Abstract
RNase H1 in mammalian cells is present in nuclei and mitochondria. Its absence in mitochondria results in embryonic lethality due to the failure to amplify mitochondrial DNA (mtDNA). Dual localization to mitochondria and nuclei results from differential translation initiation at two in-frame AUGs (M1 and M27) of a single mRNA. Here we show that expression levels of the two isoforms depend on the efficiency of translation initiation at each AUG codon and on the presence of a short upstream open reading frame (uORF) resulting in the mitochondrial isoform being about 10% as abundant as the nuclear form. Translation initiation at the M1 AUG is restricted by the uORF, while expression of the nuclear isoform requires reinitiation of ribosomes at the M27 AUG after termination of uORF translation or new initiation by ribosomes skipping the uORF and the M1 AUG. Such translational organization of RNase H1 allows tight control of expression of RNase H1 in mitochondria, where its excess or absence can lead to cell death, without affecting the expression of the nuclear RNase H1.
Proteins from a single gene are sometimes targeted to more than one subcellular compartment (17). In some cases, a single protein can be directed to two different locations. In other cases, translation of a single mRNA with two initiation codons results in proteins with different targeting signals. Alternatively spliced mRNAs can also have different coding sequences that target the proteins to different locations.
Mammalian RNases H1 (enzymes that cleave RNA in RNA-DNA hybrids) are present in mitochondria and nuclei (7). Mouse and human mRNAs encoding RNase H1 have two in-frame AUG codons with translation initiation from the first AUG producing a protein with a mitochondrial targeting sequence (MTS) (5, 36). Rnaseh1-null mouse embryos arrest development at embryonic day 8.5 (E8.5) due to failure to amplify mitochondrial DNA (mtDNA), without any apparent deficiency in chromosomal replication (7). This suggests that RNase H1 is necessary for mitochondrial DNA replication. RNase H1 is also required during early development in Drosophila melanogaster (12). In contrast, the RNH1 gene (encoding RNase H1) of Saccharomyces cerevisiae does not have a canonical N-terminal MTS, and the gene can be deleted without loss of mitochondrial function. The deletion of yeast RNH1 causes modest increases in sensitivity to DNA-damaging agents (2), suggesting a role for nuclear RNase H1 in maintaining genome integrity in yeasts and perhaps in mammals.
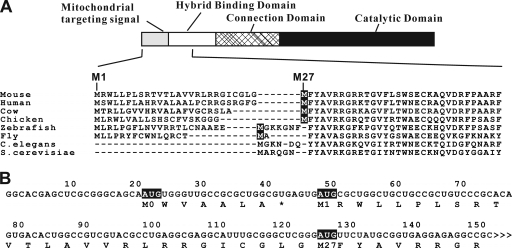
Eukaryotic RNases H1 consist of one or two hybrid binding domains (HBD) connected to the catalytic domain by a nonconserved number and sequence of amino acids (Fig. 1 A) (6). Most eukaryotic organisms, including S. cerevisiae, have single genes encoding RNase H1, whereas three related RNase H1 proteins are present in Caenorhabditis elegans (3, 18). RNase H1.0, a homolog of mouse RNase H1, does not have a canonical N-terminal MTS (3, 18), but another gene encoding a unique form of RNase H1 (RNase H1.1) has no HBD but does have an MTS (5, 18). Interestingly, regardless of the presence or absence of the extra N-terminal sequences, an AUG codon is present immediately prior to the HBD. Since the first 26-amino-acid sequence of mouse RNase H1 was shown to be a functional MTS, it was proposed that translation initiation at the M1 AUG codon produces the mitochondrial isoform and translation initiation at the M27 AUG codon produces the nuclear protein (7). M27 RNase H1 has been extensively characterized, and the importance of the HBD in enzyme function has been described (6, 13).
FIG. 1.
Organization of mouse Rnaseh1 mRNA. (A) Two in-frame AUGs are conserved in RNase H1 from flies to mammals. The domain organization of mouse RNase H1 is illustrated as a bar model. The amino acid sequence of N-terminal part of mouse RNase H1 is aligned with those of humans, cows, chickens, zebrafish, fruit flies, C. elegans (RNase H1.0), and S. cerevisiae. The second in-frame methionine residues are indicated as white letters on a dark background. The first and 27th residues (methionines) of mouse RNase H1 are indicated above the sequence. (B) Mouse Rnaseh1 mRNA has a 7-amino-acid upstream ORF. The nucleotide sequence of 5′ region of mouse Rnaseh1 mRNA is shown. AUG triplets are indicated in white letters on a dark background. Deduced amino acid sequences of ORFs are shown below the mRNA sequence. The numbering refers to the number of nucleotides from the 5′ end.
In addition to the two in-frame AUG codons present in mouse Rnaseh1 mRNA, we noted a short upstream open reading frame (uORF) (Fig. 1B). A recent study (4) showed that nearly 50% of human and mouse transcripts contain at least one uORF and the existence of uORFs generally reduces protein expression by 30 to 80%. Often uORFs are involved in posttranscriptional regulation (10, 32). The first example of a defect in a uORF causing a human disease was recently reported. Wen et al. (35) identified a mutation in a uORF of the gene encoding the human hairless homolog (HR). The mutation at the initiation codon of the uORF results in greater expression of the HR protein causing Marie Unna hereditary hypotrichosis. The best-studied examples of uORFs are the genes coding for GCN4 of yeast and ATF4 of mammals. Expression of these two genes increases under several stress conditions in which general translation is decreased. Expression of GCN4 and ATF4 occurs by a well-studied mechanism involving translational effects of multiple uORFs and phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) (16).
Employing a single transcript to produce essentially the same protein in mitochondria and nuclei has several consequences, among which are the following: (i) expression of an individual isoform is tightly linked with expression of the other changes in relative amounts of proteins, (ii) these other changes may require special control mechanisms, and (iii) loss of one activity most often would result in inactivation of both isoforms.
Here, we show that translation initiates at each of the two in-frame AUGs of the Rnaseh1 mRNA, with the longer form being imported into mitochondria. Moreover, we show that the levels of mitochondrial RNase H1 are modulated by the uORF, providing a means to express different levels of mitochondrial RNase H1 in various tissues or under different stress conditions. Interestingly, expression of excessive amounts of RNase H1 in mitochondria causes a growth defect in cultured cells. No obvious deleterious effect was observed when the nuclear RNase H1 was present in large amounts. We propose a mechanism whereby the level of RNase H1 in mitochondria is modulated without altering the level of RNase H1 in nuclei. Such regulation may be more generally employed, particularly for other mitochondrion/nucleus-targeted proteins.
MATERIALS AND METHODS
Cell lines and cultures.
Human osteosarcoma (HOS) cells were obtained from ATCC and cultured in Eagle's minimum essential medium (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and penicillin and streptomycin (Invitrogen). Flp-In T-Rex-293 (FT293) cells were obtained from Invitrogen and cultured in Dulbecco's modified Eagle's medium (DMEM; (Invitrogen) supplemented with 10% Tet system approved fetal bovine serum (Clontech, Mountain View, CA), 2 mM l-glutamine (Invitrogen), penicillin and streptomycin (Invitrogen), and 15 μg/ml blasticidin (InvivoGen, San Diego, CA). For stably transfected FT293 cells, 50 μg/ml hygromycin B (Invitrogen) was added. Doxycycline was obtained from Invitrogen and used as indicated.
Subcellular fractionation and mitochondria isolation.
Subcellular fractionation of minced mouse liver carried out using the ProteoExtract subcellular proteome extraction kit (Calbiochem, San Diego, CA). This technique employs three different detergents. The first detergent is added then the sample is centrifuged to generate a supernatant (fraction 1) and pellet. The pellet is treated with the second detergent and then the third with intervening centrifugations to yield fractions 2 and 3. The final pellet was resuspended in extraction buffer IV to yield fraction number 4.
For mitochondrial isolation by differential centrifugation, mouse liver was homogenized at 4°C with a Potter-Elvehjem homogenizer in a buffer consisting of 320 mM sucrose, 1 mM EDTA, 10 mM Tris-HCl, pH 7.4. Mitochondria were isolated as described by Fernández-Vizarra et al. (11). The homogenate was centrifuged at 1,000 × g for 5 min at 4°C. The pellet consisting of nuclei and unbroken cells was discarded, and the supernatant was centrifuged at 12,000 × g for 10 min at 4°C and washed four times with homogenizing buffer. The pellet was resuspended in homogenization buffer and was centrifuged at 1,000 × g for 5 min at 4°C and immediately incubated at room temperature for 15 min with 10 μg/ml proteinase K ± 1% Triton X-100. Protease digestion was stopped by the addition of 3 mM phenylmethylsulfonyl fluoride (PMSF), and incubation continued for 5 min at room temperature. For the RNase H1 activity gel assay, 12% SDS-PAGE with 32P-labeled poly(rA)/poly(dT) substrate (107 cpm/gel) polymerized in the running gel was run and processed in renaturation buffer (6, 14).
Antibodies.
Anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase), anti-cytochrome c oxidase IV (anti-COX IV), and antifibrillarin antibodies were purchased from Abcam (Cambridge, MA). Anti-Tom20 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Flag antibody was purchased from Sigma (St. Louis, MO). Anti-hemagglutinin (anti-HA) high-affinity rat monoclonal antibody (clone 3F10) was obtained from Roche Applied Science (Indianapolis, IN).
Construction of plasmids.
The DNA containing the complete 5′ untranslated region (UTR) and RNase H1 coding region of mouse Rnaseh1 cDNA was amplified from a cDNA clone (5), using the following primers (with the indicated restriction sites underlined and the Flag tag coding sequence in boldface): 5′-AGCTCGCGGGCAGCAATGTGGGTTGCCGCG-3′ and 5′-CCGGATCCAGTCTTGTCGTCGTCATCCTTGTAATCTGCAGCTCCACCGTCCTCAGACTGCTTCGCTCC-3′ (BamHI) (PstI). The amplified DNA was inserted once in pCRII-TOPO (invitrogen) by TA cloning, and the EcoRI-BamHI-digested fragment was then ligated into EcoRI-BamHI-cut pEGFP-N2 (Clontech, Mountain View, CA). The enhanced green fluorescent protein (EGFP) coding region was removed, and Strep-Tag II) (31) was added by PCR, using the following primers (with the indicated restriction sites underlined and the Strep-Tag II coding sequence in boldface) and BamHI digestion followed by self-ligation: 5′-CGGGATCCCACTGGAGCCACCCGCAGTTCGAAAAGTGAGGTACCATCACTCTCGGCATGGACGAGCTGT-3′ (BamHI and KpnI, respectively) and 5′-CCGGATCCAGTCTTGTCGTCGTCATCCTTGTAATCTGCAGCTCCACCGTCCTCAGACTGCTTCGCTCC-3′ (BamHI and PstI, respectivley). The resulting plasmid is pMMM-FLST. AUG-to-AUC mutations were introduced using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the following primers: 5′-CGCGGCAACCCAGATTGCTGCCCGCGAGCC-3′ and 5′-GGCTCGCGGGCAGCAATCTGGGTTGCCGCG-3′ for the M0 AUG, 5′-CGGCAGCAGCCAGCGGATCACTCACGCCAGCGC-3′ and 5′-GCGCTGGCGTGAGTGATCCGCTGGCTGCTGCCG-3′ for the M1 AUG, and 5′-CCTCACCGCATAGAAGATCCCGAGCCCGCAAATGCC-3′ and 5′-GGCATTTGCGGGCTCGGGATCTTCTATGCGGTGAGG-3′ for the M27 AUG. These plasmids were used to transfect HOS cells and to check the localization of RNase H1-Flag protein by confocal microscopy.
For luciferase assays, the Renilla luciferase gene was amplified from pΔEMCV (a gift from P. Sarnow and M. S. Carter Stanford) using the following primers (with the indicated restriction sites underlined) and inserted into the PstI site of Flag tag-Strep-Tag II plasmids: 5′-AAAGCTGCAGGTTCGAAAGTTTATGATCCAGAACAAAGG-3′ (PstI) and 5′-TTTCTGCAGCTCCACCTTGTTCATTTTTGAGAACTCGCTCAACGAA-3′ (PstI). For AUG-to-AUC mutations, we used the same sets of primers as Flag tag-Strep-Tag II plasmids, and for frameshift mutations, we used the following primers: 5′-CGCCTGAGGCGAGGCATTGCGGGCTC-3′ and 5′-GAGCCCGCAATGCCTCGCCTCAGGCG-3′ for 1-base deletion and 5′-CGGGCTCGGGATGTTCTATTGCGGTGAGGA-3′ and 5′-TCCTCACCGCAATAGAACATCCCGAGCCCG-3′ for 1-base insertion. For deletion of the termination codon of uORF, we used 5′-CCGCGCTGGCGGGAGTGATGCGC-3′ and5′-GCGCATCACTCCCGCCAGCGCGG-3′.
For the construction of FT293 cells stably overproducing RNase H1 from M27 AUG initiation, HindIII-KpnI fragment of pMMM-FLST was ligated into the HindIII-KpnI-cut pcDNA5/FRT/TO (Invitrogen) and AUG-to-AUC mutations were introduced at the M0 and M1 AUG's as described above. This plasmid is pcDNA5-M27. For the construction of FT293 cells stably overproducing RNase H1 from M1 AUG initiation, the DNA containing complete Kozak sequence and RNase H1 coding region with Flag tag and Strep-Tag II was amplified from pMMM-FLST using the following primers (with the indicated restriction sites underlined and the Kozak sequence in boldface): 5′-GCCGCGCTGGCGTGAGTGGCCACCATGCGCTGGCTGCTGCCG-3′ and 5′-CCGGATCCAGTCTTGTCGTCGTCATCCTTGTAATCTGCAGCTCCACCGTCCTCAGACTGCTTCGCTCC-3′ (BamHI and PstI, respectively). The amplified DNA was inserted in pCRII-TOPO (Invitrogen) by TA cloning, and the HindIII-BamHI-digested fragment was then inserted into HindIII plus BamHI sites of pcDNA5-M27. This resulting plasmid is pcDNA-M1. An AUG-to-AUC mutation was introduced to the M27 AUG of pcDNA-M1 by the QuikChange II site-directed mutagenesis kit (Stratagene) and with the primers described above. The resulting plasmid is pcDNA5-M1-M27I.
For the construction of plasmids for the expression and localization of the HA-tagged RNase H1 with or without the nuclear export signal (NESi), the DNA containing the RNase H1 coding region with HA tag was amplified using the following primers (with the indicated restriction sites underlined and the HA tag coding sequence in boldface): 5′-ACCTCTAGAATGAGCTGGCTTCTGTTCCTGGCCCA-3′ (XbaI) and 5′-GGTGAATTCGGCGTAGTCGGGCACGTCGTAGGGGTAGTCTTCCGATTGTTTAGCTCCTTCTCTGG-3′ (EcoRI). The amplified DNA was inserted into XbaI and EcoRI sites of either the original pcDNA3.1(−) (Invitrogen) or pcDNA3.1(−) containing NES sequence from the NS2 protein of minute virus of mice (VDEMTKKFGTLTIHDTEK), as reported by Minczuk et al. (21). For the construction of FT293 cells stably overproducing RNase H1-HA-NESi protein, the PmeI fragment of pcDNA3.1(−) + RNase H1-HA-NESi was ligated into PmeI-cut pcDNA5/FRT/TO.
Construction of stably transfected cell lines.
Flp-In T-Rex-293 cells were transfected with 900 ng pOG44 plasmid (Invitrogen) and 100 ng pcDNA5-M27, pcDNA5-M1, pcDNA5-M1-M27I, pcDNA5-CAT (Invitrogen), or RNase H1-HA-NESi using the Amaxa Nucleofector kit V (Köln, Germany) to produce FT293-M27, FT293-M1, FT293-M1-M27I, FT293-CAT, and FT293-RNase H1-HA-NESi, respectively. Cells were selected in the media containing 500 μg/ml hygromycin B. Protein production was induced by addition of 1 μg/ml doxycycline (Sigma). For growth rate measurements, cells were transferred on day 4 after addition of doxycycline to 25-cm2 flasks with 4 ml medium at 8 × 103 cells/ml. Medium was changed every other day. Cell number was counted using a hemocytometer. For mitochondrial DNA (mtDNA) copy number analysis, cells were maintained in 9-cm2 dishes (Falcon) and protein expression was induced with the appropriate dose of doxycycline. Cells were passed every 3 days, and fresh drug was added at the time of passage.
mtDNA copy number analysis.
mtDNA copy number of control and doxycycline-treated FT293 cells was estimated by amplifying a portion of the cytochrome c oxidase II (COX II) gene of mtDNA and comparing it to the nuclear single-copy gene, amyloid precursor protein, or APP, as previously described (33). Total DNA samples were prepared from cells lysed in 1 ml 75 mM NaCl, 50 mM EDTA, 0.2% SDS (pH 8.0). Cell lysates were incubated at 50°C overnight with 400 μg/ml proteinase K. DNA was precipitated by addition of an equal volume of isopropanol, pelleted by centrifugation at 20,000 × g for 30 min, and dissolved in 300 μl of Tris-EDTA (TE) buffer (pH 8.0) at 50°C overnight. The DNA concentration was adjusted to 4 ng/μl, and 20 ng was used per real-time PCR. The amplification cycle conditions on the Roche LightCycler 480 system were 95°C for 10 min and then 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 20 s. The primer sequences were as follows: COXII forward, 5′-CGTCTGAACTATCCTGCCCG-3′, COXII reverse, 5′-TGGTAAGGGAGGGATCGTTG-3′, and COXII probe, 5′-CGCCCTCCCATCCCTACGCATC-3′; and APP forward, 5′-TTTTTGTGTGCTCTCCCAGGTCT-3′, APP reverse, 5′-TGGTCACTGGTTGGTTGGC-3′, and APP probe 5′-CCCTGAACTGCAGATCACCAATGCGGTAG-3′.
COX II probes have a Quasar 670 (Q670) fluor attached to the 5′ end and a Black Hole quencher 2 (BHQ2) quencher at the 3′ end. APP probes have a 6-carboxyfluorescein (FAM) probe at the 5′ end and a BHQ1 quencher at the 3′ end (Biosearch Technologies). Different probes allowed for multiplexing. The quantitative PCR (qPCR) cycle threshold (ΔΔCT) method was employed to calculate relative levels of either mtDNA (20). Briefly, for each well, a ΔCT was calculated between the standard (APP gene) and target (COX II gene). Each was then subtracted from the average ΔCT of the control sample (parental cells) to produce the ΔΔCT. A relative copy number value was determined by calculating 2−ΔΔCT. These values were averaged (mathematically, the mean 2−ΔΔCT for control samples will be 1.0), and the outlier of the four values was dropped. The remaining three were reaveraged, and if the standard deviation was less than 10%, the value was accepted and considered as one trial. Repeated experiments began with freshly plated cells.
Confocal microscopy.
HOS cells were grown on glass coverslips and transfected with 2 μg of various pMMM-FLST-related plasmids, using 10 μl of Lipofectamine 2000 (Invitrogen) in 6-well dishes. After 24 h of incubation, 40 nM MitoTracker Red CMXRos (Invitrogen) was added to media and the cultures were incubated for an additional hour. Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 10 min and permeabilized in 0.2% Triton X-100-PBS for 5 min. After blocking with Image-iT FX signal enhancer (Invitrogen) for 30 min, fixed cells were treated with anti-Flag antibody (5 μl/ml) for 1 h followed by incubation with Alexa fluor 488 anti-IgG (5 μl/ml) for 30 min. Slides were mounted using ProLong Gold antifade with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen). Stably transfected FT293 cells were grown on polylysine-coated coverslips (BD Bioscience, Bedford, MA) in medium containing 1 μg/ml doxycycline for 24 h. Cells were fixed and immunostained as for HOS cells. Images were acquired with LSM 510 Meta confocal microscope (Zeiss) and Plan-Apochromat 100×/1.4 oil immersion objective (Zeiss).
In vitro translation.
The in vitro translation experiment was performed using TnT T7 Quick for PCR DNA (Promega, Madison, WI). Templates for transcription were amplified from pMMM-FLST, pMMM-FLST with the M1 AUC mutation, pMMM-FLST with the M27 AUC mutation, or pcDNA-M1 plasmids by PCR using the following primers: 5′-TGAATTGTAATACGACTCACTATAGGGTGGGTTGCCGCGCTGGCGTG-3′ and 5′-TTTTTTTTTTTTTTTTTTTTAAAGCAAGTAAAACCTCTACAAATGTGGTATGG-3′. Amplified DNA was purified using the MinElute PCR purification kit (Qiagen, Valencia, CA). Eighty nanograms of DNA was mixed with 40 μl of TnT T7 Quick master mix and 10 μCi of [35S]methionine for 90 min at 30°C. The translation products are analyzed by SDS-PAGE (12%) followed by autoradiography.
Luciferase assay.
HOS cells were grown in 96-well plates and transfected with 70 ng of RNase H1-luciferase fusion plasmids along with 70 ng of pGL3-control plasmid (Promega) using 0.35 μl of Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were lysed and assayed for luciferase activity following the protocol of the dual-luciferase reporter assay system (Promega). Renilla luciferase activity data were normalized using firefly luciferase activity of the cotransfected cells to measure relative transfection efficiencies. Transfection with the RNase H1-luciferase fusion plasmid having a 1-base insertion after the M27 AUG, thereby shifting translation out of frame, was used as a background control.
Quantitative RT-PCR.
RNA was isolated from the transfected HOS cells using Trizol (Invitrogen) and Purelink RNA minikit (Invitrogen). cDNA was synthesized using Omniscript reverse transcription (RT) kit (Qiagen). Quantitative PCR was performed using LightCycler 480 SYBR green I master mix (Roche) and LightCycler 480 real-time PCR instrument (Roche). The primers for firefly luciferase cDNA were 5′-GCCTGAAGTCTCTGATTAAGT-3′ and 5′-ACACCTGCGTCGAAGATGT-3′. The primers for Renilla luciferase cDNA were 5′-ATGGGATGAATGGCCTGATA-3′ and 5′-CAACATGGTTTCCACGAAGA-3′.
For RNase H1-HA-NESi, RNA was isolated from FT293 cells, using Trizol (Invitrogen). Five microliters of RNA (about 2 μg) was used to generate cDNA, using the Omniscript reverse transcription kit (Qiagen). Random hexamers and oligo(dT) were used as primers in a final reaction volume of 20 μl. Extension was carried out for 1 h at 37°C, and the enzyme was inactivated by being heated to 95°C for 5 min. Reaction mixtures were diluted by the addition of 80 μl of nuclease-free water, and 5 μl of this was used for each qPCR.
Five microliters of total cDNA was added to 15 μl of qPCR master mix (Roche) containing 10 μl qPCR mix, 1 μM GAPDH primers, 1 μM RNase H1 primers, 0.125 μM GAPDH probe, and 0.125 μM RNase H1 probe. Each sample was run in quadruplet. The amplification cycle conditions on the Roche LightCycler 480 system were 95°C for 10 min and then 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 20 s. GAPDH probes have a Q670 fluor attached to the 5′ end and a BHQ2 quencher at the 3′ end. RNase H1 probes have a FAM probe at the 5′ end and a BHQ1 quencher at the 3′ end (Biosearch Technologies). Different probes allowed for multiplexing. The primer sequences were as follows: RNase H1 forward, 5′-GACACGTTTTCCTACATGGGAGAC-3′, RNase H1 reverse, 5′-CAGTAAACGCCGATTCCTGCT-3′, and RNase H1 probe, 5′-CTACACTGATGGCTGCTGCTCCAGTAATGG-3′; and GAPDH forward, 5′-GGTGAAGGTCGGAGTCAACG-3′, GAPDH reverse, 5′-CAGAGTTAAAAGCAGCCCTGGT-3′, and GAPDH probe, 5′-ATTTGGTCGTATTGGGCGCCTGGT-3′.
In-gel activity assay.
The renaturation gel assay was performed using the method described previously (9).
RESULTS
RNase H1 in mouse liver cells is more abundant in nuclei than in mitochondria.
To examine the relative abundance of the two isoforms of RNase H1, we prepared subcellular fractions of fresh mouse liver cells by a detergent-based method (see Materials and Methods). In this experiment, a cytosolic marker (GAPDH), mitochondrial markers (COX IV and Tom20), and a nuclear marker (fibrillarin) were found in different fractions as shown by Western analysis (Fig. 2 A). To detect RNase H1 in each fraction, we performed gel renaturation assays (9). RNases H digest the radiolabeled substrates, as shown in Fig. 2A and B. We show a negative image for clarity. In our fractionated samples, an ∼30-kDa signal was detected in the fraction containing fibrillarin, indicating nuclear localization of RNase H1 (Fig. 2A, lane 3). No RNase H activity was detected from other fractions, including the mitochondrial fraction (Fig. 2A, lane 2). This result indicates that the majority of RNase H1 is present in nuclei. We also performed mitochondrial isolation based on differential centrifugation (11). By this method, we obtained a more concentrated mitochondrial fraction than that from the detergent-based method. When we used the concentrated mitochondrial sample, we detected an ∼30-kDa RNase H activity (Fig. 2B). This activity was still present after proteinase K treatment, in contrast to Tom20, a mitochondrial outer membrane protein, which was completely digested by proteinase K. Upon solubilizing the mitochondrial membrane by adding Triton X-100, RNase H activity became susceptible to proteinase K. These results demonstrate that the majority of RNase H1 in mouse liver localizes to nuclei, with only a minor portion present in mitochondria.
FIG. 2.
Subcellular fractionation of mouse liver cells. (A) The contents of fresh mouse liver cells were fractionated by the detergent-based method (see Materials and Methods). The numbers refer to the fraction numbers. The distribution of RNase H1 was analyzed by gel renaturation assay and is presented in a negative image. Distributions of GAPDH, COX IV, Tom20, and fibrillarin were analyzed by immunoblotting. (B) Intact mitochondria were isolated from mouse liver and incubated with proteinase K and/or Triton X-100. RNase H1 was detected by gel renaturation assay and is presented in a negative image. Tom20 was detected by immunoblotting.
Altering the AUG context of M1 can increase accumulation of RNase H1 in mitochondria.
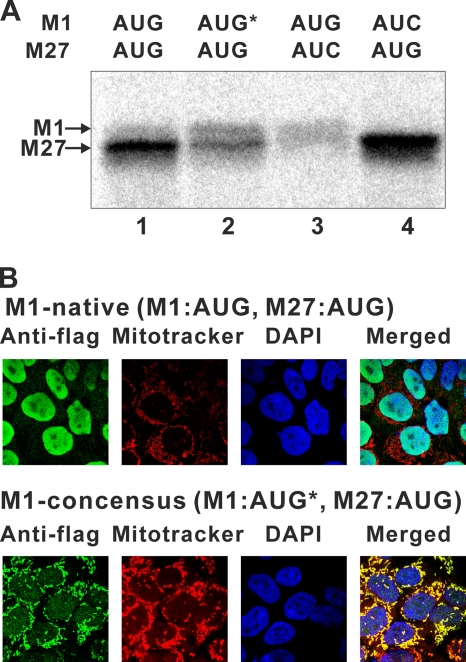
We found that all RNase H1-encoding genes in organisms from flies to mammals have two in-frame AUGs (Fig. 1) with putative MTSs at their N termini. The MTS is notably absent in yeast and Caenorhabditis elegans RNases H1 (Fig. 1A). All eukaryotic RNases H1 have highly conserved domains at their N- and C-termini: the hybrid binding domain (HBD) and the catalytic domain. The N-terminal amino acid sequences of RNases H1 of various organisms are shown in Fig. 1A. It was suggested that the second methionine codon could be an alternative initiation codon (7). In vitro translation of mouse Rnaseh1 mRNA, using rabbit reticulocyte lysate, shows two distinct proteins, with the slower-migrating band being less abundant than the faster-migrating species (Fig. 3 A, lane 1). We suspected that the low abundance of mitochondrial RNase H1 was due in part to the context of the M1 AUG codon. To test this, we inserted nucleotides immediately upstream of the M1 AUG codon to closely resemble the consensus Kozak sequence (19). In vitro translation of the RNA produced significantly more of the slower-migrating product with concomitant loss of the faster-migrating band (Fig. 3A, lane 2). Translation products of mRNAs in which the M27 and M1 AUGs were individually changed to AUC resulted in the disappearance of the faster-migrating band (Fig. 3, lane 4) and the slower-migrating species (Fig. 3A, lane 4), confirming the M1 and M27 codons are used as initiation codons.
FIG. 3.
Abundance of mitochondrial RNase H1 is affected by the context of the M1 AUG codon. (A) [35S]Met-labeled in vitro translation products of mouse Rnaseh1 with the native context of the M1 AUG (lane 1), the consensus context of the M1 AUG (AUG*) (lane 2), the native context of the M1 AUG with the M27 AUG-to-AUC mutation (lane 3), and the native context of the M1 AUG with the M1 AUG-to-AUC mutation (lane 4) were separated by SDS-PAGE and visualized by autoradiography. The bands correspond to the products indicated by arrows. (B) HEK293 cells producing Flag-tagged mouse RNase H1 were analyzed by confocal microscopy. Cells were incubated with primary antibodies to Flag tag and secondary antibodies emitting green. Mitochondria and nuclei were stained with MitoTracker (red) and DAPI (blue), respectively. Flag-tagged mouse RNase H1 was expressed with either the native context of the M1 AUG (M1-native) or the consensus context of the M1 AUG (M1-consensus, AUG*).
We also generated an HEK293 cell line having an exogenous mouse RNase H1 expressed with the context of the M1 AUG optimized for translation (19), with transcription driven by the cytomegalovirus (CMV) promoter in a doxycycline-inducible manner. The exogenous RNases H1 in this cell line contain a Flag tag at their carboxy termini. By microscopic analysis, we observed a high Flag-tagged signal in mitochondria (Fig. 3B; M1-consensus). On the other hand, a HEK293 cell line having an exogenous mouse RNase H1 in its natural context of the M1 AUG showed a high Flag-tagged signal in nuclei (Fig. 3B; M1-native). Thus, inefficient translation initiation at the M1 AUG is an important factor in limiting the level of RNase H1 in mitochondria.
An upstream AUG and two in-frame AUGs are active translation initiation sites.
In addition to two in-frame AUG codons, mouse Rnaseh1 mRNA has an AUG codon (M0) upstream of the M1 AUG (Fig. 1B). Translation initiation at the M0 AUG will produce a 7-amino-acid polypeptide that terminates 3 nucleotides (nt) prior to the M1 AUG. Upstream ORFs are present in RNase H1-encoding genes in 23 mammalian genomes as shown in Table 1. The M0 AUG codons are located between 17 and 53 bases upstream of the M1 AUG. Termination codons are located from −4 to +22 nt with respect to the start A (+1) residue of the M1 AUG codon and are at least 56 nt upstream of the second in-frame AUG. For human RNASEH1, the termination of the uORF overlaps with the M1 AUG codon (AUGA). Peptides predicted from the mRNA sequences have no obvious common properties and range in length from 6 nt in dogs to 18 nt in other organisms, including cows (see Table S1 in the supplemental material).
TABLE 1.
Positions and sizes of the conserved uORFs of RNase H1 genes
| Mammal(s) | nt position |
uORF length (aa) | 2nd Met position (aa) | Length from end to 2nd Met (nt) | |
|---|---|---|---|---|---|
| Start | End | ||||
| Cow | −53 | 4 | 18 | 23 | 62 |
| Dolphin | −53 | 4 | 18 | 28 | 77 |
| Megabat | −53 | 4 | 18 | 27 | 74 |
| Microbat | −53 | 4 | 18 | 25 | 68 |
| Mouse, rat, rabbit, pika, mouse lemur, guinea pig | −27 | −4 | 7 | 27 | 81 |
| Lesser hedgehog tenrec, hyrax, alpaca, elephant | −27 | −4 | 7 | 28 | 84 |
| Human, gorilla, chimpanzee, macaque, orangutan | −26 | 4 | 9 | 27 | 74 |
| Tarsier | −26 | 22 | 15 | 27 | 56 |
| Armadillo | −25 | −2 | 7 | 28 | 82 |
| Dog | −17 | 4 | 6 | 28 | 77 |
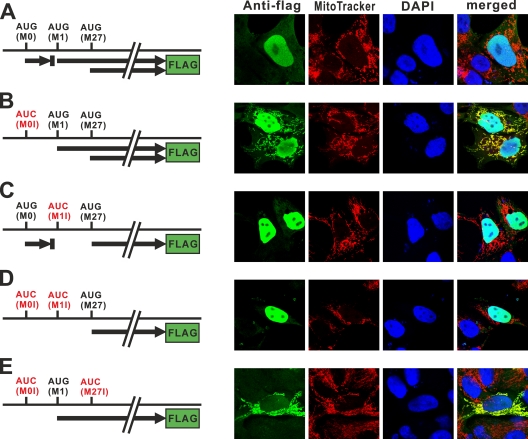
To investigate the effect of the uORF on the level of expression and subcellular localization of RNase H1, we transiently transfected the human osteosarcoma (HOS) cell line with various plasmid DNAs producing Flag-tagged RNase H1. The plasmids contain the full-length mouse RNase H1 coding sequence and complete 5′ region of the mRNA with each of the AUG codons present or changed to AUC. In the situation in which all three AUG codons are present, Flag tag signal was detected only in nuclei (Fig. 4 A). However, when we changed the AUG of the upstream ORF (uORF) from the M0 AUG to AUC, we were able to easily observe the Flag tag signal in both mitochondria and nuclei (Fig. 4B). This result suggests that the uORF decreases the mitochondrial RNase H1 to a level that is undetectable in this assay. Changing the M1 AUG to AUC results in complete loss of a signal in mitochondria (Fig. 4C). In the absence of the M0 and M1 AUGs, only nuclear localization is observed (Fig. 4D). This indicates that the appearance of mitochondrial localization by the removal of the M0 AUG (Fig. 4B) is due to an increase in the level of translation from the M1 AUG. When we altered the M27 AUG codon, we observed mitochondrial protein only when there is no uORF (Fig. 4E) (data not shown), consistent with the effects of changing the uORF to AUC in the normal arrangement of translation (Fig. 4A and B). Taken together, we conclude that ribosomes initiate translation at the M0 AUG, M1 AUG, and M27 AUG. Moreover, the uORF affects the production of mitochondrial isoform of RNase H1 by decreasing translation initiation at the M1 AUG.
FIG. 4.
Subcellular localization of Flag-tagged mouse RNase H1 in HOS cells. HOS cells transfected with plasmids (A to E) producing Flag-tagged mouse RNase H1 were analyzed by confocal microscopy. Plasmids used in panels B to E contain AUG-to-AUC mutations, as indicated by the red color. Thick arrows represent ORFs. Cells were incubated with primary antibodies to Flag tag and secondary antibodies emitting green. Mitochondria and nuclei were stained with MitoTracker (red) and DAPI (blue), respectively. We also examined constructs in which all three AUG codons were changed to AUG, and as expected, we observed no Flag signal.
Elimination of the uORF enhances production of mitochondrial RNase H1 but not nuclear RNase H1.
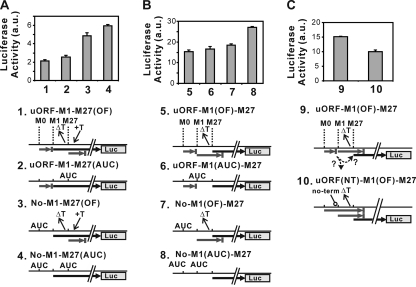
To obtain quantitative estimates of the translational effects imposed by the uORF on expression of each isoform of RNase H1, we created plasmids which express an RNase H1-luciferase fusion protein (Fig. 5). Renilla luciferase reporter assays were performed after transient transfection of HOS cells with any one of several plasmids. All data were analyzed after subtraction of background and normalization to firefly luciferase activity from a cotransfected control plasmid (see Materials and Methods).
FIG. 5.
uORF represses translation initiation at the M1 AUG. HOS cells were transiently transfected with plasmids producing mouse RNase H1 fused to Renilla luciferase. Luciferase activity is detected only from proteins initiated at the M1 AUG (A) or at the M27 AUG (B and C). Plasmids contain mutations as indicated by ΔT (one-thymine-base deletion), +T (one-thymine-base insertion), AUC (AUG-to-AUC substitution), and “no-term” (in which the termination codon of uORF, UGA, was changed to GGA). Black thick arrows represent ORFs fused to the luciferase gene. ORFs shown by gray arrows have termination codons before the luciferase gene. Bar graphs represents the mean ± standard deviation (SD) of luciferase activity (n = 3) of HOS cells transfected with the indicated plasmid.
To determine the effect that translation from M0 and M27 has on translation from M1, we changed the M27 AUG codon to AUC or made a single-nucleotide deletion between the M1 and M27 AUGs accompanied by a 1-base insertion after the M27 AUG in the presence or absence of uORF (Fig. 5A). The plasmid carrying the uORF-M1-M27OF version permits detection of luciferase activity only from M1 AUG translation initiation, since starting at the M27 AUG has a different reading frame (out of frame [OF]) terminating translation after incorporation of only 18 amino acids. Two other plasmids in which the uORF is absent were generated (substitution of AUC for AUG at M0) (Fig. 5A; No-M1-M27OF/No M1-M27AUC). In both instances, elimination of the uORF from these plasmids resulted in a doubling of luciferase activity. Of the ribosomes that can initiate at the M1 AUG, about half are prevented from doing so by the uORF. These data also indicate, as expected, that initiation at the downstream M27 AUG has no effect on upstream initiation at the M1 AUG.
We followed the same scheme to examine the M27 AUG-directed product (Fig. 5B). To observe luciferase activity only from M27 AUG translation initiation, we made a single-nucleotide deletion between the M1 and M27 AUGs (uORF-M1OF-M27). Owing to the frameshift, translation initiated at the M1 AUG produces a peptide of 31 amino acids terminating downstream of the M27 AUG, whereas translation starting at the M27 AUG produces a full-length RNase H1-luciferase fusion protein. uORF-M1OF-M27 (Fig. 5B) produced about 7 times more luciferase activity than its M1 counterpart, uORF-M1-M27OF (Fig. 5A), which is indicative of the ratio of production of nuclear and mitochondrial RNase H1. In contrast to No-M1-M27OF (Fig. 5A), elimination of the uORF from uORF-M1OF-M27 resulted in only a small increase (No-M1OF-M27) (Fig. 5B). Furthermore, elimination of translation initiation at the M1 AUG also resulted in only a slight increase in luciferase activity (uORF-M1AUC-M27) (Fig. 5B). However, elimination of both the M0 AUG and M1 AUG by AUG-to-AUC mutations (No-M1AUC-M27) significantly increased the M27 initiation product. These data indicate that both the uORF and M1 initiation have an effect on M27 translation initiation.
Ribosomes reinitiate translation at M27 when they translate the uORF.
The level of luciferase activity increasing when changing the M0 AUG to AUC (Fig. 5B) could indicate that initiation at the M27 AUG is limited by the number of ribosomes that skip both the M0 and M1 AUG codons. We showed that the absence of the M0 AUG permits more ribosomes to reach the M1 AUG codon, and this increase of translation initiation at the M1 AUG could limit the number of ribosomes reaching the M27 AUG codon. Were this to be the case, ribosomes reaching the uORF termination codon would leave the mRNA and be unable to translate at either of the downstream ORFs. However, if ribosomes remain on the mRNA after reaching the termination codon of the uORF, they could move past the nearby M1 AUG codon and reinitiate at the M27 AUG codon. To determine what effect the uORF termination codon has on expression of M27 RNase H1, we changed the UGA termination codon of the uORF to GGA in a plasmid with a 1-base deletion between the M1 and M27 AUG codons. Only the protein initiating at the M27 AUG codon will be fused to luciferase (Fig. 5C). As expected if reinitiation is important for expressing the M27 RNase H1, elimination of the termination codon decreased the amount of fusion protein to about 60% that seen when translation of the uORF terminates at its normal site (Fig. 5C).
Reinitiation requires the eIF2α/Met-tRNA/GTP complex to be reloaded on ribosomes as they scan for another initiating AUG codon (23). It is known that such reinitiation requires a minimal distance between termination and reinitiation sites. In mouse Rnaseh1, the distance from the termination codon of the uORF to the M1 AUG is only 3 nucleotides, but to the M27 AUG there are 81 nucleotides (Fig. 1B). In RNase H1 mRNAs from other mammals, the positions of termination sites of the uORF are not a conserved distance from the start of the M1 AUG codon, and in some cases, the uORF termination codon follows the M1 AUG codon. However, there are always at least 56 nucleotides between the uORF termination site and the M27 AUG initiation codon (Table 1). For the mouse, the M1 AUG is too close to the termination site, but the M27 AUG may be far enough to permit reinitiation.
Our conclusion that translational regulation accounts for the amount and types of RNase H1 levels could be attributed to increases or decreases in mRNA levels. To exclude this possibility, we performed qPCR analyses and observed small differences in mRNA level, but the changes were modest at best and in no way correlated with the increases and decreases in protein levels we have observed (Table 2 ).
TABLE 2.
Ratio of Renilla to firefly luciferase mRNA
| Plasmid type | Renilla/firefly luciferase mRNA ratioa | SE (n = 4) |
|---|---|---|
| uORF-M1-M27OF | 0.53 | 0.22 |
| uORF-M1-M27AUC | 0.77 | 0.17 |
| No-M1-M27OF | 0.86 | 0.21 |
| No-M1-M27AUC | 0.68 | 0.24 |
| uORF-M1AUC-M27 | 0.64 | 0.07 |
| No-M1AUC-M27 | 0.59 | 0.10 |
| uORF-M1OF-M27 | 0.52 | 0.11 |
| No-M1OF-M27 | 0.45 | 0.09 |
| uORFNT-M1OF-M27 | 0.60 | 0.12 |
The mRNA levels of HOS cells cotransfected with pGL3 and one of the plasmids in Fig. 5 were measured by real-time quantitative PCR.
Model of translation regulation.
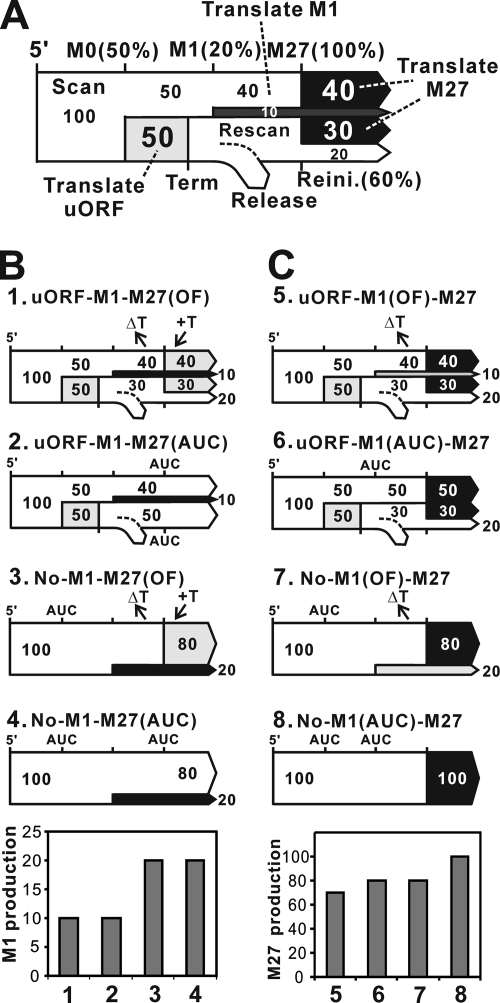
Using the data generated in the luciferase reporter assays, we propose a model to account for the relative expression of the different constructs as measured by luciferase activity (Fig. 6 A). This model has four parameters: translational initiation efficiencies at the M0 AUG, M1 AUG, and M27 AUG and reinitiation efficiency at the M27 AUG. Initiation efficiency is defined as the probability of a scanning ribosome initiating translation after encountering an AUG codon. Initiation efficiency and reinitiation efficiency could be different because a rescanning ribosome may not have all of the necessary initiation factors when it encounters the AUG site. Luciferase activity measures the fraction of ribosomes reaching the AUG codon multiplied by the initiation frequency. To make the model, we arbitrarily set the initiation efficiency at the M27 AUG as 100%, although the actual frequency must be substantial lower than 100% (Fig. 6A; M27). Then the translation initiation efficiency at the M1 AUG (Fig. 6A; M1) becomes 20% because translation initiation efficiency at M1 is ∼20% of M27 (Fig. 5A, No-M1-M27AUC; and B, No-M1AUC-M27) and the translation initiation efficiency at the M0 AUG (Fig. 6A; M0) becomes 50% because removal of the uORF from uORF-M1-M27AUC increased luciferase activity two times (No-M1-M27AUC) (Fig. 5A). If we now consider 100 events in which a ribosome is loaded to the 5′ cap of mouse Rnaseh1, you can calculate the product of initiation at M1 is 10 (20% of the 50 ribosomes bypassing the uORF) and the product of initiation at M27 is 40 (the number of ribosomes not initiating at either the uORF or M1). These 40 products of M27 initiation represent ∼60% of the total M27 initiation product (Fig. 6C). The 50 ribosomes that initiated and terminated at the uORF must then resume scanning to produce the remaining M27 fusion protein. These rescanning ribosomes must account for the remaining ∼40% of the total M27 protein. Therefore, the reinitiation efficiency at the M27 AUG must then be ∼60%. In this example, 30 of the ribosomes reinitiating would provide the remaining ∼40% (30/70) (Fig. 6A). Once the four parameters are set, we can calculate the probability of events that may occur on the mRNA in the presence of the mutations we introduced in Fig. 5A and B. Figure 6B and C show the result of such calculation. The experimental and calculated data match very well.
FIG. 6.
A model for translation of Rnaseh1 mRNA production of mitochondrial and nuclear RNase H1. (A) Possible statuses of a ribosome on Rnaseh1 mRNA are depicted on a “traffic roadmap.” Ribosomes move left to right from the 5′ end of mRNA in “scanning mode.” A ribosome in the scanning mode may change its status to “translating mode” at AUG codons with certain probabilities indicated on the top of the road. After termination of uORF translation, the ribosome may enter the “rescanning mode” and generate a 60% chance of reinitiating (Reini.) translation at the M27 AUG. The white parts of the road represent the “scanning mode” and the light gray, dark gray, and black parts of the road represent the “translating mode” of uORF, M1 AUG initiation, and M27 AUG initiation, respectively. Numbers in the road represent the probability (%) that a ribosome at the positions of the mRNA is in that status. (B) Using the model, production levels of M1 AUG initiation in Fig. 5A were calculated. Black represents M1 translation, and gray represents nonproductive translation. The bar graph represents calculated results. The graph is comparable to that in Fig. 5A. (C) Likewise, production levels of M27 AUG initiation in Fig. 5B were calculated. Black represents M27 translation, and gray represents nonproductive translation. The bar graph represents calculated results. The graph is comparable to that in Fig. 5B.
Overproduction of mitochondrial RNase H1 inhibits cell growth and causes profound mtDNA depletion.
Because the mitochondrial isoform of RNase H1 is normally expressed at much lower levels than the nuclear isoform, we wanted to know if expression of high levels of mtRNase H1 would affect cell growth. We constructed doxycycline-inducible stable FT293 cell lines, FT293-M1 (expressing both isoforms of RNase H1 but, due to the change in consensus around the M1 AUG, with excess RNase H1 seen only in mitochondria) (Fig. 3B), FT293-M1-M27I (producing only the mitochondrial form of RNase H1, in which M27 is changed to I), and FT293-M27 (producing only the nuclear version of RNase H1). When 1 μg/ml doxycycline was added to FT293-M1, growth continued for several days but started to decrease after 7 to 8 days (Fig. 7 A). FT293-M1-M27I cells also stop growing 7 or 8 days after addition of doxycycline. In contrast, FT293-M27 cells continue to multiply (Fig. 7A) and grow normally for at least 30 days after induction (data not shown). These findings indicate that excess amounts of RNase H1 in mitochondria result in cell death due to loss of mitochondrial DNA. To confirm this observation, we used a stable FT293 cell line that expressed human RNase H1 in a doxycycline-inducible manner. To limit accumulation of RNase H1 to mitochondria, the protein contained a nuclear export sequence. Analysis by confocal microscopy demonstrates that the excess RNase H1 accumulates extensively in mitochondria and is undetectable in nuclei. The RNase H1 mRNA, HA-tagged RNase H1 protein, and RNase H activity all increased after induction (Fig. 7B). This cell line responded in a dose- and time-dependent manner to RNase H1 overexpression (Fig. 7B). In as little as 3 days after induction, mtDNA copy numbers decreased to <20% of normal, and by day 9 mtDNA copy number decreased to less than 1% (Fig. 7B). Cells were no longer growing at day 12.
FIG. 7.
Overexpression of RNase H1 in mitochondria impairs growth and results in mtDNA loss. (A) FT293-CAT, FT293-M27, FT293-M1-M27I, and FT293-M1 cells were grown in the media with (filled squares) or without (open squares) 1 μg/ml doxycycline. The FT293-CAT cell line has the CAT gene inserted at the flip-in site. Likewise, the other FT293 cell lines have the cDNA for the Rnaseh1 mRNA, starting at M1 (FT293-M1), M27 (FT293-M27), or M1 with the M27 AUG changed to AUC (FT293-M1-M27I). The numbers of cells were counted with a hemocytometer. (B) RNase H1-HA-NESi is expressed, activated, and localized to mitochondria in 293 cells. RNase H1 mRNA levels were assayed by RT-qPCR at different time points and dosages of doxycycline. The initial black bars are the parental FT293 cell line lacking RNase H1-HA-NESi. Subsequent bars from left to right in each set are cells with RNase H-HA-NESi expressed at 0 (red), 1 (blue), 3 (yellow), and 10 (green) ng/ml doxycycline. Protein contents of cell lysates were analyzed by Coomassie brilliant blue (CBB) stain, Western blotting for the HA epitope, and the RNase H1 gel activity assay with poly(rA)/poly(dT) as the substrate. Equal amounts of total protein (as assessed from the CBB staining) were loaded in each lane. For clarity, activity is shown as a negative image. Expression of RNase H1-HA-NESi was induced for 6 days using 0, 3, 30, and 60 ng/ml doxycycline. At the bottom and to the right is shown confocal microscopy of FT293-RNase H1-HA-NESi. After 24 h of induction using 100 ng/ml doxycycline, the entire signal detected with the anti-HA antibody (green) colocalized with mitochondria (MitoTracker orange). mtDNA copy number decreases as the dosage of doxycycline and days of incubation increase. For each day, the first bar (black) is FT293 without the cDNA for human RNase H1 treated with 10 ng/ml doxycycline. Subsequent bars from left to right in each set contain 0 (violet), 0.1 (red), 1 (blue), 3 (yellow), or 10 (green) ng/ml doxycycline. At day 6, the values for 1, 3, and 10 ng/ml doxycycline are 0.036, 0.032, and 0.027, respectively. At day 9, the values for 1, 3, and 10 ng/ml doxycycline are 0.018, 0.011, and 0.010, respectively.
DISCUSSION
It was a major surprise when the 14.8-billion-bp DNA sequence of the human genome indicated the presence of only slightly more genes than seen in D. melanogaster despite a 25-fold difference in DNA size (28). It is now clear that there is not a direct correlation between genome size and gene number due in part to the economic use of genes via alternative splicing and synthesis of two or more versions of the same protein that will reside in different subcellular compartments (17, 27).
Here, we show that a single mouse Rnaseh1 mRNA is translated to make proteins with or without an N-terminal mitochondrial targeting sequence (MTS) by initiating translation at two AUG codons that should produce essentially the same proteins in mitochondria and nuclei. Which in-frame codon is used is governed by the context of the AUG codon and the presence of a short uORF. Translation of mouse Rnaseh1 mRNA in rabbit reticulocyte lysates (Fig. 3A) and expression in tissue culture cells (Fig. 3B and 4A) mimicked the distribution and relative abundance of the two isoforms present in mouse liver (Fig. 2). This translation control results in a 7:1 ratio of nuclear to mitochondrial RNase H1 (Fig. 2).
The distribution of RNase H1 (higher abundance in nuclei than in mitochondria) is primarily achieved by two factors: translation initiation efficiency at the M1 and M27 AUGs and the presence of an uORF. Translation initiation efficiency at the M1 AUG start codon is very low because of its context around the AUG codon. We showed that altering the sequence immediately in front of the M1 AUG start codon to more closely resemble the consensus Kozak sequence significantly increased production of mitochondrial RNase H1 with a concomitant decrease in synthesis of the nuclear RNase H1 (Fig. 3B). The short uORF also contributes to the abundance of RNase H1 isoforms. Our evidence shows that translation to produce the nuclear enzyme requires either leaky scanning of the ribosome, bypassing the uORF and mitochondrial RNase H1 start codons, or reinitiation after synthesis of the uORF peptide. The absence of the uORF increases the amount of the mitochondrial but not the nuclear isoform. Presumably, the absence of the uORF increases the frequency of initiation at the M1 AUG, which keeps the number of initiations at the M27 AUG the same as if the uORF were present (Fig. 5). Using data from expression of RNase H1-luciferase fusion proteins, we have constructed a model relating the frequency of initiation at each of the three sites (Fig. 6).
The arrangement of uORF-mitochondrial AUG-nuclear AUG is conserved in all 23 mammalian genomes we examined, suggesting the use of translational regulation for synthesis of RNase H1 in mammals and perhaps other species. The peptide coding region of the uORF is not conserved in length and sometimes overlaps the first AUG codon, but it always terminates very close to the initiating AUG codon of the mitochondrion-targeted protein and prior to the nucleus-targeted protein initiating AUG codon (Table 1). The effects of uORFs can be complicated. The best-studied examples of regulation by uORFs are those in which skipping the uORF permits increased synthesis of proteins. Regulation occurs in response to stress that is mediated by phosphorylation of eukaryotic initiation factor eIF2α (10, 16, 32). Most uORFs are thought to simply alter levels of expression of downstream translation by acting as initiation sites. However, there are more complex situations such as the S. cerevisiae GCN4 mRNA, which has four uORFs. Translation of the first uORF allows efficient reinitiation at downstream AUG codons and sensitizes reinitiation to the level of eIF2α phosphorylation. The idea that eIF2α phosphorylation affects RNase H1 regulation is appealing. However, our attempts to see changes in expression of RNase H1 when eIF2α is phosphorylated have all produced negative results (data not shown). Studies on mutations in the uORF of the hairless (HR) transcription factor gene suggest that the 34-amino-acid peptide of the uORF plays a direct cis-acting role to maintain the optimal level of the HR protein, not simply acting as a site to decrease synthesis of a downstream ORF (35). Because the length and composition of the uORFs of RNases H1 in numerous mammalian genomes are not conserved, it is unlikely that the uORF peptide is directly involved in regulation, and most probably the uORF acts in a more typical manner.
Why is regulation at the translation level required to balance the distribution of RNase H1 to mitochondria and nuclei? One probable answer is because cells require mitochondrial RNase H1 for mitochondrial DNA replication but at the same time have to limit production of mitochondrial RNase H1 to avoid toxicity. Variation in mtDNA copy number is known to change during development, in different tissues, and during the general proliferative state of cells (15, 22, 30). Even modest deviations from normal amounts of mitochondrial DNA trans-acting enzymes can affect mtDNA copy number. For example, Pohjoismaki et al. have reported that increases or decreases in the levels of the mitochondrial transcription factor A, TFAM, result in mtDNA depletion (25). The effect on mtDNA of overexpressing RNase H1 was even more dramatic than that of TFAM. Transgene induction with 1 ng/ml doxycycline produced a 95% decrease in mtDNA copy number within 6 days (Fig. 7B), whereas a 10-fold-higher dose for 10 days produced less-marked mtDNA depletion in the case of TFAM (25). Mitochondrial DNA replication appears to involve incorporation of RNA on the lagging strand (26, 37), which would be expected to be highly vulnerable to an excess of RNase H1. Alternatively, the RNA primers for leading- and lagging-strand DNA synthesis may be destroyed by RNase H1, and so the replication process may never be able to begin when the amount of RNase H1 in mitochondria exceeds strict limits. In any case, mtDNA depletion and cell death result from imbalances in RNase H1 (this report and see reference 7), supporting the idea that maintaining the fine balance of RNase H1 is critical for mtDNA copy number stability. Increasing expression of the mitochondrial isoform of RNase H1 may not drastically alter the amount of nuclear RNase H1, as was the case when we eliminated translation initiation at the uORF codon (Fig. 5B). This leads us to suggest that regulation of the amount of mitochondrial RNase H1 is the major function of the uORF-M1-M27 transcript organization.
Upstream ORFs could well play a role in determining the level of expression of a number of other genes whose products are targeted to mitochondria and the nucleus.
The Top3α and Trnt1 genes have conserved uORFs and two in-frame AUGs in at least 11 mammalian genomes (see Tables S2 and S3 in the supplemental material). Top3α encodes DNA topoisomerase IIIα, which has been shown to be present in mitochondria and nuclei (34). Trnt1 encodes tRNA-nucleotidyltransferase 1, which is an enzyme that adds the CCA triplet to the 3′ end of tRNAs. In mammals, this protein has been found in mitochondria, but so far there is no report of its presence in the cytoplasm (24). However, in S. cerevisiae, the CCA1 gene has been shown to produce both mitochondrial and cytoplasmic enzymes from different in-frame AUGs (8). We do not know the number of genes with mRNAs having the uORF-AUG-AUG organization, but it could be that such an arrangement could be used for coordinated expression of critical DNA-transacting enzymes.
The AUG codon used for initiation of translation appears to be the critical step to direct RNase H1 either to mitochondria or to nuclei, even though both proteins must possess a nuclear localization sequence. Initiation of translation at the M1 AUG produces RNase H1 localized specifically in mitochondria, whereas proteins without the MTS are present in nuclei and absent in mitochondria (Fig. 4E). A single mRNA thus has the potential to simultaneously produce both mitochondrial and nuclear isoforms. The simplest solution for delivering each form of RNase H1 to their sites is to travel by independent mechanisms after completion of synthesis: for example, via interaction of the MTS with heat shock protein 70 (HSP70) followed by transport of the protein from the site of synthesis to the surface of mitochondria. RNase H1 destined for mitochondria represents only a small fraction of that present in the nucleus. Given the overall preference to produce the nuclear isoform of RNase H1, it might be beneficial for translation of Rnaseh1 mRNA to occur near mitochondria leading to cotranslational import, another mechanism for directing nuclearly encoded mitochondrial proteins (1, 29). Cotranslational import makes little sense if the mRNA is translated to make the nuclear isoform. Because of the great distances between mitochondria and nuclei in certain neurons, and possibly other cell types, there might be a benefit to preferentially synthesize the protein with an MTS once the mRNA was to reach the mitochondria. Perhaps enhanced translation of mRNAs for RNase H1, Top3α, Trn1, and any other dually targeted mRNAs with uORF mitochondrial enzyme could be influenced by the presence of the uORF, either by aiding delivery of the mRNA to mitochondria or due to some effects of translation near mitochondria. We are currently testing this possibility.
Supplementary Material
Acknowledgments
We thank Tom Dever (NICHD) for helpful comments and suggestions on the manuscript. Nicole Follmer initiated some of these experiments. Xiongfong (Jack) Chen provided help in database searches. We thank P. Sarnow and M. S. Carter, Stanford University, for luciferase plasmids.
This work was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health. Yutaka Suzuki was a Japan Society for the Promotion of Science Research Fellow in Biomedical and Behavioral Research at the National Institutes of Health. J. Bradley Holmes is a National Institutes of Health-Cambridge University Biomedical Research Scholar. Michal Minczuk and Ian Holt are supported by the U.K. Medical Research Council.
Footnotes
Published ahead of print on 7 September 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmed, A. U., and P. R. Fisher. 2009. Import of nuclear-encoded mitochondrial proteins: a cotranslational perspective. Int. Rev. Cell Mol. Biol. 273:49-68. [DOI] [PubMed] [Google Scholar]
- 2.Arudchandran, A., S. Cerritelli, S. Narimatsu, M. Itaya, D. Y. Shin, Y. Shimada, and R. J. Crouch. 2000. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells 5:789-802. [DOI] [PubMed] [Google Scholar]
- 3.Arudchandran, A., S. M. Cerritelli, N. J. Bowen, X. Chen, M. W. Krause, and R. J. Crouch. 2002. Multiple ribonuclease H-encoding genes in the Caenorhabditis elegans genome contrasts with the two typical ribonuclease H-encoding genes in the human genome. Mol. Biol. Evol. 19:1910-1919. [DOI] [PubMed] [Google Scholar]
- 4.Calvo, S. E., D. J. Pagliarini, and V. K. Mootha. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U. S. A. 106:7507-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerritelli, S. M., and R. J. Crouch. 1998. Cloning, expression, and mapping of ribonucleases H of human and mouse related to bacterial RNase HI. Genomics 53:300-307. [DOI] [PubMed] [Google Scholar]
- 6.Cerritelli, S. M., and R. J. Crouch. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerritelli, S. M., E. G. Frolova, C. Feng, A. Grinberg, P. E. Love, and R. J. Crouch. 2003. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 11:807-815. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. Y., P. B. Joyce, C. L. Wolfe, M. C. Steffen, and N. C. Martin. 1992. Cytoplasmic and mitochondrial tRNA nucleotidyltransferase activities are derived from the same gene in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 267:14879-14883. [PubMed] [Google Scholar]
- 9.Crouch, R. J., A. Arudchandran, and S. M. Cerritelli. 2001. RNase H1 of Saccharomyces cerevisiae: methods and nomenclature. Methods Enzymol. 341:395-413. [DOI] [PubMed] [Google Scholar]
- 10.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Vizarra, E., M. J. Lopez-Perez, and J. A. Enriquez. 2002. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods 26:292-297. [DOI] [PubMed] [Google Scholar]
- 12.Filippov, V., M. Filippov, and S. S. Gill. 2001. Drosophila RNase H1 is essential for development but not for proliferation. Mol. Genet. Genomics 265:771-777. [DOI] [PubMed] [Google Scholar]
- 13.Gaidamakov, S. A., I. I. Gorshkova, P. Schuck, P. J. Steinbach, H. Yamada, R. J. Crouch, and S. M. Cerritelli. 2005. Eukaryotic RNases H1 act processively by interactions through the duplex RNA-binding domain. Nucleic Acids Res. 33:2166-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, L. Y., W. P. Ma, and R. J. Crouch. 1997. Ribonuclease H renaturation gel assay using a fluorescent-labeled substrate. Biotechniques 23:920-926. [DOI] [PubMed] [Google Scholar]
- 15.Heerdt, B. G., and L. H. Augenlicht. 1990. Changes in the number of mitochondrial genomes during human development. Exp. Cell Res. 186:54-59. [DOI] [PubMed] [Google Scholar]
- 16.Holcik, M., and N. Sonenberg. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6:318-327. [DOI] [PubMed] [Google Scholar]
- 17.Karniely, S., and O. Pines. 2005. Single translation—dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 6:420-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kochiwa, H., M. Itaya, M. Tomita, and A. Kanai. 2006. Stage-specific expression of Caenorhabditis elegans ribonuclease H1 enzymes with different substrate specificities and bivalent cation requirements. FEBS J. 273:420-429. [DOI] [PubMed] [Google Scholar]
- 19.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 21.Minczuk, M., P. Kolasinska-Zwierz, M. P. Murphy, and M. A. Papworth. 2010. Construction and testing of engineered zinc-finger proteins for sequence-specific modification of mtDNA. Nat. Protoc. 5:342-356. [DOI] [PubMed] [Google Scholar]
- 22.Moraes, C. T. 2001. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 17:199-205. [DOI] [PubMed] [Google Scholar]
- 23.Morris, D. R., and A. P. Geballe. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20:8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaike, T., T. Suzuki, Y. Tomari, C. Takemoto-Hori, F. Negayama, K. Watanabe, and T. Ueda. 2001. Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J. Biol. Chem. 276:40041-40049. [DOI] [PubMed] [Google Scholar]
- 25.Pohjoismaki, J. L., S. Wanrooij, A. K. Hyvarinen, S. Goffart, I. J. Holt, J. N. Spelbrink, and H. T. Jacobs. 2006. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 34:5815-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pohjoismäki, J. L. O., J. B. Holmes, S. R. Wood, M. Y. Yang, T. Yasukawa, A. Reyes, L. J. Bailey, T. J. Cluett, S. Goffart, S. Willcox, R. E. Rigby, A. P. Jackson, J. N. Spelbrink, J. D. Griffith, R. J. Crouch, H. T. Jacobs, and I. J. Holt. 2010. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 397:1144-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regev-Rudzki, N., and O. Pines. 2007. Eclipsed distribution: a phenomenon of dual targeting of protein and its significance. Bioessays 29:772-782. [DOI] [PubMed] [Google Scholar]
- 28.Rubin, G. M. 2001. The draft sequences: comparing species. Nature 409:820-821. [DOI] [PubMed] [Google Scholar]
- 29.Russo, A., G. Russo, M. Cuccurese, C. Garbi, and C. Pietropaolo. 2006. The 3′-untranslated region directs ribosomal protein-encoding mRNAs to specific cytoplasmic regions. Biochim. Biophys. Acta 1763:833-843. [DOI] [PubMed] [Google Scholar]
- 30.Scarpulla, R. C. 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88:611-638. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, T. G., and A. Skerra. 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2:1528-1535. [DOI] [PubMed] [Google Scholar]
- 32.Sonenberg, N., and A. G. Hinnebusch. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyynismaa, H., H. Sembongi, M. Bokori-Brown, C. Granycome, N. Ashley, J. Poulton, A. Jalanko, J. N. Spelbrink, I. J. Holt, and A. Suomalainen. 2004. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 13:3219-3227. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Y., Y. L. Lyu, and J. C. Wang. 2002. Dual localization of human DNA topoisomerase IIIalpha to mitochondria and nucleus. Proc. Natl. Acad. Sci. U. S. A. 99:12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen, Y., Y. Liu, Y. Xu, Y. Zhao, R. Hua, K. Wang, M. Sun, Y. Li, S. Yang, X. J. Zhang, R. Kruse, S. Cichon, R. C. Betz, M. M. Nothen, M. A. van Steensel, M. van Geel, P. M. Steijlen, D. Hohl, M. Huber, G. S. Dunnill, C. Kennedy, A. Messenger, C. S. Munro, A. Terrinoni, A. Hovnanian, C. Bodemer, Y. de Prost, A. S. Paller, A. D. Irvine, R. Sinclair, J. Green, D. Shang, Q. Liu, Y. Luo, L. Jiang, H. D. Chen, W. H. Lo, W. H. McLean, C. D. He, and X. Zhang. 2009. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie Unna hereditary hypotrichosis. Nat. Genet. 41:228-233. [DOI] [PubMed] [Google Scholar]
- 36.Wu, H., W. F. Lima, and S. T. Crooke. 1998. Molecular cloning and expression of cDNA for human RNase H. Antisense Nucleic Acid Drug Dev. 8:53-61. [DOI] [PubMed] [Google Scholar]
- 37.Yasukawa, T., A. Reyes, T. J. Cluett, M. Y. Yang, M. Bowmaker, H. T. Jacobs, and I. J. Holt. 2006. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 25:5358-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.