Abstract
The m7G cap binding protein eukaryotic initiation factor 4E (eIF4E) is a rate-limiting determinant of protein synthesis. Elevated eIF4E levels, commonly associated with neoplasia, promote oncogenesis, and phosphorylation of eIF4E at Ser209 is critical for its tumorigenic potential. eIF4E phosphorylation is catalyzed by mitogen-activated protein kinase (MAPK)-interacting serine/threonine kinase (Mnk), a substrate of Erk1/2 and p38 MAPKs. Interaction with the scaffolding protein eIF4G, which also binds eIF4E, brings Mnk and its substrate into physical proximity. Thus, Mnk-eIF4G interaction is important for eIF4E phosphorylation. Through coimmunoprecipitation assays, we showed that MAPK-mediated phosphorylation of the Mnk1 active site controls eIF4G binding. Utilizing a naturally occurring splice variant, we demonstrated that the C-terminal domain of Mnk1 restricts its interaction with eIF4G, preventing eIF4E phosphorylation in the absence of MAPK signaling. Furthermore, using a small-molecule Mnk1 inhibitor and kinase-dead mutant, we established that Mnk1 autoregulates its interaction with eIF4G, releasing itself from the scaffold after phosphorylation of its substrate. Our findings indicate tight control of eIF4E phosphorylation through modulation of Mnk1-eIF4G interaction.
In eukaryotes, initiation of protein synthesis is facilitated by eukaryotic initiation factor 4F (eIF4F), a complex consisting of the scaffolding factor eIF4G and its interaction partners, the m7G cap binding protein eIF4E and the helicase eIF4A. In the preinitiation complex, eIF4G serves as a central ribosome adaptor module, attracting 40S ribosomal subunits to the 5′ end of mRNAs via direct association with eIF3 (4). Interactions of eIF4E with the m7G cap and eIF4G are recognized as rate-limiting steps in translation. They are tightly controlled by key mitogenic signals, such as the phosphoinositide-3-kinase/mammalian target of rapamycin (PI3K/mTOR) and Ras/mitogen-activated protein kinase (MAPK) signal transduction pathways. A myriad of effects of PI3K/mTOR and MAPK mitogenic signals on translation of discrete mRNA subsets and the global transcriptome have been reported (15).
Dysregulated translational control is a significant factor in tumorigenesis and constitutes a prominent target for therapy. Thus, it is of central interest to mechanistically unravel the effects of mitogenic signals on the translation apparatus. A signature oncogenic signaling effect is eIF4E phosphorylation at Ser209 upon activation of Erk1/2 or p38 MAPKs (2, 22). Erk1/2 and p38 MAPK signals converge on Mnk, which is uniquely capable of catalyzing eIF4E Ser209 phosphorylation (21). eIF4E has been implicated in tumorigenesis (9, 10), and Ser209 phosphorylation has been shown to be required for eIF4E's oncogenic potential (3, 20, 23). Unraveling mechanisms of protein synthesis modulation due to eIF4E phosphorylation has been pursued intensely, but the consequences of eIF4E phosphorylation for the regulation of translation initiation remain a matter of debate (19).
The Mnk proteins are serine/threonine kinases encoded by two distinct genes, Mnk1 and Mnk2 (17). Both Mnk1 and Mnk2 transcripts are subject to alternative splicing, giving rise to full-length versions (Mnk1a/2a) as well as truncated versions (Mnk1b/2b) lacking the MAPK binding domain (11). Mnk1a and Mnk2a are activated by p38 and/or Erk1/2 MAPKs. However, Mnk2 has a high basal level of catalytic activity toward eIF4E phosphorylation and can be active even in unstimulated cells (17). Interestingly, Mnks do not form a stable binary complex with eIF4E to achieve Ser209 phosphorylation. Instead, they interact with the scaffolding protein eIF4G, bringing the kinase and its substrate into physical proximity (14). Consequently, association of Mnk with eIF4G is essential for eIF4E Ser209 phosphorylation. We report here that phosphorylation of Mnk1 by p38 or Erk1/2 MAPKs not only activates its kinase activity but also modulates Mnk1 interaction with eIF4G, thereby facilitating eIF4E phosphorylation. MAPK-mediated control over Mnk1-eIF4G binding constitutes an added level of regulation over eIF4E phosphorylation.
MATERIALS AND METHODS
Cloning of expression plasmids.
pcDNA5/FRT/TO myc-eIF4GI-flag was generated by modifying pcDNA5/FRT/TO myc-eIF4GI-b (7) with a C-terminal Flag tag. Briefly, a C-terminal eIF4G fragment fused to Flag protein was generated by PCR using primers 1 and 2 (Table 1), digested with NheI-NotI, and inserted into the pcDNA5/FRT/TO myc-eIF4GI-b backbone. pcDNA5/FRT/TO-HA was created by inserting a hemagglutinin (HA) epitope, generated with complementary oligonucleotides 3 and 4, into Acc65I-BamHI-digested pcDNA5/FRT/TO. pcDNA5/FRT/TO-HA-Mnk1a, -Mnk1b, and -Mnk2a expression constructs were generated as follows. Fragments encoding Mnk1a, Mnk1b, and Mnk2a were reverse transcription-PCR (RT-PCR) amplified from HEK-293 total RNA by using primer pairs 5/6 (Mnk1a and Mnk1b) and 7/8 for Mnk2a. The PCR fragments were digested with BamHI-NotI and inserted into the pcDNA5/FRT/TO-HA backbone. Mnk1a mutants with T2A2, D191A, and A362P mutations and a deletion of the C-terminal 24 amino acids containing the MAPK binding site (Δ24) were generated by overlapping PCR using primer pair sets 5/10 and 6/9, 5/14 and 6/13, 5/18 and 6/17, and 5/12 and 6/11, respectively. The overlapping fragments were fused in the second PCR with primer pair 5/6 and inserted into pcDNA5/FRT/TO-HA. To generate the HA-Mnk1aΔ12 variant, which lacks the first 12 codons of Mnk1a, a Mnk1a N-terminal fragment was PCR amplified from full-length HA-Mnk1a by using primer pair 15/16, digested with BamHI-Bsu36I, and inserted into pcDNA5/FRT/TO-HA-Mnk1a. To create HA-Mnk1a with eIF4G deleted (Δ4G), the mutant pcDNA5/FRT/TO-HA-Mnk1a backbone was digested with BamHI-BsrGI, gel purified, filled in with Klenow fragment, and religated. pcDNA3.1 HA-Mnk1a, HA-Mnk1b, HA-Mnk2a, and HA-Mnk1a mutants were generated by digesting the corresponding pcDNA5/FRT/TO expression vectors with Acc65I-NotI and ligating with the pcDNA3.1 backbone. pcDNA3.1 myc-eIF4E has been described previously (1). To create pGEX4T-1 glutathione S-transferase (GST)-eIF4E, the eIF4E coding region was PCR amplified from pcDNA3.1 myc-eIF4E by using primers 17 and 18, digested with BamHI-NotI and inserted into the pGEX4T-1 backbone.
TABLE 1.
List of primers used
| Primer no. | Primer name | Sequence (5′-3′)a |
|---|---|---|
| 1 | 5′flag-tag | CGG AGC TTC AGC AAG GAA GTG G |
| 2 | 3′flag-tag | GTG CGG CCG CTC ACT TAT CGT CGT CAT CCT TGT AAT CGT TGT GGT CAG ACT CCT CCT CTG C |
| 3 | 5′HA-tag | GT ACC ATG TAC CCA TAC GAC GTC CCA GAC TAC GCT G |
| 4 | 3′HA-tag | GA TCC AGC GTA GTC TGG GAC GTC GTA TGG GTA CAT G |
| 5 | 5′BamHI-Mnk1a | GC GGATCC GTA TCT TCT CAA AAG TTG GAA AAA CCT ATA GAG ATG GGC |
| 6 | 3′NotI-Mnk1a | GAG CGG CCG CTC AGA GTG CTG TGG GCG GGC TCC T |
| 7 | 5′BamHI-Mnk2a | GC GGATCC GTG CAG AAG AAA CCA GCC GAA CTT CAG |
| 8 | 3′NotI-Mnk2a | GA GCGGCCGC TCA GGC GTG GTC TCC CAC CAG GAC |
| 9 | 5′hT2A2 | ATA ACC GCA CCA GAG CTG ACC GCC CCA TGT |
| 10 | 3′hT2A2 | ACA TGG GGC GGT CAG CTC TGG TGC GGT TAT |
| 11 | 5′Mnk1Δ 24 | CTC CCT GCA AGT GAC GCC TGG CCC |
| 12 | 3′Mnk1Δ 24 | GGG CCA GGC GTC ACT TGC AGG GAG |
| 13 | 5′D191A | CAG TGA AAA TCT GTG CCT TTG ACT TGG GCA |
| 14 | 3′D191A | TGC CCA AGT CAA AGG CAC AGA TTT TCA CTG |
| 15 | 5′BamHI-Mnk1-Δ12 | GC GGATCC GGC AGT AGC GAA CCC CTT |
| 16 | 3′Bsu36I-Mnk1-Δ12 | GC CCTCAGG GGC CAT GTA TTC TGC AGA |
| 17 | 5′A362P | ACG CTC TTC GCA CCT GAG GCC ATC |
| 18 | 3′A362P | GAT GGC CTC AGG TGC GAA GAG CGT |
| 19 | 5′BamHI-eIF4E | GC GGATCC GCG ACT GTC GAA CCG G |
| 20 | 3′NotI-eIF4E | GA GCGGCCGC TTA AAC AAC AAA CCT ATT TTT AGT GGT GG |
Restriction sites used for cloning are underlined.
Cell lines, stable cell lines, and DNA transfections.
HEK-293 cells and immortalized fibroblasts from Mnk1/Mnk2 double-knockout (DKO) or wild-type (WT) mice (21) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and nonessential amino acids (all from Invitrogen, Carlsbad, CA). Stable, tetracycline (Tet)-inducible cell lines were established using the Flp-In T-Rex system (Invitrogen) according to the manufacturer's instructions. Transfection of pcDNA3.1 expression plasmids for immunoprecipitation (IP) experiments was performed with HEK-293 Tet-inducible cells grown in 10-cm dishes. Cells were transfected with 2.5 μg DNA and 6.5 μl Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. We used relatively small quantities of plasmid cDNA for transfections, since high levels of transfection reagent are toxic to cells and lead to undesirable activation of stress pathways and eIF4E phosphorylation (data not shown). Six hours posttransfection, the media were changed to the starvation media (without FBS) containing Tet. After 18 h of incubation, the cells were stimulated with the indicated compounds, harvested, and lysed as described below.
Kinase inhibitors and activators.
Inhibitors of MEK (UO126) (Cell Signaling, Danvers, MA), Mnk (CGP57380) (Sigma, St. Louis, MO), and p38 (SB203580) (Sigma) were dissolved in dimethyl sulfoxide (DMSO) and used at the concentrations indicated. 12-O-Tetradecanoylphorbol-13-acetate (TPA), anisomycin, and sodium arsenite (Sigma) were dissolved in DMSO or distilled water (sodium arsenite) and used at concentrations as indicated.
Immunoprecipitation, m7G cap pulldown, and immunoblotting.
Cell lysates were prepared using low-salt lysis buffer (10 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 2 mM dithiothreitol [DTT]) as described previously (7). Prior to IP or cap pulldown, Flag- or HA-Sepharose (Sigma) or 7-methyl-GTP Sepharose 4B (GE Healthcare, Piscataway, NJ) was blocked with 1% bovine serum albumin (BSA) for 30 min. Reaction mixtures were incubated for 2 to 4 h for IP or for 1 h for cap pulldown and contained 25 μl of Sepharose slurry and 1.5 mg of protein lysate. Precipitates were washed with a buffer identical to lysis buffer but containing 0.05% NP-40 and 1 mM MgCl2. Beads were resuspended in either sample buffer for SDS-PAGE or wash buffer with 0.1 mg/ml Flag peptide or 0.1 mg/ml HA peptide (Sigma) for elution of Flag- or HA-tagged proteins, respectively. Immunoblotting was performed as described previously (7). Antibodies used were against eIF4G1, Mnk1, eIF4E, HA, Erk1/2, eIF4A, eIF4E-P, Mnk1-P, Erk1/2-P (Cell Signaling), c-myc, and poly(A) binding protein (PABP) (Sigma). Phospho-eIF4E immunoblot signals were quantified by densitometry using the FluorChem FC2 imaging system (Cell Biosciences, Santa Clara, CA) and analyzed using the AlphaEase FC program. The phospho-eIF4E signal in the vector control lane was set as the background and subtracted from the values for the WT Mnk1a or Δ4G Mnk1a lane. The resulting values were normalized to the values of the respective immunoblot signals for unphosphorylated eIF4E in the same samples.
In vitro kinase assay and recombinant eIF4E.
HEK-293 cells were transfected with various HA-Mnk1a constructs and lysed as described above. HA-Mnk1 was immunoprecipitated as described above with the following modifications: the last wash and elution with HA peptide were performed in kinase buffer (20 mM HEPES-KOH, pH 7.4, 10 mM MgCl2, 1 mM DTT, 25 μM ATP). Eluates were either assayed by immunoblotting to analyze equal loadings of HA-Mnk1 kinase or incubated with 3 μg of recombinant purified eIF4E and 10 μCi of [γ-33P]ATP at 30°C for 1 h. Reactions were stopped by the addition of sample buffer, the volumes were subjected to SDS-PAGE, and proteins were visualized by autoradiography. Purification of recombinant GST-eIF4E protein was performed as described previously (7). The GST tag was removed with PreScission protease (GE Healthcare) according to the manufacturer's instructions.
RESULTS
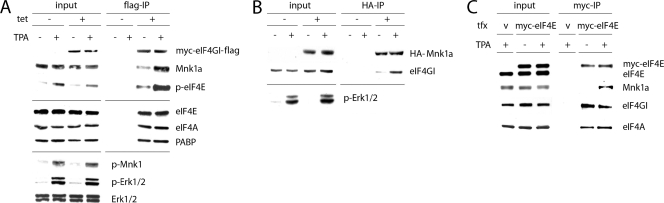
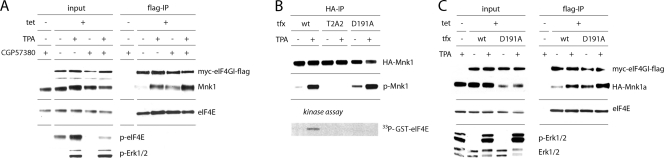
Stimulation of the Erk1/2 or p38 MAPK signaling pathways produces Mnk1 activation and eIF4E phosphorylation at Ser209. Since eIF4E phosphorylation is dependent on Mnk1 binding to eIF4G1 (14), we investigated whether the upstream MAPK signal controls Mnk1-eIF4G interaction. To this end, we generated a stable HEK-293 cell line (HEK-293eIF4G) that upon induction with tetracycline (Tet) expresses an N-terminally (myc) and C-terminally (Flag) tagged form of eIF4G1 (referred to as eIF4G from here on) (Fig. 1 A). Treatment of these cells with 12-O-tetradecanoylphorbol-13-acetate (TPA) produced robust activation of Erk1/2, which led to the phosphorylation of Thr209/Thr214 in the Mnk1 active site and eIF4E phosphorylation (Fig. 1A).
FIG. 1.
Stimulation of cells with TPA results in increased binding of Mnk1 to eIF4G. (A) HEK-293eIF4G cells were uninduced (−) or Tet induced (+), serum starved for 18 h, and treated for 15 min with DMSO (−) or 100 nM TPA (+). Cell lysates were either analyzed by immunoblotting or subjected to anti-Flag IP. Dividers separate immunoblot results obtained with different gels from the same experiment contained within the panels. (B) Cell extracts of HEK-293Mnk1 cells were prepared as described for panel A and subjected to anti-HA IP. (C) HEK-293 cells were transiently transfected with myc-eIF4E, and cell lysates were used for anti-myc IP and analysis of the co-IP complex. v, vector; tfx, transfection.
We used HEK-293eIF4G lysates for anti-Flag immunoprecipitation (IP) of overexpressed eIF4G and analyzed eIF4G binding partners by immunoblotting. TPA treatment increased coimmunoprecipitation (co-IP) of Mnk1 with eIF4G substantially (Fig. 1A), while binding of other known interaction partners, such as the poly(A) binding protein (PABP), eIF4A, and eIF4E, was not affected (Fig. 1A). We also observed increased phospho-eIF4E coprecipitating with eIF4G, in step with enhanced eIF4E Ser209 phosphorylation as a consequence of TPA treatment. Next, we tested if a similar effect would be evident with overexpressed Mnk1 binding to endogenous eIF4G (Fig. 1B). We generated stable HEK-293 cells (HEK-293Mnk1) expressing hemagglutinin (HA)-tagged human Mnk1a in a Tet-inducible fashion. Lysates from HEK-293Mnk1 cells upon TPA treatment exhibited enhanced binding of endogenous eIF4G to HA-Mnk1 (Fig. 1B). To test if MAPK activation enhances physical proximity of Mnk1 and its substrate, we overexpressed myc-eIF4E and performed anti-myc IP (Fig. 1C). TPA exposure resulted in enhanced co-IP of Mnk1 with eIF4E, while eIF4E-eIF4G interaction was unchanged (Fig. 1C). Our results suggest that MAPK activation stimulates Mnk1 binding to eIF4G and, thus, interaction with its substrate eIF4E.
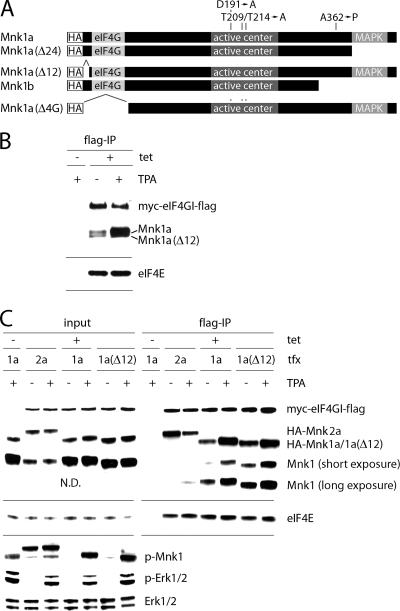
There are two isoforms of Mnk1a that have been reported to arise from alternate use of AUGs 12 codons apart (Mnk1a and -1aΔ12; Fig. 2 A), and they may exhibit intrinsically distinct eIF4G binding activities (13). Using anti-Mnk1 antibody, we consistently detected two distinct bands in total cell lysates, both of which decreased in mobility upon TPA stimulation (data not shown). Using electrophoresis conditions that achieve higher resolution, we observed two bands in Flag-eIF4G precipitates, likely representing both isoforms (Fig. 2B). When overexpressed, the two Mnk1a isoforms exhibited similar eIF4G binding responses upon TPA treatment of cells (Fig. 2C). We also investigated if Mnk2-eIF4G interaction is regulated by MAPK signals. Mnk2 shares 71% sequence identity with Mnk1 and phosphorylates eIF4E in vivo (17). Since we lack reliable anti-Mnk2 antibodies, we overexpressed HA-tagged Mnk2. Binding of HA-Mnk2 to eIF4G did not increase upon the addition of TPA but, instead, was slightly diminished (Fig. 2C). We attribute this to competition of HA-Mnk2 with endogenous Mnk1, which upon activation with TPA binds more strongly to eIF4G in the same lysate (Fig. 2C). These data suggest that Mnk2-eIF4G binding is secondary to Mnk1-eIF4G interaction. Thus, Mnk2-eIF4G binding does not appear to be directly regulated by MAPK signal transduction but responds indirectly to competition with Mnk1.
FIG. 2.
Differential regulation of Mnk1 and Mnk2 binding to eIF4G. (A) Schematic view of Mnk1 constructs used in this study. The HA tag, activation loop, truncation mutants, amino acids manipulated in this study, and the eIF4G and MAPK binding sites are indicated. (B) HEK-293eIF4G cells were serum starved for 18 h and treated with DMSO (−) or 100 nM TPA (+) for 15 min. Anti-Flag IPs were run on a 10% Bis-Tris gel to separate two similar-sized isoforms of Mnk1a and analyzed by immunoblotting. (C) HEK-293eIF4G cells were transfected with the indicated constructs and after 24 h stimulated for 15 min with DMSO (−) or 100 nM TPA (+) as shown. Cell extracts were subjected to anti-Flag IP. Various exposures are shown for the Mnk1 co-IP to visualize faint signal with endogenous protein. N.D., not determined; 1a, Mnk1a; 2a, Mnk2a.
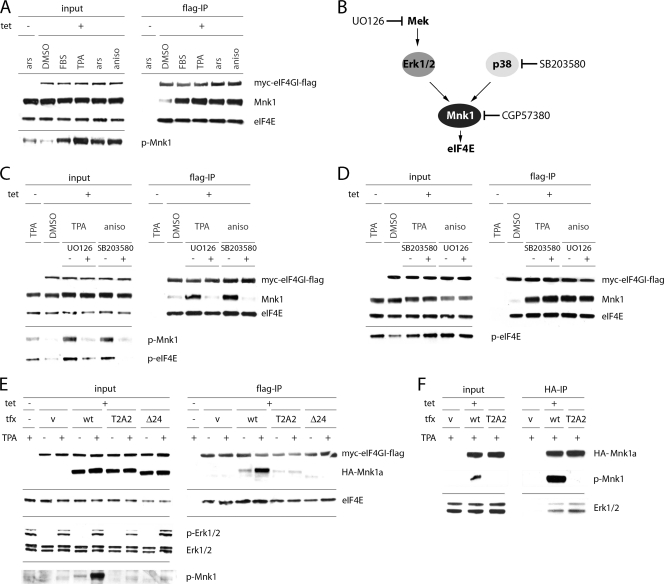
Next, we tested if Mnk1-eIF4G interaction can be increased by a variety of treatments that stimulate eIF4E phosphorylation. Since both p38 and Erk1/2 MAPKs can activate eIF4E phosphorylation, we employed multiple treatments to induce the two signaling pathways (Fig. 3 A). All conditions that stimulated Mnk1 phosphorylation markedly increased Mnk1-eIF4G association (Fig. 3A). Importantly, this effect was abrogated by the inhibition of MEK (immediately upstream of Erk1/2) with UO126 or p38 inhibition with SB203580 (Fig. 3B and C). Next, we verified if downregulation of Mnk1-eIF4G binding by inhibitors is due to decreased activity of their respective kinase targets. SB203580 and UO126 failed to inhibit Mnk1-eIF4G binding stimulated by Erk1/2 and p38 MAPK activation, respectively (Fig. 3D). This suggests that both inhibitors block Mnk1-eIF4G association through specific effects on their target kinases only.
FIG. 3.
MAPK-mediated phosphorylation of Mnk1a at T209/T214 activates Mnk1-eIF4G interaction. (A) HEK-293eIF4G cells were uninduced or Tet induced, serum starved for 18 h, and treated for 15 min with DMSO, 20% fetal bovine serum (FBS), 100 nM TPA (Erk1/2 stimulation), 0.1 mM sodium arsenite (ars; broad stimulation), or 10 μg/ml anisomycin (aniso; p38 stimulation). Cell extracts were subjected to anti-Flag IP. (B) Schematic representation of MAPK signal transduction to eIF4E and corresponding kinase inhibitors. (C and D) HEK-293eIF4G cells were serum starved for 18 h, incubated for 2 h with DMSO, 20 μM UO126 (MEK inhibitor), or 10 μM SB203580 (p38 inhibitor), and treated for 15 min with DMSO (−), 100 nM TPA, or 10 μg/ml anisomycin. Cell extracts were subjected to anti-Flag IP. (E) HEK-293eIF4G cells were transfected with the indicated HA-Mnk1a variants for 24 h and stimulated for 15 min with DMSO (−) or 100 nM TPA (+). Cell extracts were subjected to anti-Flag IP. (F) HEK-293 cells were transfected with the indicated HA-Mnk1a variants for 24 h and treated for 15 min with 100 nM TPA (+). Cell extracts were subjected to anti-HA IP.
Since Erk1/2 and p38 MAPKs phosphorylate Mnk1, the most obvious explanation for increased eIF4G binding would be conformational changes triggered by Mnk1 phosphorylation. This has been predicted based on the crystal structure of the Mnk1 catalytic domain (6). However, MAPK signals may also cause posttranslational modification of eIF4G (16), which could conceivably modulate Mnk1 binding. To distinguish these possibilities, we created a mutant of Mnk1 in which Thr209 and Thr214 were mutated to Ala (Mnk1a T2A2) and a deletion variant which lacks the C-terminal 24 amino acids containing the MAPK binding site (Mnk1aΔ24) (Fig. 2A) (20). Neither of these proteins can be phosphorylated by MAPKs. We used these expression constructs to transfect HEK-293eIF4G cells and test binding to Flag-eIF4G (Fig. 3E). In both cases, binding to eIF4G was severely impaired. It is possible that the Mnk1a T2A2 mutant did not bind eIF4G due to misfolded conformation of the mutated protein. However, the T2A2 HA-Mnk1a protein was still capable of binding Erk1/2 to the same extent that WT HA-Mnk1 could (Fig. 3F). Thus, not only is phosphorylation of the Mnk1 active site required for kinase activity but it also determines binding to eIF4G.
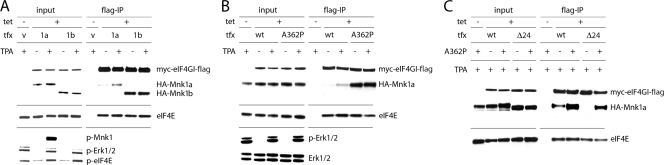
Next, we investigated Mnk-eIF4G binding with the natural splice variant Mnk1b, which lacks 84 C-terminal amino acids relative to Mnk1a (Fig. 2A) (11). Mnk1b had a substantially higher basal level of binding to eIF4G than HA-Mnk1a (Fig. 4 A) and was nonresponsive to TPA stimulation (Fig. 4A). This suggests that a specific region within the Mnk1a C terminus, upstream of the terminal 24 amino acids, interferes with binding of eIF4G, and this interference is alleviated upon stimulation by MAPK. Our results are in agreement with previous data proposing that the C-terminal region of Mnk1a occludes access to the catalytic domain (5, 18).
FIG. 4.
The C-terminal domain of unstimulated Mnk1a blocks binding to eIF4G. HEK-293eIF4G cells were uninduced or Tet induced and transfected with the following expression plasmids: control vector, HA-Mnk1a, or HA-Mnk1b (A), WT or A362P HA-Mnk1a (B), or WT or Δ24 HA-Mnk1a with the A362P mutation either absent (−) or present (+) (C). After 24 h, cells were stimulated for 15 min with DMSO (−) or 100 nM TPA (+). Cell extracts were subjected to anti-Flag IP.
To test if the Mnk1a C terminus can indeed block binding to eIF4G, we utilized an alanine-to-proline mutation (A362P) that has been proposed to disrupt the α-helical conformation of the Mnk1 C terminus (5). Indeed, A362P HA-Mnk1 exhibited significantly enhanced binding to eIF4G (which was unresponsive to TPA) (Fig. 4B), supporting the notion of an inhibitory effect of the Mnk1a C-terminal domain. The A362P mutation even rescued deficient binding of Mnk1aΔ24 (lacking the MAPK binding domain) to eIF4G (Fig. 4C). This suggests that destabilizing the inhibitory C-terminal region of Mnk1a bypasses the requirement for MAPK activation of Mnk1a to induce eIF4G binding.
Next, we investigated why phosphorylation of Thr209/Thr214 is important for eIF4G binding. Since Mnk1 may be capable of autophosphorylation (2), the active kinase may assume an altered conformation favoring eIF4G binding upon phosphorylation of a secondary site(s). To investigate this possibility, we used a competitive inhibitor of ATP (CGP57380) to suppress Mnk1 kinase activity. Surprisingly, while this inhibitor diminished the levels of phospho-eIF4E, it strongly stimulated Mnk1 binding to eIF4G in the absence or presence of TPA (Fig. 5 A). This finding indicates that catalytic activity of Mnk1 is not required for eIF4G binding and that inhibiting Mnk1 enzymatic activity increases eIF4G interaction.
FIG. 5.
Mnk1 kinase activity limits its binding to eIF4G. (A) HEK-293eIF4G cells were uninduced or Tet induced, serum starved for 18 h, and incubated for 2 h with 10 μM CGP57380 (Mnk1 inhibitor) and then stimulated for 15 min with DMSO (−) or 100 nM TPA (+). Cell extracts were subjected to anti-Flag IP. (B) HEK-293eIF4G cells were uninduced or Tet induced and transfected with WT, T2A2, or D191A forms of HA-Mnk1a for 24 h and stimulated for 15 min with DMSO or 100 nM TPA. Cell extracts were subjected to anti-HA IP. Precipitates were either analyzed by immunoblotting with HA- or p-Mnk1-specific antibodies (top panels) or used in in vitro kinase reactions with recombinant eIF4E as a substrate (bottom panel). (C) Cell extracts from panel B were subjected to anti-Flag IP, and precipitates were analyzed by immunoblotting.
CGP57380 is a compound with a broad inhibitory spectrum. Thus, it could indirectly affect Mnk1-eIF4G interaction via secondary effects on kinases other than Mnk1. To exclude this possibility, we confirmed our findings by using a kinase-dead version of Mnk1. To this end, we established an Asp191→Ala (D191A) mutant which disrupts the metal-coordinating site in Mnk1 and abolishes kinase activity of the enzyme (8). We confirmed that the D191A mutant was efficiently activated by TPA because it was readily recognized by the phosphospecific anti-Mnk1 antibody (Fig. 5B). However, the D191A mutant was unable to phosphorylate recombinant GST-eIF4E in an in vitro kinase reaction as expected (Fig. 5B). Co-IP with eIF4G revealed that despite the expression level of the D191A mutant being significantly lower than that of WT HA-Mnk1a, the former exhibited substantially enhanced binding to eIF4G in the absence or presence of TPA (Fig. 5C). Thus, considering these two independent lines of investigation, we conclude that inhibiting Mnk1 catalytic activity results in elevated interactions between the kinase and the eIF4G scaffold. One plausible explanation for these findings could be that blocking phosphoryl transfer with CGP57380 or by the D191A mutation prevents Mnk1 from disassociating from eIF4G. Thus, Mnk1 may change its conformation after it completes phosphorylation of its substrate and subsequently dissociate from eIF4G.
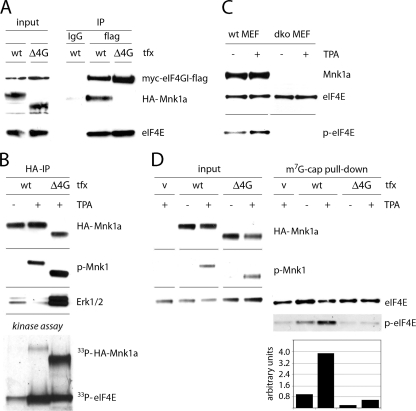
Ultimately, it was critical to demonstrate what contribution Mnk1-eIF4G binding makes to eIF4E phosphorylation independently of Mnk1 catalytic activity. To address this problem, we created a deletion mutant of Mnk1 that lacked the eIF4G binding site (Mnk1aΔ4G) (Fig. 2A) and investigated whether it could phosphorylate eIF4E in vivo. First, using a co-IP approach, we validated that HA-Mnk1aΔ4G did not bind eIF4G (Fig. 6 A). Next, we examined if deletion of the eIF4G binding domain impairs the kinase activity of Mnk1. This is unlikely, given that the eIF4G binding site lies outside the catalytic domain (6). We immunoprecipitated either WT or Δ4G Mnk1a proteins and tested their ability to phosphorylate recombinant eIF4E in an in vitro kinase reaction. WT Mnk1a and mutant Mnk1a phosphorylated eIF4E equally well (Fig. 6B). Interestingly, we consistently detected significantly enhanced Mnk1a phosphorylation with the Δ4G mutant, either by immunoblotting with phospho-Mnk1-specific antibody or with an in vitro kinase assay (Fig. 6B). Also, HA-Mnk1aΔ4G coimmunoprecipitated significantly higher levels of Erk1/2 than WT HA-Mnk1a, which probably explains the higher levels of phosphorylation (Fig. 6B). Since the MAPK binding site lies at the end of the C terminus of Mnk1a, this phenomenon further indicates that the N terminus of Mnk1a functionally interacts with the C terminus.
FIG. 6.
Mnk1-eIF4G binding is required for efficient eIF4E phosphorylation. (A) HEK-293eIF4G cells were Tet induced, transfected for 24 h with either WT or Δ4G HA-Mnk1a, and stimulated for 15 min with 100 nM TPA. Cell extracts were subjected to IP with either nonspecific IgG (anti-HA-Sepharose) or anti-Flag-Sepharose. (B) HEK-293 cells were transfected for 24 h with either WT or Δ4G HA-Mnk1a and stimulated for 15 min with DMSO (−) or 100 nM TPA (+). Cell extracts were subjected to anti-HA IP. Precipitates were either analyzed by immunoblotting with HA-, p-Mnk1-, or Erk1/2-specific antibodies (top panels) or used in in vitro kinase reactions with recombinant eIF4E as a substrate (bottom panel). (C) Immortalized fibroblasts from Mnk1/Mnk2 double-knockout (DKO) and WT mice were serum starved for 18 h and then stimulated for 15 min with DMSO (−) or 100 nM TPA (+). Cell extracts were analyzed by immunoblotting. (D) Mnk1/Mnk2 DKO cells were transfected for 48 h with the indicated constructs and stimulated for 30 min with 100 nM TPA. Cell extracts were subjected to cap-Sepharose pulldown to analyze eIF4E phosphorylation levels (right panel). eIF4E phosphorylation was assessed by immunoblotting (top panels) and by densitometry as described in Materials and Methods (bottom panel).
To investigate the inherent catalytic activity of Mnk1aΔ4G in vivo, we used a system in which eIF4E phosphorylation is categorically absent. Immortalized mouse embryonic fibroblasts (MEF) from Mnk1/Mnk2 double-knockout cells (DKO MEFs) (21) lack any activity that can phosphorylate eIF4E (Fig. 6C). We utilized these cells to transfect either the WT or HA-Mnk1aΔ4G. TPA stimulation of these cells produced efficient phosphorylation of both Mnk1a variants (Fig. 6D). To enrich phospho-eIF4E, we performed m7G cap pulldowns (Fig. 6D). Since Mnk1/Mnk2 DKO MEFs lack inherent eIF4E phosphorylation (21), weak signal with the phospho-eIF4E-specific antibody in the vector control lane likely represents cross-reactivity with unphosphorylated eIF4E. WT Mnk1 expression alone produced very modest eIF4E phosphorylation, which was substantially elevated after TPA stimulation of transfected cells. In contrast, HA-Mnk1aΔ4G failed to raise eIF4E phosphorylation levels significantly above the background, even after TPA stimulation (Fig. 6D). This indicates that Mnk1a-eIF4G binding is indeed required for efficient eIF4E phosphorylation in vivo.
DISCUSSION
In this study, we investigated how MAPK signaling pathways control Mnk1 binding to eIF4G and regulate phosphorylation of the principal Mnk substrate eIF4E. We demonstrated that activation of either p38 or Erk1/2 MAPKs enhances Mnk1 binding to eIF4G. Our results are at variance with previously published work. In studies with mouse Mnk1, Scheper et al. reported decreased Mnk1 interaction with eIF4G upon TPA stimulation (17). Several factors may explain these divergent findings. First, Scheper et al. ectopically expressed mouse Mnk1 in HEK-293 cells. There are important differences between murine and human Mnk1 proteins; the former lacks the N-terminal 12 amino acids and features distinct amino acid sequences in the eIF4G binding site and C-terminal portion of the protein. Since our data suggest that intramolecular interactions regulate Mnk1-eIF4G binding, a variant primary sequence in the mouse protein may alter regulation of Mnk1-eIF4G interaction. Second, Orton et al. showed that interaction of mouse Mnk1 with eIF4G is negatively regulated by the Pak2 kinase (12). However, in human Mnk1, a corresponding Pak2 phosphorylation site is replaced with glycine, thus eliminating this mode of regulation. Lastly, overexpressed murine Mnk1 in human cells may compete with endogenous Mnk1 for interaction with eIF4G. If human Mnk1 has higher affinity for eIF4G, it may displace exogenous mouse Mnk1 subsequent to TPA stimulation, similar to the apparent competition between Mnk1 and Mnk2 (Fig. 2C).
Since activation of either p38 or Erk1/2 MAPKs stimulates Mnk1-eIF4G binding, we speculate that phosphorylation of Thr209/Thr214 triggers conformational changes in Mnk1 that coordinately promote both Mnk1-eIF4G interaction and Mnk1 kinase activity. Accordingly, neither the T2A2 mutant nor the Δ24 MAPK binding site deletion mutant responded to TPA stimulation with enhanced eIF4G binding. In the absence of MAPK signaling, the C-terminal part of Mnk1 abrogates Mnk1-eIF4G interaction. Upon disruption of the structural integrity of the C-terminal domain (the A362P mutant) or in its absence (Mnk1b), eIF4G binding is constitutive and no longer requires MAPK activation.
An additional level of regulation of Mnk1-eIF4G binding appears to directly involve Mnk1 enzymatic activity, since blocking Mnk1 either by the D191A mutation or with CGP57380 increased Mnk1-eIF4G interaction. It could be argued that the D191A mutation or binding of CGP57380 could simply alter the conformation of Mnk1 so that it constitutively binds eIF4G, as is the case for the A362P mutant. However, TPA stimulated eIF4G association with the D191A mutant protein or with wild-type Mnk1 in the presence of CGP57380, suggesting that at least two factors may influence eIF4G interaction: MAPK-mediated conformational changes of Mnk1 and dissociation of ADP-bound Mnk1 after phosphoryl transfer is complete. The latter may be physiologically significant, since it would enable Mnk1 to bind successive eIF4F complexes, thus allowing rapid turnaround of the substrate.
Our findings suggest that after MAPK activation, Mnk1 not only becomes catalytically active but is also actively recruited to the vicinity of eIF4E through increased interaction with the scaffolding protein eIF4G. The latter is biologically significant, because the Δ4G Mnk1 mutant, which is a fully active kinase but cannot bind eIF4G, is unable to reconstitute an adequate eIF4E phosphorylation response to TPA stimulation in Mnk1/2 DKO MEFs.
The data presented here uncover an additional level of control over Mnk1 activity and consequently eIF4E phosphorylation. This complex regulation is likely influenced by other aspects not addressed in this study. For example, it is possible that phosphorylation of eIF4G by signal transduction events elicited by TPA may also play a role in regulation of eIF4E phosphorylation, e.g., by modulating Mnk1 association. Considering that phosphorylation of eIF4E by Mnk proteins appears to be critical for malignant transformation (3, 20, 23), it will be of interest to fully define both the events that govern this posttranslational modification and the consequences it has for control of mRNA translation.
Acknowledgments
We thank E. Y. Dobrikova for technical assistance, C. Goetz for providing recombinant eIF4E, and R. Fukunaga for generously sharing Mnk1/2 DKO MEFs.
This work was supported by PHS grant CA140510 (to M.G.) and a grant from the Susan G. Komen Foundation.
Footnotes
Published ahead of print on 7 September 2010.
REFERENCES
- 1.Bradrick, S. S., and M. Gromeier. 2009. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One 4:e7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukunaga, R., and T. Hunter. 1997. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16:1921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furic, L., L. Rong, O. Larsson, I. H. Koumakpayi, K. Yoshida, A. Brueschke, E. Petroulakis, N. Robichaud, M. Pollak, L. A. Gaboury, P. P. Pandolfi, F. Saad, and N. Sonenberg. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. U. S. A. 107:14134-14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 5.Goto, S., Z. Yao, and C. G. Proud. 2009. The C-terminal domain of Mnk1a plays a dual role in tightly regulating its activity. Biochem. J. 423:279-290. [DOI] [PubMed] [Google Scholar]
- 6.Jauch, R., M. K. Cho, S. Jakel, C. Netter, K. Schreiter, B. Aicher, M. Zweckstetter, H. Jackle, and M. C. Wahl. 2006. Mitogen-activated protein kinases interacting kinases are autoinhibited by a reprogrammed activation segment. EMBO J. 25:4020-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaiser, C., E. Y. Dobrikova, S. S. Bradrick, M. Shveygert, J. T. Herbert, and M. Gromeier. 2008. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA 14:2170-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knauf, U., C. Tschopp, and H. Gram. 2001. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol. Cell. Biol. 21:5500-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544-547. [DOI] [PubMed] [Google Scholar]
- 10.Mamane, Y., E. Petroulakis, L. Rong, K. Yoshida, L. W. Ler, and N. Sonenberg. 2004. eIF4E—from translation to transformation. Oncogene 23:3172-3179. [DOI] [PubMed] [Google Scholar]
- 11.O'Loghlen, A., V. M. Gonzalez, D. Pineiro, M. I. Perez-Morgado, M. Salinas, and M. E. Martin. 2004. Identification and molecular characterization of Mnk1b, a splice variant of human MAP kinase-interacting kinase Mnk1. Exp. Cell Res. 299:343-355. [DOI] [PubMed] [Google Scholar]
- 12.Orton, K. C., J. Ling, A. J. Waskiewicz, J. A. Cooper, W. C. Merrick, N. L. Korneeva, R. E. Rhoads, N. Sonenberg, and J. A. Traugh. 2004. Phosphorylation of Mnk1 by caspase-activated Pak2/gamma-PAK inhibits phosphorylation and interaction of eIF4G with Mnk. J. Biol. Chem. 279:38649-38657. [DOI] [PubMed] [Google Scholar]
- 13.Pyronnet, S. 2000. Phosphorylation of the cap-binding protein eIF4E by the MAPK-activated protein kinase Mnk1. Biochem. Pharmacol. 60:1237-1243. [DOI] [PubMed] [Google Scholar]
- 14.Pyronnet, S., H. Imataka, A. C. Gingras, R. Fukunaga, T. Hunter, and N. Sonenberg. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajasekhar, V. K., A. Viale, N. D. Socci, M. Wiedmann, X. Hu, and E. C. Holland. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12:889-901. [DOI] [PubMed] [Google Scholar]
- 16.Raught, B., A. C. Gingras, S. P. Gygi, H. Imataka, S. Morino, A. Gradi, R. Aebersold, and N. Sonenberg. 2000. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 19:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheper, G. C., N. A. Morrice, M. Kleijn, and C. G. Proud. 2001. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 21:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheper, G. C., J. L. Parra, M. Wilson, B. Van Kollenburg, A. C. Vertegaal, Z. G. Han, and C. G. Proud. 2003. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol. Cell. Biol. 23:5692-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheper, G. C., and C. G. Proud. 2002. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 269:5350-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueda, T., M. Sasaki, A. J. Elia, I. I. Chio, K. Hamada, R. Fukunaga, and T. W. Mak. 2010. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc. Natl. Acad. Sci. U. S. A. 107:13984-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda, T., R. Watanabe-Fukunaga, H. Fukuyama, S. Nagata, and R. Fukunaga. 2004. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24:6539-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waskiewicz, A. J., A. Flynn, C. G. Proud, and J. A. Cooper. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wendel, H. G., R. L. Silva, A. Malina, J. R. Mills, H. Zhu, T. Ueda, R. Watanabe-Fukunaga, R. Fukunaga, J. Teruya-Feldstein, J. Pelletier, and S. W. Lowe. 2007. Dissecting eIF4E action in tumorigenesis. Genes Dev. 21:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]