi, S., S. Ottosen, A. Izrael-Tomasevic, D. Arnott, M. Lamkanfi, J. Lee, J. Liu, K. O'Rourke, V. M. Dixit, and A. C. Wilson. 2009. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 29:2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mokrani, H., O. Sharaf el Dein, Z. Mansuroglu, and E. Bonnefoy. 2006. Binding of YY1 to the proximal region of the murine beta interferon promoter is essential to allow CBP recruitment and K8H4/K14H3 acetylation on the promoter region after virus infection. Mol. Cell. Biol. 26:8551-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatani, Y., and V. Ogryzko. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430-444. [DOI] [PubMed] [Google Scholar]
- 39.Narayanan, A., W. T. Ruyechan, and T. M. Kristie. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 104:10835-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773-786. [DOI] [PubMed] [Google Scholar]
- 41.Nishikawa, H., W. Wu, A. Koike, R. Kojima, H. Gomi, M. Fukuda, and T. Ohta. 2009. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 69:111-119. [DOI] [PubMed] [Google Scholar]
- 42.Nowak, D. E., B. Tian, and A. R. Brasier. 2005. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. Biotechniques 39:715-725. [DOI] [PubMed] [Google Scholar]
- 43.Ozcan, S., S. S. Andrali, and J. E. Cantrell. 2010. Modulation of transcription factor function by O-GlcNAc modification. Biochim. Biophys. Acta 1799:353-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pickart, C. M., and M. J. Eddins. 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695:55-72. [DOI] [PubMed] [Google Scholar]
- 45.Pickart, C. M., and D. Fushman. 2004. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8:610-616. [DOI] [PubMed] [Google Scholar]
- 46.Reyes-Turcu, F. E., K. H. Ventii, and K. D. Wilkinson. 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78:363-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheuermann, J. C., A. G. de Ayala Alonso, K. Oktaba, N. Ly-Hartig, R. K. McGinty, S. Fraterman, M. Wilm, T. W. Muir, and J. Muller. 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465:243-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlisio, S., T. Halperin, M. Vidal, and J. R. Nevins. 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21:5775-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuettengruber, B., D. Chourrout, M. Vervoort, B. Leblanc, and G. Cavalli. 2007. Genome regulation by polycomb and trithorax proteins. Cell 128:735-745. [DOI] [PubMed] [Google Scholar]
- 50.Seelan, R. S., and L. I. Grossman. 1997. Structural organization and promoter analysis of the bovine cytochrome c oxidase subunit VIIc gene. A functional role for YY1. J. Biol. Chem. 272:10175-10181. [DOI] [PubMed] [Google Scholar]
- 51.Shi, Y., J. S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed] [Google Scholar]
- 52.Shi, Y., J. Sawada, G. Sui, E. B. Affar, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, and Y. Nakatani. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 53.Sinclair, D. A., M. Syrzycka, M. S. Macauley, T. Rastgardani, I. Komljenovic, D. J. Vocadlo, H. W. Brock, and B. M. Honda. 2009. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U. S. A. 106:13427-13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sowa, M. E., E. J. Bennett, S. P. Gygi, and J. W. Harper. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stokes, M. P., J. Rush, J. Macneill, J. M. Ren, K. Sprott, J. Nardone, V. Yang, S. A. Beausoleil, S. P. Gygi, M. Livingstone, H. Zhang, R. D. Polakiewicz, and M. J. Comb. 2007. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 104:19855-19860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sui, G., E. B. Affar, Y. Shi, C. Brignone, N. R. Wall, P. Yin, M. Donohoe, M. P. Luke, D. Calvo, S. R. Grossman, and Y. Shi. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117:859-872. [DOI] [PubMed] [Google Scholar]
- 57.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyagi, S., A. L. Chabes, J. Wysocka, and W. Herr. 2007. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell 27:107-119. [DOI] [PubMed] [Google Scholar]
- 59.van Waveren, C., and C. T. Moraes. 2008. Transcriptional co-expression and co-regulation of genes coding for components of the oxidative phosphorylation system. BMC Genomics 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ventii, K. H., N. S. Devi, K. L. Friedrich, T. A. Chernova, M. Tighiouart, E. G. Van Meir, and K. D. Wilkinson. 2008. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 68:6953-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vercauteren, K., N. Gleyzer, and R. C. Scarpulla. 2008. PGC-1-related coactivator complexes with HCF-1 and NRF-2β in mediating NRF-2(GABP)-dependent respiratory gene expression. J. Biol. Chem. 283:12102-12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vogel, J. L., and T. M. Kristie. 2000. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 19:683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weill, L., E. Shestakova, and E. Bonnefoy. 2003. Transcription factor YY1 binds to the murine beta interferon promoter and regulates its transcriptional capacity with a dual activator/repressor role. J. Virol. 77:2903-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welchman, R. L., C. Gordon, and R. J. Mayer. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6:599-609. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, A. C., K. LaMarco, M. G. Peterson, and W. Herr. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115-125. [DOI] [PubMed] [Google Scholar]
- 66.Wilson, A. C., J. E. Parrish, H. F. Massa, D. L. Nelson, B. J. Trask, and W. Herr. 1995. The gene encoding the VP16-accessory protein HCF (HCFC1) resides in human Xq28 and is highly expressed in fetal tissues and the adult kidney. Genomics 25:462-468. [DOI] [PubMed] [Google Scholar]
- 67.Wood, L. D., D. W. Parsons, S. Jones, J. Lin, T. Sjoblom, R. J. Leary, D. Shen, S. M. Boca, T. Barber, J. Ptak, N. Silliman, S. Szabo, Z. Dezso, V. Ustyanksky, T. Nikolskaya, Y. Nikolsky, R. Karchin, P. A. Wilson, J. S. Kaminker, Z. Zhang, R. Croshaw, J. Willis, D. Dawson, M. Shipitsin, J. K. Willson, S. Sukumar, K. Polyak, B. H. Park, C. L. Pethiyagoda, P. V. Pant, D. G. Ballinger, A. B. Sparks, J. Hartigan, D. R. Smith, E. Suh, N. Papadopoulos, P. Buckhaults, S. D. Markowitz, G. Parmigiani, K. W. Kinzler, V. E. Velculescu, and B. Vogelstein. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318:1108-1113. [DOI] [PubMed] [Google Scholar]
- 68.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xi, H., Y. Yu, Y. Fu, J. Foley, A. Halees, and Z. Weng. 2007. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res. 17:798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang, W. M., C. Inouye, Y. Zeng, D. Bearss, and E. Seto. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. U. S. A. 93:12845-12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, X., F. Zhang, and J. E. Kudlow. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110:69-80. [DOI] [PubMed] [Google Scholar]
- 72.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng, P. Y., C. R. Vakoc, Z. C. Chen, G. A. Blobel, and S. L. Berger. 2006. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41:694, 696, 698. [DOI] [PubMed] [Google Scholar]
- 74.Zhao, J. H., T. Inoue, W. Shoji, Y. Nemoto, and M. Obinata. 1998. Direct association of YY-1 with c-Myc and the E-box binding protein in regulation of glycophorin gene expression. Oncogene 17:1009-1017. [DOI] [PubMed] [Google Scholar]