Abstract
Targeted disruption of the Fli1 gene results in embryonic lethality. To dissect the roles of functional domains in Fli1, we recently generated mutant Fli1 mice that express a truncated Fli1 protein (Fli1ΔCTA) that lacks the carboxy-terminal regulatory (CTA) domain. Heterozygous Fli1ΔCTA mice are viable, while homozygous mice have reduced viability. Early postnatal lethality accounts for 30% survival of homozygotes to adulthood. The peripheral blood of these viable Fli1ΔCTA/Fli1ΔCTA homozygous mice has reduced platelet numbers. Platelet aggregation and activation were also impaired and bleeding times significantly prolonged in these mutant mice. Analysis of mRNA from total bone marrow and purified megakaryocytes from Fli1ΔCTA/Fli1ΔCTA mice revealed downregulation of genes associated with megakaroyctic development, including c-mpl, gpIIb, gpIV, gpIX, PF4, NF-E2, MafG, and Rab27B. While Fli1 and GATA-1 synergistically regulate the expression of multiple megakaryocytic genes, the level of GATA-1 present on a subset of these promoters is reduced in vivo in the Fli1ΔCTA/Fli1ΔCTA mice, providing a possible mechanism for the impared transcription observed. Collectively, these data showed for the first time a hemostatic defect associated with the loss of a specific functional domain of the transcription factor Fli1 and suggest previously unknown in vivo roles in megakaryocytic cell differentiation.
The Friend leukemia integration 1 (Fli1) gene is a member of the Ets gene family of transcription factors, which bind to the DNA consensus sequence GGA(A/T) (20, 54, 66). Fli1 is preferentially expressed in cells of hematopoietic lineages and vascular endothelial cells and has been shown to transactivate megakaryocytic (MK) genes, including those encoding GATA-1 (53, 66), glycoprotein IIb (gpIIb) (29, 64, 69), gpVI (19), gpIX, gpIb (14), and c-mpl (13, 23). It has been demonstrated that Fli1 expression promotes megakaryocytic differentiation in K562 cells (3). Viral integration and insertional activation of Fli-1 are associated with hematological cancers, including erythroleukemia induced by Friend murine leukemia virus (Friend-MuLV) (4), granulocytic leukemia induced by the Graffi virus (12), primitive stem cell tumors induced by the 10A1 isolate of MuLV (42), and non-T, non-B lymphomas induced by the Cas-Br virus (6). Overexpression of Fli1 results in an increased number of mature B cells, which have a reduced activation-induced apoptotic response compared to B cells from wild-type animals (68). Taken together, these results suggested that Fli1 plays significant roles in hematopoiesis. To clarify the physiological role of Fli1 in hematopoiesis, we (58) and others (18) generated mice with a targeted disruption of Fli1. Fli1 homozygous mutant (Fli1−/−) embryos showed hemorrhage from the dorsal aorta into the lumen of the neural tube and the ventricles of the brain beginning on embryonic day 11.0 (E11.0) and were dead on or before day E12.0. In addition, severe dysmegakaryopoiesis (18, 25) and vascular defects (18) were found. We also noted that livers of the E11.0 Fli1−/− embryos were pale and contained primarily polychromatophilic and orthochromatic normoblasts (58). Subsequent analyses of Fli1 chimeric mice revealed cell-autonomous defects consistent with important roles of the Fli1 gene in myelomonocytic, erythroid, and NK cell development (36).
The Ets (winged helix-turn-helix [wHTH] DNA binding) domain of human FLI1 is located between amino acids 277 and 361. Deletion analysis has identified two possible transcriptional activation domains, designated ATA and CTA (49), for amino-terminal transcriptional and carboxy-terminal transcriptional activation domains, respectively. We recently generated a new Fli1 allele that expresses a truncated Fli1 protein that lacks the carboxy-terminal amino acids, including the CTA domain. In this study, we describe in vivo characterization of the biological significance of this activation domain, providing novel evidence for the importance of Fli1 in megakaryocytic differentiation and platelet function.
MATERIALS AND METHODS
Generation and identification of mouse lines carrying the recombined Fli1 allele.
The original vector we utilized to target Fli1 contained a selectable neomycin gene under the transcriptional control of the RNA polymerase II (Pol II) promoter flanked by the loxP Cre recombinase recognition sequences. This “floxed” neo cassette was inserted into the unique EcoRV site present in exon IX between the ETS DNA binding domain and the carboxy-terminal transactivation domain (CTA). The resultant targeted allele has a termination signal provided by the loxP sequence located between the DNA binding domain and the CTA domain.
Fli1 heterozygous (Fli1−/Fli1+) mice were crossed with cytomegalovirus (CMV) Cre transgenic mice that express Cre recombinase in all tissues, including germ cells [BALB/c-TgN(CMV-Cre)#Cgn; Jackson Laboratory, Bar Harbor, ME]. Mice harboring the Cre transgene and recombined Fli1 allele were identified by PCR (described below), and those capable of germ line transmission of the Fli1ΔCTA allele were identified by crossing them with wild-type C57/BL6 mice. To minimize possible effects of the genetic background, Fli1ΔCTA/Fli1+ mice were backcrossed at least 10 generations prior to performing the studies described here.
Genotyping.
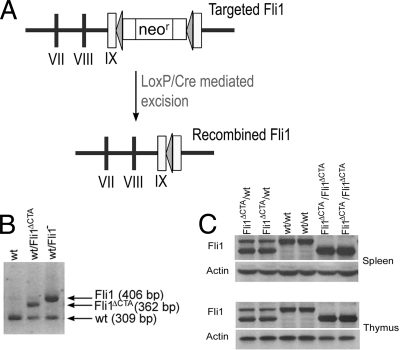
For genotyping of mice and embryos, we used PCR to detect fragments of the wild-type Fli1 and targeted Fli1 alleles. The PCR was carried out as follows: 94°C for 2 min, followed by 35 cycles (94°C for 1 min, 68°C for 1 min, and 72°C for 1 min). A 309-bp fragment indicates the presence of the wild-type allele. After Cre-mediated excision of the floxed neomycin cassette, the recombined Fli1ΔCTA allele (Fli1ΔCTA) retains a single loxP element as well as sequences derived from cloning, resulting in a 362-bp (309 + 53 bp) fragment. A 406-bp fragment is amplified from the original targeted allele (Fli1−) (58).
Primers used were as follows: Fli1 Exon IX/Forward primer (1156 to 1180), GACCAACGGGGAGTTCAAAATGACG; Fli1 Exon IX/Reverse primer (1441 to 1465), GGAGGATGGGTGAGACGGGACAAAG; Pol II/Reverse primer, GGAAGTAGCCGTTATTAGTGGAGAGG.
Western blotting.
Homogenates from wild-type, heterozygous, and homozygous mutant Fli1 embryos were prepared by lyses in radioimmunoprecipitation assay (RIPA) buffer in the presence of protease inhibitors (Sigma-Aldrich). Sonicated lysates were clarified by centrifugation, and the protein concentration of supernatants was determined using the Bio-Rad assay (Bio-Rad Laboratories, Hercules, CA). Thirty micrograms of each protein extract was separated on an SDS-12.5% polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were incubated for 2 h with Fli1 polyclonal antibody (made against full-length human FLI1) and then with horseradish hydroperoxidase-labeled secondary antibody (Amersham, Piscataway, NJ). Antibody was detected using ECL (Amersham). The presence of equivalent protein loading was verified by reprobing stripped membranes with β-actin (Sigma) antibody.
Antibodies.
The following monoclonal antibodies were purchased from BD Pharmingen (San Diego, CA): purified anti-CD42d (1C2; hamster IgG3), phycoerythrin (PE)-conjugated anti-CD41 (MWReg 30; rat IgG1k), fluorescein isothiocyanate (FITC)-conjugated anti-CD41 (MWReg 30; rat IgG1k), FITC-conjugated anti-CD62P (P-selectin; RB40.34; rat IgG1λ), and allophycocyanin (APC)-conjugated anti-CD34 (RAM34; rat IgG2a). Isotype controls were also purchased from BD Pharmingen. FITC-conjugated anti-CD61 (2C9.G3; Armenian hamster IgG) antibodies were purchased from eBiosciences (San Diego, CA). Secondary FITC-conjugated antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Cell preparation.
(i) PB and BM isolation. Peripheral blood (PB) was obtained from anesthetized mice either by retro-orbital plexus puncture using heparin-coated micropipettes (Drummond Scientific Co, Broomall, PA) or by terminal cardiac puncture. Red blood cells were lysed with 1× PharM Lyse lysing buffer (BD Pharmingen) and washed twice with Ca2-, Mg2-free phosphate-buffered saline (PBS) (Life Technologies, Grand Island, NY) containing 0.1% bovine serum albumin (BSA) (Sigma-Aldrich, St. Louis, MO). Bone marrow (BM) cells were flushed from femurs and tibiae, pooled, and washed twice with PBS containing 0.1% BSA. BM samples were made into single-cell suspensions by triturating and filtering through a 40-μm nylon mesh.
(ii) MK isolation. For isolation of CD34+ CD41+ CD42− megakaryocytic cells, single-cell suspensions of BM cells were prepared and stained as described above, and CD34+ CD41+ CD42− cells were sorted using a MoFlo cell sorter by gating on the CD41+ population of cells and sorting the CD34+ and CD42− population. The purity of the cells was confirmed by flow cytometry and found to be >93% in each sort.
Cytochemical studies.
BM cells were flushed from femurs and tibiae of mice, washed in PBS containing 0.1% BSA, and processed to single-cell suspension. BM cells were then centrifuged in a Cytospin 2 centrifuge (Shandon, Pittsburgh, PA), dried, and processed for acetylcholinesterase staining (24). The procedure for quantification of megakaryocytes was adapted from the manufacturer's instructions (MegaCult-C: assay for the quantitation of human and murine megakaryocytic progenitors, a technical manual; StemCell Technologies, Vancouver, Canada). Based on this protocol, cytospun samples were incubated in staining solution for 6 h. This allowed immature megakaryocytes containing low levels of acetylcholinesterase and mature megakaryocytes with high acetylcholinesterase content to be differentiated based on their staining intensity. The total number of cells staining red-brown (immature MK) and those staining dark brown-black (mature MK) were counted for each slide. These values were also expressed as percentages of total cells examined.
Flow cytometry.
For platelet (PLT) analysis, PB cells were stained with FITC-conjugated CD41. For megakaryocyte analysis, BM cells were stained with PE-conjugated anti-CD41, purified anti-CD42d (followed by FITC-conjugated secondary antibody), and APC-conjugated anti-CD34. Platelet surface expression of CD41 and CD42d or CD41 and CD61 was measured using PE-conjugated anti-CD41, purified anti-CD42d (followed by FITC-conjugated secondary), and FITC-conjugated anti-CD61 antibodies. Platelet activation was examined using PE-conjugated anti-CD41 and FITC-conjugated anti-CD62P.
All cells were stained with propidium iodide at a concentration of 1 μg/ml, and the cells were washed twice and resuspended in PBS containing 0.1% BSA for analysis using a FACSCalibur (Becton Dickinson Bioscience, San Jose, CA) or MoFlo (Dako Cytomation, Ft. Collins, CO) cell sorter.
Analysis and isolation of megakaryocyte subpopulations in bone marrow.
Single-cell suspensions of BM cells were prepared as described above and then immunolabeled with the following antibodies: PE-conjugated anti-mouse CD41, APC-conjugated anti-mouse CD34, and purified anti-mouse CD42d, followed by FITC-conjugated anti-Armenian hamster IgG(H+L). Cells from wild-type and Fli1ΔCTA/Fli1ΔCTA mice were then analyzed by flow cytometry (FACSCalibur) to determine the percentage of bone marrow cells that were CD41 positive. CD41+ cells were then analyzed for their expression pattern of CD34 and CD42.
Bleeding time assays.
Mouse tail bleeding times were determined as previously described (30). Briefly, a 2-mm portion of distal tail was removed from anesthetized 4-week-old mice at weaning, and the tail was immersed in isotonic saline (37°C). A complete cessation of blood flow was defined as the bleeding time. When bleeding time exceeded 4 min or bleeding was so extensive as to risk terminal bleeding, measurements were stopped.
Preparation of platelet-poor plasma and platelet-rich plasma.
Whole blood was obtained by puncturing the right ventricle of anesthetized mice. Blood was anticoagulated by collecting into a 1/10 volume of a solution containing 130 mmol/liter of citric acid, 125 mmol/liter of trisodium citrate, and 110 mmol/liter of glucose. Manual (microscopic) cell counts of platelets were performed using a Nikon microscope (magnification, ×100). Platelet-rich plasma (PRP) was isolated by centrifuging the whole blood at a low speed (100 × g, 8 min, 22°C). Platelet-poor plasma (PPP) was prepared by centrifuging the whole blood at 2,000 × g for 30 min (22°C). Both platelet-poor plasma and platelet-rich plasma were used within 1 h after preparation.
Platelet aggregation and activation.
PRP from 3 to 5 mice was pooled to measure platelet aggregation. To determine platelet aggregation, light transmission was measured using PRP diluted to 5 × 105 cells/ml with PPP. Transmission was recorded in a 4-channel aggregometer (whole-blood lumi-aggregometer [Ca2+]; Chrono-log Corporation, Havertown, PA) over 5 min and was expressed as arbitrary units with 100% transmission adjusted with PPP. To measure changes in transmission, PRP samples (450 ml) were incubated with stirring for 2 min at 37°C prior to induction of platelet aggregation by the addition of ADP (1 μM) or thrombin (0.5 U) (Chrono-log Corporation). The baseline was set with PRP, and the maximal possible light transmission was set with PPP.
Flow cytometry to evaluate activated platelets.
Platelet-rich plasma samples were incubated in the presence of 0.1 U thrombin for 10 min at 37°C. Samples were then fixed in 2% formalin for 30 min at 37°C, washed in PBS, and stained for 20 min with phycoerythrin-conjugated anti-CD41 (to label total platelets) and with FITC-conjugated anti-CD62P (P-selectin) (to label activated platelets). Analysis by flow cytometry was then conducted on the MoFlo cell sorter.
Real-time PCR.
Total RNA was extracted from BM nuclear cells, and megakaryocytes were prepared from wild-type Fli1 and Fli1ΔCTA/Fli1ΔCTA homozygous mice using the Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. One μg of total RNA was reversed transcribed using Superscript II single-strand synthesis reverse transcriptase [RT] (Invitrogen). Aliquots from this reaction were subjected to real-time PCR using the indicated gene-specific primers (Table 1). The Fli1 primers are directed toward a region present in both wild-type and mutant alleles. Hypoxanthine phosphoribosyl transferase (HPRT) served as the control. The reaction mixture contained 1.5 mM Mg2+, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1× Taq Gold buffer, 0.88 pmol/ml primers, and 0.02 U/ml Taq Gold.
TABLE 1.
PCR primers used for real time RT-PCR
| Gene name | Forward primer | Reverse primer | Amplicon length (bp) | Accession no | Ta (°C)a |
|---|---|---|---|---|---|
| gpIIb | CGGTGCTGACAATGTGTTGGA | GGATTCTGGCTGTTCTTGCTCC | 318 | NM_010575 | 57 |
| gpV | GCGTTCATCGTGTTTGCCAT | TAACCCAGGCCAGGCTAGAAAG | 280 | NM_008148.3 | 60 |
| gpIX | TGTACTGTGCCAGCCCTGACTT | AAGTGTCTTTGCCCATGAGGC | 320 | NM_018762 | 58 |
| CD41 | TGCCGTGGTATTGCATGGA | CAGACAAGCCTCTCAAAGCCCT | 267 | NM_010575 | 58 |
| CD61 | CCTCCAGCTCATTGTTGATGCT | ACGGTGAGGCTGTCCTTAAAGC | 261 | NM_016780 | 60 |
| P-selectin | GGATTGATGAACCGTTGCACA | CCAAACCCTGAGAGTGGCATCT | 252 | NM_011347 | 60 |
| PECAM-1 | CTTCGGTGCTCTGTGAAAGAGG | CCAATGACAACCACCGCAAT | 269 | NM_001032378 | 59 |
| NFE2 | AACAAGGTGGCAGCCCAAA | GCCAGATTCATCCCGAAGATG | 305 | NM_008685 | 58 |
| Rab27B | AGCAGATGGAGCGTCAGGAAA | TGTCCTGTAGCTGCGCTTGTTT | 333 | NM_030554 | 59 |
| MAFG | CACGCCTGTGTTTCGTACGTT | GGAAAGCGCTCAAATCCCA | 202 | NM_010756 | 61 |
| MAFK | CTAATCCCAAGCCCAACAAGG | TCTGTGTCACACGCTTGATGC | 219 | NM_010757 | 61 |
| RUNX1 | TCATGGCAGGCAACGATGA | CGTCCACTGTGATTTTGATGGC | 201 | NM_009821 | 60 |
| Tubulin β1 | ATGAGCGGAATCACGACTTCAC | TCTGAATGGACAGCAGCTGTTG | 307 | XM_988085 | 60 |
| PU.1 | AGGCGTGCAAAATGGAAGG | TCTCACCCTCCTCCTCATCTGA | 411 | M38252 | 60 |
| GATA-1 | TAAGGTGGCTGAATCCTCTGCATC | CCGTCTTCAAGGTGTCCAAGAACGT | 445 | NM_008089 | 58 |
| c-mpl | ATGCCTACCGAGGAGAGAAGCC | GCCATAGCGGAGTTCATGCC | 317 | NM_010823 | 60 |
| ITGB1 | CGCATTGGCTTTGGCTCAT | AAGTGAAACCCAGCATCCGTG | 311 | NM_010578 | 58 |
| PF4 | CCGAAGAAAGCGATGGAGATCT | CCAGGCAAATTTTCCTCCCA | 162 | NM_019932 | 60 |
| Fli1 | TGCTGTTGTCGCACCTCAGTTA | TGTTCTTGCCCATGGTCTGTG | 204 | NM_008026 | 60 |
| HPRT | GCTGGTGAAAAGGACCTCTC | ATGGCCACAGGACTAGAACAC | 254 | J00423 | 62 |
Ta, annealing temperature.
Real-time PCR was performed with the Platinum SYBR green qPCR SuperMix UDG (Invitrogen) using a LightCycler 2.0 PCR system (Roche, Nutley, NJ). The cycling conditions for all genes were as follows: preincubation at 50°C for 2 min and 95°C for 2 min, followed by 30 to 50 cycles of denaturation at 94°C for 5 s, annealing at 1 degree below the lowest melting temperature (Tm) for a given primer pair (Table 1) for 5 s, and extension for 45 s at 72°C, with a single acquisition of data at the end of each extension. All ramping was done at 20°C per second. Relative expression analysis was conducted using the software program LinRegPCR (48).
Cell culture, transfections, and transient luciferase assays.
HeLa cells were maintained at 37°C under 5% CO2 in Dulbecco's modified Eagle minimum essential medium supplemented with 10% fetal bovine serum (GibcoBRL, Carlsbad, CA). Transient transfections were carried out using Fugene VI reagent (Roche) as per the manufacturer's directions. Green fluorescent protein (GFP) reporters used under these conditions showed 70% to 80% transfection efficiency in these cells. For reporter assays, cells were seeded the day before transfection in 6-well plates. Equal amounts of DNA were maintained in each transfection by addition of the pSG5 and pcDNA3.1 vectors. Cell lysates were prepared and reporter assays carried out as per the manufacturer's protocols. Cells were harvested 48 h after transfection, and luciferase activity was measured using a luciferase assay kit (Promega) using equivalent volumes of protein extract. All transient reporter assays were performed in triplicate, and results were normalized to the total protein concentration because dual-reporter vectors are modulated by transcription factors and are Ets responsive (5, 40, 67).
Plasmid constructs.
A 1,026-bp fragment of the human c-mpl promoter (−922/+104) was amplified from normal human placenta genomic DNA by PCR (13, 38). The 5′ primer (5′-CGGAATTCGGCAGGGTAAGGAGTGTGAG-3′) and the 3′ primer (5′-CGGAATTCTCACCTTGGCTGCTGACTTG-3′) contain the recognition sequence for EcoRI. The resultant PCR product was isolated, digested with EcoRI, and cloned into EcoRI sites of the pBluescript II KS vector (Stratagene) to generate the pBluescript c-mpl vector, and the sequence was validated by automated sequencing (MUSC Sequencing Facility). The human c-mpl reporter plasmid (−218/+46) was constructed using the pBluescript c-mpl plasmid described above by digestion with StuI and BstEII. After filling in with Klenow polymerase, the resultant fragment was cloned at the Asp718 site of the pGL2 basic vector (Promega, Madison, WI) to generate pGL2 c-mpl −218/+46. The insert ends were sequenced to identify clones with proper orientation.
Full-length mouse Fli1 (mFli1) cDNA in the pGEM7 vector was generated as previously described. mFli1ΔCTA was generated by ligation of a duplex oligonucleotide (5′ CTC GAG CTC GCG AAA GCT TAT AAC TTC GTA TAG) with mFli1 previously linearized with EcoRV, providing the sequence and termination codon from the loxP cassette in the original targeting vector. mFli1 and mFli1ΔCTA inserts were released by digestion with EcoRI and recloned into pSG5 linearized with EcoRI.
ChIP.
MK cells (4 × 106) were isolated from wild-type and Fli1ΔCTA/Fli1ΔCTA mice. Chromatin immunoprecipitation (ChIP) assays were adapted from the method of Boyd et al. (8) with modifications as described previously (23) using Fli1 rabbit polyclonal antibody, described above (Western), or a commercially available GATA-1 antibody (Santa Cruz, Santa Cruz, CA). DNA isolated using the ChIP assay was linearly amplified using the REPLI-g Mini whole-genome amplification system (Qiagen), following the manufacturer's instructions. For the total input control, 20 ng of input DNA was used for the whole-genome amplification in a 50-μl total reaction mixture. Following the whole-genome amplification process, total input control amplified DNA samples were diluted (1:10) in Tris-EDTA (TE) buffer. Immunoprecipitated amplified DNA samples were diluted 1:2 in TE. Two microliters of the diluted DNA samples were used for PCR. The specific primers for the examined promoters are indicated in Table 2.
TABLE 2.
Primers for real-time PCR ChIP analyses
| Primer name | Sequence (5′-3′) |
|---|---|
| GPIIb F | GCCATGAGCTCCAGTCTGATAA |
| GPIIb R | AGCTCTTTCCCTTTCCCTGAA |
| c-mpl F | CTGCCAACAGAAGGCTCATG |
| c-mpl R | CTGTCAGATACAGCCCCACGT |
| GPIX F | GCCTCCTGGCCCTGACA |
| GPIX R | TGTGGCTGCTGCCTGACA |
| GPIb F | TGGTGGCTAGTAGCTGCAAAGTC |
| GPIb R | TTATCAGCTCTCTGCACAGCATTC |
| PF4 F | GCTGCTGGCCTGCACTTAAG |
| PF4 R | GCCACTGGACCCAAAGATAAAG |
| PECAM1-P4F | TATTGTCGGAGAAGCTGGCC |
| PECAM1-P4R | GGACAAGGAGGTTTTATTTCCACTC |
| mCD61-P216F | TTCCACACCCTTCCTCCAGA |
| mCD61-P366R | CTCCATCCCCAGCACTACGT |
| mgpV-P433F | CTAGGACAGCAGAAAGAACCCTTG |
| mgpV-P584R | TTTGTGGACGGAGTCACTGACT |
| Negative Cont Fa | AAATAGATGTCAAGTTGGCATAAACCT |
| Negative Cont R | TGCCAGCGTTCAAGTACAAAA |
Negative control: upstream gpIIb region which lacks a functional Ets site.
Computational analysis.
Putative transcription factor binding sites were predicted using the MatInspector professional software program (Genomatix) (46).
Statistical analysis.
Student's paired t test was used to analyze the statistical significance of the differences between values obtained for wild-type and Fli1ΔCTA/Fli1ΔCTA mice. A P value of ≤0.05 was considered statistically significant. For flow cytometric studies, the two-sample equal-variance t test was used to analyze the statistical significance of the differences between values obtained for wild-type and Fli1ΔCTA/Fli1ΔCTA mice except in cases where results of an F test indicated unequal variance. A P value of ≤0.05 was considered statistically significant.
RESULTS
Generation and identification of mice carrying the recombined Fli1 allele.
To define the in vivo function of the CTA domain and as a possible means to further assess the hematopoietic defects in the Fli1 null mice, we generated a new mutant Fli1 line by loxP/Cre-mediated removal of the neomycin gene present in the Fli1 locus of our original targeted mice. The resultant targeted allele also contains termination signals provided by sequences flanking the loxP element located between the DNA binding domain and the CTA domain. Thus, this new Fli1 allele is predicted to express a truncated Fli1 protein (amino acids 1 to 384) that lacks the CTA domain (amino acids 402 to 452) (Fig. 1 A).
FIG. 1.
Generation of mice with a mutant Fli1 allele. (A) Schematic diagram of the targeted Fli1 and Fli1ΔCTA alleles. (B) PCR analysis of DNA from heterozygous (Fli1ΔCTA/Fli1+ and Fli1−/Fli1+) and wild-type (+/+) mice. The wild-type, targeted Fli1−, and Fli1ΔCTA alleles are identified by the 309-, 406-, and 362-bp PCR products, respectively. (C) Western blot of spleen and thymus tissue extracts prepared from wild-type, heterozygous Fli1ΔCTA/Fli1+, and homozygous Fli1ΔCTA/Fli1ΔCTA mice. Thirty μg of protein extract was resolved on a 12.5% acrylamide gel and probed with rabbit anti-Fli1 polyclonal antibody. Blots were reprobed with anti-β-actin as a loading control.
We obtained heterozygous Fli1ΔCTA (Fli1+/Fli1ΔCTA) mice capable of germ line transmission of the truncated Fli1 gene via in vivo Cre recombination in germ cells (Fig. 1B). Total protein was prepared from spleen and thymus tissue from wild-type mice and heterozygous and homozygous mutant mice. Mice harboring this recombined Fli1 (Fli1ΔCTA) allele express the expected-size truncated Fli1 protein that lacks the CTA domain (Fig. 1C).
Reduction of platelets in Fli1ΔCTA/Fli1ΔCTA mice.
Heterozygous Fli1ΔCTA mice are viable, while homozygous mice have reduced viability. At 4 weeks of age, Fli1ΔCTA/Fli1ΔCTA mice accounted for less than 9% of the offspring (Fli1+/+, Fli1+/Fli1ΔCTA, and Fli1ΔCTA/Fli1ΔCTA, 31.2%, 59.8%, and 9.0%, respectively; n = 1,559), indicating ∼70% lethality. A retrograde genotypic analysis was conducted. Genotypes of embryos at E18 showed a normal Mendelian ratio (Fli1+/+, Fli1+/Fli1ΔCTA, and Fli1ΔCTA/Fli1ΔCTA, 24%, 55%, and 22%, respectively; n = 29), supporting the model that the homozygous mice have partial perinatal lethality. Surviving Fli1ΔCTA/Fli1ΔCTA mice are smaller, weighing 15% (females) to 20% (males) less than their wild-type littermates at 6 weeks (P < 0.01).
The peripheral blood (PB) of viable Fli1ΔCTA/Fli1ΔCTA homozygous mice demonstrates a statistically significant ∼3-fold reduction in circulating platelet (PLT) numbers (Table 3) (P = 0.006). The results obtained by quantifying CD41-positive cells by flow cytometry were confirmed by direct microscopic counting (98.5 × 104 ± 10.3 × 104/μl and 32.9 × 104 ± 3.9 × 104/μl) for wild-type and Fli1ΔCTA/Fli1ΔCTA homozygous mice, respectively; P < 0.01). These findings support the model that the absence of the CTA domain affects platelet development. There was no significant difference in circulating platelet numbers between wild-type and heterozygous mice (Table 3). We next examined whether a difference in the platelet half-life contributes to the thrombocytopenia of mutant mice. Analysis showed that there was no statistically significant difference between wild-type and Fli1ΔCTA/Fli1ΔCTA mice throughout the 72 h of study (P ≥ 0.05 in all cases). The platelet half-life was shown to be ∼48 h for both genotypes (data not shown).
TABLE 3.
Flow cytometric analysis of PB from wild-type, Fli1ΔCTA/+ heterozygous, and Fli1ΔCTA/Fli1ΔCTA homozygous mice
| Factor | Cell counta |
||
|---|---|---|---|
| Wild type (n = 5) | Fli1ΔCTA/+ (n = 5) | Fli1ΔCTA/Fli1ΔCTAb | |
| RBC | 965.60 ± 110.67 | 948.00 ± 219.36 | 883.00 ± 253.00 |
| WBC | 10,930.00 ± 1,944.09 | 11,510.00 ± 2,633.30 | 11,770.00 ± 1,698.75 |
| PLT | 69.55 ± 18.88 | 57.06 ± 13.77 | 19.24 ± 8.84*† |
n = 5 mice for all groups. Units: RBC, ×104/μl; WBC, cells/μl; PLT, ×104/μl.Values represent means ± SD.
*, P = 0.006 when compared to results for the wild type, Student's t test; †, P = 0.009 compared to results for heterozygous mice, Student's t test.
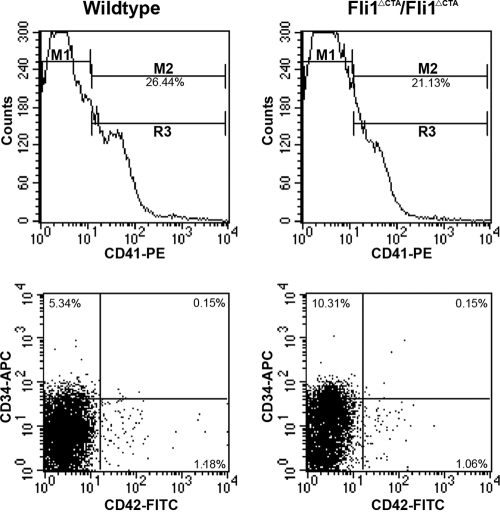
The reduced platelet number could be due to a reduced megakaryocyte (MK) number or a decrease in the breakdown of MKs to form platelets. The total numbers of bone marrow (BM) MKs were similar between the wild-type and Fli1ΔCTA/Fli1ΔCTA homozygous mice (Table 4), suggesting that Fli1 regulates the release of platelets from MKs. Since we did not find a significant difference in acetylcholinesterase-positive cells from BM (P > 0.679), flow analyses were performed to more carefully examine whether megakaryocytic development was affected in the Fli1ΔCTA/Fli1ΔCTA mice. The numbers of more-mature (CD34+ CD41+ CD42+ and CD34− CD41+ CD42+) MKs were less affected than those of the intermediately mature (CD34+ CD41+ CD42−) cells, which were elevated ∼2-fold in the Fli1ΔCTA/Fli1ΔCTA mice (Table 5 and Fig. 2), suggesting that CTA domain-associated Fli1 function may be especially important for the maturation of this subpopulation. CD41+ BM cells were analyzed by flow cytometry for the percentage of megakaryocytes in each ploidy class. No statistical difference was found (P ≥ 0.05) in megakaryocyte ploidy between wild-type and Fli1ΔCTA/Fli1ΔCTA mice (data not shown).
TABLE 4.
Analysis of acetylcholine esterase-positive MKs in BM from wild-type and Fli1ΔCTA/Fli1ΔCTA B6 micea
| Miceb | MK count |
Total cells counted | % Total MKs | ||
|---|---|---|---|---|---|
| Immature | Mature | Total | |||
| Wild type | 4.00 ± 4.00 | 3.60 ± 1.67 | 7.60 ± 4.56 | 14,644.80 ± 7884.59 | 0.056 ± 0.022 |
| Fli1ΔCTA/Fli1ΔCTA | 4.60 ± 1.52 | 4.00 ± 1.58 | 8.60 ± 2.51 | 19,764.00 ± 7,466.07 | 0.051 ± 0.028 |
Values represent means ± SD. Student's t test shows no statistical difference in total numbers of MKs (P > 0.679) or % total MKs (P > 0.765).
n = 5 mice for both groups.
TABLE 5.
Analysis of megakaryocytic subpopulations in BM from wild-type and Fli1ΔCTA/Fli1ΔCTA B6 micea
| Mice | % CD41+ cells | % Gated on CD41+ cells |
||
|---|---|---|---|---|
| CD34+ CD42− | CD34+ CD42+ | CD34− CD42+ | ||
| Wild type (n = 5) | 22.82 ± 6.87 | 5.06 ± 1.16* | 0.19 ± 0.08 | 1.37 ± 0.32 |
| Fli1ΔCTA/Fli1ΔCTA (n = 5) | 17.28 ± 3.98 | 11.34 ± 2.64* | 0.18 ± 0.02 | 1.37 ± 0.33 |
Values represent means ± SD. *, P < 0.001, Student's t test.
FIG. 2.
Representative flow cytometric analysis of bone marrow megakaryocytic populations from wild-type and Fli1ΔCTA/Fli1ΔCTA mice. Bone marrow samples from wild-type (left panels) or Fli1ΔCTA/Fli1ΔCTA (right panels) mice were analyzed for the total percentage of cells expressing CD41, denoted by M2 (upper panels). Samples were then gated on CD41+ cells (M2) and analyzed for percentages of cells in three subpopulations (lower panels): CD34+/CD42− (upper left quadrant), CD34+/CD42+ (upper right quadrant), and CD34−/CD42+ (lower right quadrant). All samples were analyzed using established gates and uniform quadrant parameters. Percentages in each population for representative wild-type and Fli1ΔCTA/Fli1ΔCTA mice are given in each quadrant.
Prolonged bleeding time in Fli1ΔCTA/Fli1ΔCTA mice.
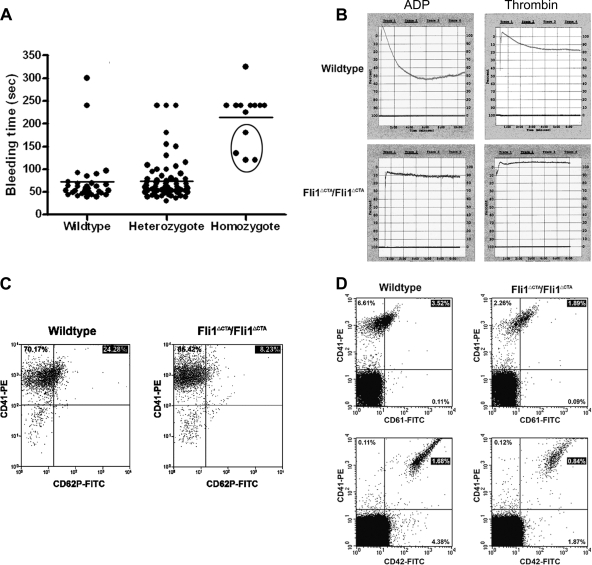
To define the role of Fli1 in hemostasis, tail bleeding time assays were performed (Fig. 3 A). In wild-type and heterozygous mice, bleeding was typically arrested within 1 min (mean, 72.1 ±10 s) after the tail tip segment was cut, whereas bleeding times in Fli1ΔCTA/Fli1ΔCTA mice was significantly prolonged (214.2 ±16 s) (P < 0.001). Indeed, the severity of bleeding often necessitated termination of the assay prior to 4 min (in 4 of 13 homozygous mice analyzed).
FIG. 3.
Prolonged bleeding times and defective platelet aggregation and activation in Fli1ΔCTA/Fli1ΔCTA mice. (A) Bleeding time measurements: mice were anesthetized, and a 2-mm segment of the tail tip was cut off with a scalpel. Tail bleeding times of wild-type (n = 31), heterozygous (n = 73), and homozygous (n = 13) mice were monitored visually. Each circle represents one mouse, and horizontal lines indicate averages of bleeding times. The experiment was stopped after 4 min or when bleeding was too severe (circled homozygous n = 4 mice). (B) Platelet aggregation was used to monitor responses of wild-type and Fli1ΔCTA/Fli1ΔCTA platelets to ADP and thrombin. (C) Platelet activation: PRP samples from wild-type (left panel) or Fli1ΔCTA/Fli1ΔCTA (right panel) mice were analyzed for the percentages of cells expressing CD41 and/or CD62P. All samples were analyzed using established gates and uniform quadrant parameters on a MoFlo cell sorter. Percentages in each population for representative wild-type and Fli1ΔCTA/Fli1ΔCTA mice are given in each quadrant. (D) Measurement of platelet surface glycoprotein levels. Peripheral blood was obtained by cardiac puncture from wild-type (left panels) or Fli1ΔCTA/Fli1ΔCTA (right panels) anesthetized mice. The percentage of CD41+ cells that coexpressed CD61 (upper panels) or CD42d (lower panels) was then examined. All samples were analyzed using established gates and uniform quadrant parameters on a MoFlo cell sorter. Percentages in each population for representative wild-type and Fli1ΔCTA/Fli1ΔCTA mice are given in each quadrant.
Platelet aggregation and activation are reduced in Fli1ΔCTA/Fli1ΔCTA mice.
Platelet adhesion, activation, and aggregation are critical for control of blood loss and coagulation following vascular injury. Platelet aggregation studies were performed with Fli1ΔCTA/Fli1ΔCTA and wild-type mice (Fig. 3B). After exposure to ADP, platelets purified from Fli1ΔCTA/Fli1ΔCTA mice failed to aggregate. Closer examination of the data supports the model that platelets from the Fli1ΔCTA/Fli1ΔCTA mice undergo agonist-dependent shape change but do not show the subsequent aggregation observed in wild-type mice. A similar phenotype was observed in response to another agonist, thrombin.
We performed platelet activation studies with five Fli1ΔCTA/Fli1ΔCTA mice and six normal control mice by measuring the appearance of P-selectin on the platelet cell surface in response to thrombin. This analysis showed a nearly 3-fold reduction in activation of mutant platelets by thrombin (Table 6 and Fig. 3C).
TABLE 6.
Flow cytometric analysis of platelet activation and surface marker expression of PB from wild-type and Fli1ΔCTA/Fli1ΔCTA homozygous micea
| Platelet characteristic analyzed | Population | Value for mice |
P value | |
|---|---|---|---|---|
| Wild type (n = 6) | Fli1ΔCTA/Fli1ΔCTA (n = 5) | |||
| Activation | CD41+ total | 90.16 ± 10.18 | 93.17 ± 2.02 | 0.51 |
| CD41+ pSel− | 69.87 ± 7.15 | 85.19 ± 2.58 | 0.001* | |
| CD41+ pSel+ | 20.29 ± 6.14 | 7.98 ± 1.52 | 0.004* | |
| Surface marker expression | CD41+ CD61+ | 2.08 ± 0.84 | 1.19 ± 0.44 | 0.03* |
| CD41+ CD42d+ | 1.93 ± 0.64 | 0.59 ± 0.22 | 0.001* | |
Values represent mean % of total population ± SD. n = 6 wild-type and 5 Fli1ΔCTA/Fli1ΔCTA mice. *, P ≤ 0.05, Student's t test.
Fli1 contributes to surface expression of membrane receptors, CD42 and CD61, on murine platelets.
Platelet adhesion and activation require proper expression of surface glycoproteins. Fli1 DNA-binding sites are present in the regulatory sequences of multiple MK glycoprotein-encoding genes, and a reduction in expression of these glycoproteins may therefore account for the inhibition of the response of platelets to ADP and thrombin. Surface expression of CD42 (gpIb, the and von Willebrand factor [vWF] receptor) and CD61 (integrin β3) was measured using FITC-conjugated murine-specific antibodies and demonstrated a 3-fold and 1.7-fold reduction in expression of CD42 and CD61, respectively, relative to results for the wild-type platelets (Table 6 and Fig. 3D).
Regulation of megakaryocytic development genes in Fli1ΔCTA/Fli1ΔCTA and wild-type mice.
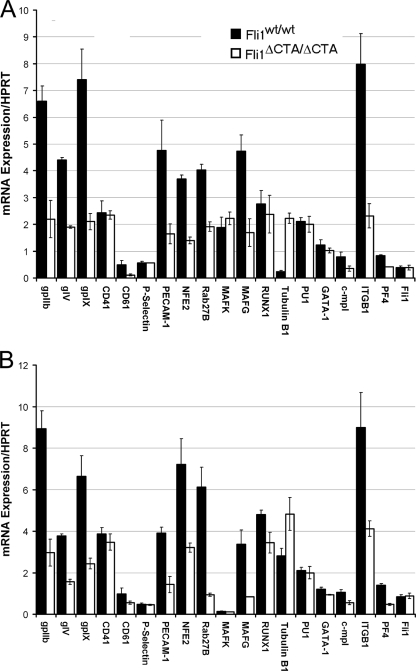
The defective megakaryocytic phenotypes identified in this mouse model support the model that absence of the CTA domain of Fli1 contributes to altered MK differentiation and reduced platelet numbers. Based upon these novel observations, we hypothesized that the identification of differentially expressed transcripts in Fli1ΔCTA/Fli1ΔCTA and wild-type mice would define genes that are regulated by Fli1 and play important roles in megakarypoiesis. To test this hypothesis, real-time RT-PCR analyses of potential targets of Fli1 that may contribute to the altered megakaryopoiesis seen in Fli1ΔCTA/Fli1ΔCTA cells were performed, and multiple transcripts demonstrated statistically significant changes (Fig. 4 and Table 7). It is important to note that the Fli1 mRNA levels were not significantly different in BM from those for Fli1ΔCTA/Fli1ΔCTA and wild-type mice (Fig. 4A and Table 6). Fli1 is required for expression of c-mpl (25), and consistent with the observed MK and platelet phenotypes, real-time quantitative RT-PCR analysis of mRNA from total BM cells from Fli1ΔCTA/Fli1ΔCTA mice revealed a statistically significant 1.6-fold downregulation of c-mpl. Platelet glycoprotein IIb (gpIIb) and the glycoprotein IX (gpIX) were 3-fold and 3.5-fold downregulated in BM cells obtained from Fli1ΔCTA/Fli1ΔCTA mice. Similarly, a 2.3-fold reduction in the levels of glycoprotein V (gpV) was observed. We next examined the level of gene transcripts for those with altered expression during normal terminal differentiation of megakaryocytes to platelets, including NF-E2 and its cofactors, MafG, MafK, Runx1, RAB27B, and tubulin β1. The mRNA levels for NF-E2, MafG, and two NF-E2 targets, RAB27B and tubulin β1, were significantly changed (2.1- to 9.3-fold) in BM in the Fli1ΔCTA/Fli1ΔCTA mice. We also noted that β1-integrin mRNA is significantly reduced (3.4-fold) in Fli1ΔCTA/Fli1ΔCTA mice.
FIG. 4.
Altered expression of potential Fli-1 target genes in cells of Fli1ΔCTA/Fli1ΔCTA mutant mice. Real-time PCR quantitation of mRNA expression. Expression levels were normalized relative to those of the hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene. Expression of megakaryocyte-related genes in RNA prepared from total BM (A) or flow-sorted megakaryocytes (B) is shown. Each point represents the average from biological duplicates analyzed in triplicate.
TABLE 7.
Real-time PCR comparison of BM mRNA from wild-type and Fli1ΔCTA/ΔCTA mice
| Gene product | mRNA expression |
Expression ratio |
Double-sided P value | ||||
|---|---|---|---|---|---|---|---|
|
Fli1wt/wt |
Fli1ΔCTA/ΔCTA |
||||||
| Mean | SD | Mean | SD | ΔCTA/wt | wt/ΔCTA | ||
| gpIIb | 6.6 | 0.57 | 2.2 | 0.7 | 0.33 | 3 | 0.001 |
| gpV | 4.4 | 0.09 | 1.9 | 0.05 | 0.43 | 2.32 | 0.0001 |
| gpIX | 7.4 | 1.14 | 2.1 | 0.3 | 0.28 | 3.52 | 0.003 |
| CD41 | 2.43 | 0.45 | 2.35 | 0.16 | 0.97 | 1.03 | 0.787 |
| CD61 | 0.49 | 0.17 | 0.11 | 0.04 | 0.22 | 4.45 | 0.019 |
| P-Selectin | 0.57 | 0.05 | 0.56 | 0 | 0.98 | 1.02 | 0.67 |
| PECAM-1 | 4.76 | 1.13 | 1.65 | 0.37 | 0.35 | 2.88 | 0.011 |
| NFE2 | 3.7 | 0.14 | 1.4 | 0.13 | 0.38 | 2.64 | 0.005 |
| Rab27B | 4.03 | 0.22 | 1.91 | 0.18 | 0.47 | 2.11 | 0.011 |
| MAFK | 1.89 | 0.38 | 2.23 | 0.24 | 1.18 | 0.85 | 0.027 |
| MAFG | 4.73 | 0.61 | 1.69 | 0.52 | 0.36 | 2.80 | 0.043 |
| RUNX1 | 2.76 | 0.51 | 2.38 | 0.71 | 0.86 | 1.16 | 0.634 |
| Tubulin B1 | 0.24 | 0.04 | 2.23 | 0.19 | 9.29 | 0.11 | 0.003 |
| PU1 | 2.11 | 0.14 | 2 | 0.3 | 0.95 | 1.06 | 0.6 |
| GATA-1 | 1.23 | 0.2 | 1.03 | 0.09 | 0.84 | 1.19 | 0.3 |
| c-mpl | 0.78 | 0.18 | 0.35 | 0.1 | 0.45 | 2.23 | 0.038 |
| ITGB1 | 7.98 | 1.14 | 2.32 | 0.45 | 0.29 | 3.44 | 0.02 |
| PF4 | 0.83 | 0.03 | 0.41 | 0.001 | 0.5 | 2.02 | 0.05 |
| Fli1 | 0.38 | 0.07 | 0.39 | 0.09 | 1.03 | 0.97 | 0.92 |
The presence of Ets binding sites and the observed altered expression of specific genes in the Fli1ΔCTA/Fli1ΔCTA cells support the model that they may be direct Fli1 targets. However, it is possible that the block in lineage differentiation may result in an insufficient number of cells that express the gene in question. To address this possibility, we next determined whether the observed expression changes were maintained in individual cell populations. MKs were isolated from total BM by flow sorting. It is important to notice that analysis of mRNA expression in enriched MKs indicated that each of the changes observed in the BM population were retained in the MK population (Fig. 4B and Table 8). While Runx1 and GATA-1 mRNA expression were not significantly different in total BM, we note a statistically significant decrease in these levels in the MKs of Fli1ΔCTA/Fli1ΔCTA mice.
TABLE 8.
Real-time PCR comparison of Fli1 target gene mRNA of MKs from wild-type and Fli1ΔCTA/ΔCTA mice
| Gene product | mRNA expression |
Expression ratio |
Double-sided P value | ||||
|---|---|---|---|---|---|---|---|
| Fli1wt/wt |
Fli1ΔCTA/ΔCTA |
||||||
| Mean | SD | Mean | SD | ΔCTA/wt | wt/ΔCTA | ||
| gpIIb | 8.94 | 0.86 | 2.96 | 0.65 | 0.33 | 3.02 | 0.001 |
| gpV | 3.77 | 0.09 | 1.56 | 0.12 | 0.41 | 2.42 | 0.0001 |
| gpIX | 6.65 | 0.98 | 2.44 | 0.26 | 0.37 | 2.73 | 0.002 |
| CD41 | 3.86 | 0.31 | 3.47 | 0.39 | 0.9 | 1.11 | 0.309 |
| CD61 | 0.97 | 0.29 | 0.56 | 0.08 | 0.58 | 1.73 | 0.0776 |
| P-Selectin | 0.48 | 0.05 | 0.45 | 0.02 | 0.94 | 1.07 | 0.389 |
| PECAM-1 | 3.91 | 0.29 | 1.43 | 0.39 | 0.37 | 2.73 | 0.0027 |
| NFE2 | 7.22 | 1.23 | 3.21 | 0.22 | 0.44 | 2.25 | 0.0051 |
| Rab27B | 6.12 | 0.97 | 0.94 | 0.07 | 0.15 | 6.51 | 0.0008 |
| MAFK | 0.14 | 0.02 | 0.11 | 0 | 0.79 | 1.27 | 0.0602 |
| MAFG | 3.36 | 0.7 | 0.84 | 0 | 0.25 | 4.00 | 0.0042 |
| RUNX1 | 4.8 | 0.22 | 3.44 | 0.5 | 0.72 | 1.40 | 0.0125 |
| Tubulin B1 | 2.81 | 0.37 | 4.83 | 0.8 | 1.72 | 0.58 | 0.0165 |
| PU1 | 2.11 | 0.14 | 2 | 0.3 | 0.95 | 1.06 | 0.595 |
| GATA-1 | 1.21 | 0.1 | 0.93 | 0.02 | 0.77 | 1.30 | 0.0089 |
| c-mpl | 1.05 | 0.13 | 0.55 | 0.1 | 0.52 | 1.91 | 0.061 |
| ITGB1 | 9 | 1.68 | 4.12 | 0.37 | 0.46 | 2.18 | 0.011 |
| PF4 | 1.4 | 0.07 | 0.47 | 0.05 | 0.34 | 2.98 | 0.001 |
| Fli1 | 0.85 | 0.09 | 0.88 | 0.13 | 1.04 | 0.97 | 0.758 |
CTA deletion of Fli1 affects interaction with GATA-1 in vivo.
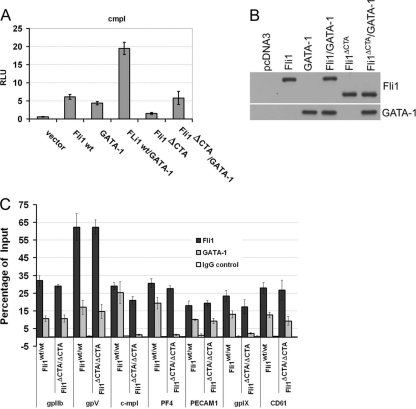
The functional activity of Ets factors is modulated by interaction with other nuclear proteins. The ability of Fli1 and GATA-1 to act synergistically to regulate gene transcription of multiple megakaryocytic genes is well established. We and others have previously demonstrated that Fli1 and GATA-1 synergistically activate the c-mpl promoter in vitro and cooccupy this promoter in vivo and chose this model promoter to compare the activities of the Fli1 and Fli1ΔCTA proteins. Transient-transfection studies indicate that the Fli1ΔCTA protein is less able to stimulate promoter transcription than the full-length wild-type Fli1 protein (Fig. 5 A, compare lanes 2 and 5). Moreover, only wild-type Fli1 can synergize with GATA-1, increasing promoter activity.
FIG. 5.
Fli1+ and Fli1ΔCTA cooperate with GATA-1 to activate transcription of the c-mpl promoter in vitro but show differential association with promoters in vivo. (A) HeLa cells were transfected with the indicated transcription factors (200 ng) and a c-mpl-luciferase reporter plasmid (200 ng). Relative activation of the reporter is shown. (B) Western blot analysis using polyclonal anti-Fli1 or anti-GATA-1 antibody on the same cell extracts. (C) Real-time PCR analysis of ChIP products. Chromatin was prepared from wild-type or Fli1ΔCTA/Fli1ΔCTA MKs, and chromatin immunoprecipitation assays were performed using anti-Fli1 or anti-GATA-1 antibodies or IgG (control). “Input” represented 5% total cross-linked, reversed chromatin before immunoprecipitation. Values represent percentages of input.
To further examine Fli1 direct target genes that may contribute to the phenotype of the Fli1ΔCTA/Fli1ΔCTA cells, we prepared chromatin from isolated MK cells and performed chromatin immunoprecipitation (ChIP) studies (Table 9). Based upon the observed reduction in the c-mpl transcript levels in RNA prepared from BM and purified MKs from Fli1ΔCTA/Fli1ΔCTA mice (Fig. 4), we investigated whether both Fli1 and Fli1ΔCTA are able to recruit GATA-1 to this promoter in vivo. ChIP analyses using anti-Fli1 and anti-GATA-1 antibodies were performed (Fig. 5C) . Real-time quantitative analysis demonstrated that the c-mpl promoter region was detected in the anti-Fli1-ChIP enriched fragments from both Fli1+/+ and Fli1ΔCTA/Fli1ΔCTA cells. In contrast, the c-mpl promoter was significantly more enriched in the anti-GATA-1-ChIP fragments prepared from the wild-type (Fli1+/+) cells. Consistent with the failure of Fli1ΔCTA and GATA-1 to synergistically activate the c-mpl promoter-luciferase reporters in vitro, Fli1ΔCTA is not able to efficiently recruit GATA-1 to this promoter in vivo.
TABLE 9.
ChIP RT-PCR analysis of Fli1 and GATA-1 bound to the indicated promoters
| Promoter | Phenotype | % bound |
|||||
|---|---|---|---|---|---|---|---|
| Fli1 |
GATA-1 |
IgG control |
|||||
| Mean | SD | Mean | SD | Mean | SD | ||
| gpIIb | Fli1wt/wt | 32.00 | 3.00 | 10.67 | 1.53 | 0.30 | 0.10 |
| Fli1ΔCTA/ΔCTA | 29.00 | 1.00 | 10.70 | 2.08 | 0.37 | 0.12 | |
| gpV | Fli1wt/wt | 62.33 | 7.57 | 17.00 | 4.00 | 0.77 | 0.21 |
| Fli1ΔCTA/ΔCTA | 62.33 | 4.16 | 14.67 | 4.04 | 0.23 | 0.06 | |
| c-mpl | Fli1wt/wt | 29.00 | 2.00 | 25.33 | 6.03 | 0.93 | 0.12 |
| Fli1ΔCTA/ΔCTA | 21.00 | 2.00 | 1.53 | 0.32 | 0.23 | 0.15 | |
| PF4 | Fli1wt/wt | 30.67 | 2.52 | 19.33 | 3.21 | 0.47 | 0.15 |
| Fli1ΔCTA/ΔCTA | 27.67 | 1.53 | 1.50 | 0.36 | 0.47 | 0.31 | |
| PECAM1 | Fli1wt/wt | 18.00 | 2.65 | 10.00 | 0.53 | 1.17 | 0.74 |
| Fli1ΔCTA/ΔCTA | 19.33 | 1.53 | 9.33 | 1.53 | 0.43 | 0.15 | |
| gpIX | Fli1wt/wt | 23.33 | 3.21 | 13.00 | 2.00 | 0.60 | 0.36 |
| Fli1ΔCTA/ΔCTA | 17.33 | 4.04 | 2.33 | 0.58 | 0.55 | 0.44 | |
| CD61 | Fli1wt/wt | 28.00 | 3.00 | 12.67 | 1.53 | 0.67 | 0.21 |
| Fli1ΔCTA/ΔCTA | 26.67 | 5.69 | 9.33 | 2.52 | 0.25 | 0.30 | |
| Negative controla | Fli1wt/wt | 0.09 | 0.01 | 0.003 | 0 | 0.001 | 0.0 |
| Fli1ΔCTA/ΔCTA | 0.03 | 0 | 0.005 | 0 | 0.0 | 0.0 | |
Negative control: upstream gpIIb region which lacks functional Ets site.
Fli1 and GATA-1 function synergistically to regulate multiple megakaryocytic genes, including those encoding gpIIb, gpVI, gpIX, gpIb, and c-mpl. The expression of multiple genes was significantly altered in Fli1ΔCTA/Fli1ΔCTA mice, and selected promoters were analyzed by ChIP using anti-Fli1 and anti-GATA-1 antibodies (Fig. 5C and Table 9). ChIP analyses of MK demonstrated that Fli1 was present on each of the promoters examined, supporting the model that they are direct transcriptional targets of Fli1 in vivo. In general, as was observed with c-mpl, Fli1 and Fli1ΔCTA were found on each promoter. As observed for c-mpl, anti-GATA-1 antibodies preferentially enriched the PF4 and gpIX promoters in the Fli1 cells compared to the Fli1ΔCTA cells. Reduced GATA-1 binding in Fli1ΔCTA/Fli1ΔCTA mice was not found for all promoters examined. For example, the gpIIb, gpV, PECAM, and CD61 promoters were bound to a similar or greater extent in the Fli1ΔCTA/Fli1ΔCTA mice. Thus, differential GATA-1 binding in vivo was promoter specific.
DISCUSSION
In this study, we demonstrated the in vivo importance of the carboxy-terminal regulatory (CTA) domain of Fli1 for normal hematopoiesis. Removal of this domain affects MK and platelet development. These observations suggest previously unknown in vivo roles of a specific functional domain of Fli1 in megakaryocytic proliferation and differentiation.
The most widely used markers for examining MK differentiation are CD61, CD41, and CD42. MK differentiation is characterized by the expression of CD61 (integrin β3) and increased expression of CD41 (integrin αIIb) (37, 50; reviewed in reference 51), which together form a heterodimeric receptor complex known as glycoprotein IIb/IIIa (gpIIb/IIIa) (9, 44). This receptor is present on the surfaces of cells of the MK lineage from progenitor cells through platelets, and their expression levels increase as cells mature (31, 47). GpIIb/IIIa functions as an adhesive for fibrinogen, fibronectin, vitronectin, and von Willebrand factor (vWF) (9, 37). The expression of CD42 (gpIb, the vWF receptor) is slightly later than that of CD41 (50); however, expression levels of the two correlate with MK maturity (50). Thus, CD34+ CD41+ CD42+ and CD34− CD41+ CD42+ represent more-mature MKs, while CD34+ CD41+ CD42− cells represent intermediately mature MKs.
Although the number of mature MKs is not affected, the number of intermediately mature CD34+ CD41+ CD42− MKs was significantly increased in the Fli1ΔCTA/Fli1ΔCTA mice. Interestingly, Fli1 expression is monoallelic in the majority (65%) of these intermediately mature cells (50), which may make this population especially sensitive to Fli1-regulated pathways. Indeed, it has been suggested that this population is affected in Jacobson syndrome (Paris-Trousseaus syndrome [PTS]) patients, who harbor hemizygous deletions in 11q, including Fli1 (50). Thus, these patients have only one copy of the Fli1 gene due to a heterozygous loss of regions in chromosome 11 and display normal and small, immature, lysing MKs. Lentivirus-mediated Fli1 expression in CD34+ cells restored normal megakaryopoiesis (50). It is significant that Fli1 expression is monoallelic in the intermediately mature CD41+ CD42− cells but biallelic before and after this stage of megakaryopoiesis (50). Mature megakaryopoiesis appears normal in Fli1ΔCTA/Fli1ΔCTA mice, perhaps due to compensation by other Ets factors, including GA-binding protein alpha (GABPα) (43), ERG (26, 33, 59), and to a lesser extent ETS2 (59).
Platelet number and function are impared in the Fli1ΔCTA/Fli1ΔCTA mice. The absence of a significant difference in circulating platelet numbers between wild-type and heterozygous mice supports the model that Fli1ΔCTA is not functioning as a dominant-negative transcription factor.
TPO is the main physiologic growth factor for MK proliferation, differentiation, and platelet production. Expression of multiple transcription factors, including Fli1, GATA-1, RUNX1, and NF-E2, is critical for normal megakaryopoiesis (43, 61). GATA-1 is a zinc finger protein expressed in erythrocytes, MKs, eosinophils, and mast cells. Homozygous null mutation of GATA-1 in mice is embryonic lethal due to anemia (17), while an MK-specific knockdown of these genes results in thrombocytopenia and accumulation of immature MKs (56). Fli1 and GATA-1 work synergistically to regulate multiple megakaryocytic genes, including those encoding gpIIb, gpVI, gpIX, and gpIb, and c-mpl (23). Although the MK population from the Fli1ΔCTA/Fli1ΔCTA mice expresses reduced GATA-1 mRNA, the most striking feature is the apparent failure of the Fli1ΔCTA protein to recruit GATA-1 to several megakaryocytic promoters. However, TPO−/− or Mpl−/− mice, even with only 10% of circulating platelets present, remained healthy and showed no signs of spontaneous hemorrhage (7). This suggested that some alternative regulators of megakaryocytopoiesis other than TPO might exist and that these regulators alone might be sufficient for maintaining hemostasis. TPO-independent megakaryocytopoiesis forms a regulatory system that includes four signals and an unknown signaling pathway(s). These four pathways are the gp130 (glycoprotein 130)-dependent signaling pathway, the Notch pathway, N-methyl-d-aspartate (NMDA) receptor-mediated signaling, and the stromal cell-derived factor 1 (SDF-1)/fibroblast growth factor 4 (FGF-4) paradigm (71).
RUNX1 (AML1) null embryos die at midgestation with hemorrhage in the central nervous system (63) similar to that observed for Fli1 null animals (58). Mice with conditional RUNX deletion have reduced platelet numbers and small (micro-MK), hypoploid MKs (22, 45). Conversely, RUNX1 overexpression in hematopoietic cells leads to megakaryocytic differentiation (39). A recent study has demonstrated direct interaction between Fli1 and RUNX1 during terminal megakaryocytic differentiation (21). The observed reduced RUNX1 expression may contribute to the observed platelet phenotype.
Fli1ΔCTA/Fli1ΔCTA mice have significantly reduced expression of NF-E2 and MafG. NF-E2 is a bZIP DNA transcription factor expressed in erythroid, megakaryocytic, and mast cells. NF-E2 has been shown to regulate transcription of MK genes, the including tubulin β1, thromboxane synthase, and Rab27b genes (51). Homozygous loss of NF-E2 leads to neonatal lethality due to hemorrhage secondary to a lack of circulating platelets (thrombocytopenia), although these mice have elevated levels of MKs that express PF4 and gpIIb that fail to mature properly (28, 57). MK progenitors from the NF-E2 null mice showed a reduced response to TPO, indicating that NF-E2 also plays a role in the proliferation of committed MK progenitors (32). NF-E2 functions as a heterodimer consisting of the hematopoietic restricted 45-kDa subunit (p45) and one of three ubiquitously expressed 18-kDa subunits (MafK and MafG in MKs). MafG homozygous mutant mice show impaired platelet formation (55), and an absence of MafK enhances the thrombocytopenia phenotype of MafG animals due to impaired proplatelet production, as well as an erythroid phenotype more severe than that observed in NF-E2 mice (55). NF-E2 null mice do not express Rab27b, and we note a significant reduction of this mRNA in Fli1ΔCTA/Fli1ΔCTA mice.
The terminal phase of MK differentiation, release of platelets from the MK, is dependent upon the formation of proplatelets. Proplatelets are cytoplasmic extensions of the MK, and this projection requires changes in the cytoskeleton, including activation of microtubules. The tubulin β1 gene, an NF-E2 target gene, constitutes the major isotype present in microtubules and is required for normal proplatelet profusion. While β1 knockout mice have reduced platelet numbers (52), β1 expression is increased in Fli1ΔCTA/Fli1ΔCTA mice. In contrast, the observed decreased integrin β1 expression may affect platelet adhesion, since integrins α2 and β form the collagen receptor.
Fli1 interaction with other proteins can either be positive (e.g., GATA-1 [14, 23], serum response factor [SRF] [10, 65], and Pax-5 [16, 34]) or antagonistic (TEL [27, 62], EKLF [60], and retinoic acid receptor [RAR] [11]). Although carboxy-terminal truncation of Fli1 (retaining positions 1 to 375) has been reported to not have a significant effect on DNA binding specificity in vitro (35), the CTA domain is required for maximal activation of promoter-reporters, as shown by in vitro transient-transfection studies (49).
We have previously shown that Fli1ΔCTA/Fli1ΔCTA mice have decreased peripheral blood B220 cells, fewer splenic follicular B cells, increased transitional and marginal zone B cells, and reduced bone marrow pre-B cells compared with those of wild-type controls (70). The CTA domain is also required for many aspects of collagen fibrillogenesis in the skin (2). In this study, we demonstrate that removal of this domain of Fli1 reduces in vitro transcriptional activation activity measured on the c-mpl promoter. We had previously determined that this truncation reduces activity on the MMP1 promoter by 40 to 50% (data not shown). Consistent with these observations, previous in vitro analyses have demonstrated that the carboxy-terminal domain contributes to transcriptional activity and transformation properties of EWS/Fli1 (1, 41). In addition to interacting with GATA-1, Fli1 binding enhances the affinity of GATA-1 for proximal binding sites (14). We found that the presence of GATA-1 on multiple promoters was significantly reduced in Fli1ΔCTA/Fli1ΔCTA BM cells in vivo, although the effect was promoter specific. Previous in vitro studies demonstrated that Fli-1 interacts with GATA-1 primarily through the Ets domain, although weak interactions were observed between GATA-1 and amino acids 363 to 452 of Fli-1 (14). This is in contrast with the results from in vitro transient-transfection studies, which demonstrated a significant effect of the CTA domain deletion on transcriptional synergy with GATA-1. However, the impact of the C-terminal region on synergy with GATA-1 in vitro and recruitment of GATA-1 in vivo is promoter dependent. Binding of other transcription factors (e.g., Runx1 [21]) may contribute to the promoter-specific effects observed. In addition, multiple other Ets factors that function during megakaryopoiesis, including GABPα (43) and ERG (26, 33, 59), may provide some redundant function of specific promoters. Collectively, our results demonstrate that the CTA-containing region of Fli1 is essential for proper transcriptional activation in vivo.
Based upon the still-limited studies of Fli1-interactive proteins, several additional properties of the CTA domain may offer additional mechanistic insights in the future. Fli1 is also a partner with EKLF via its DNA binding domain and the N- and C-terminal regions (60). Antagonistic interaction of Fli1 with RAR and TEL has been mapped to its N-terminal domain. FLI1 and EWS-FLI1 are able to form ternary complexes with SRF on the SRE elements of the Egr-1 (Early growth response 1) gene. In addition to removal of the CTA domain, the Fli1ΔCTA allele also lacks one of the two SBM domains (SBM2, located between amino acids 392 and 401) required for ternary complex formation on SRE elements (10). PAX-1 (BSAP) and Ets proteins (Fli1, Ets1, and GABPα) interact through their respective DNA binding domains (15). It remains to be determined whether possible effects on these and other Fli1 protein interactions alter transcriptional activities, contributing to Fli1ΔCTA/Fli1ΔCTA observed and yet-to-be-characterized phenotypes. The CTA domain clearly has functions essential in early postnatal development that may reveal novel functions in further investigations.
Acknowledgments
This work was supported by National Institutes of Health grants PO1-CA78582 (to D.K.W.), RO1 HL69123 (to M.O.), and K01 AR051385 (to X.K.Z.) and the Office of Research and Development, Medical Research Services, Department of Veterans Affairs (to M.O., A.C.L., X.K.Z., and G.G.). The Flow Cytometry & Cell Sorting Shared Resource of the Hollings Cancer Center is supported in part by a Cancer Center Support Grant (P30 CA 138313).
We thank Perry V. Halushka for discussion of platelet aggregation and function. We also thank Elizabeth Kruse (the Walter and Eliza Hall Institute of Medical Research, Australia) for providing advice on platelet half-life studies. We acknowledge technical assistance from Haiqun Zeng and James Klein. We thank John Lazarchick and Joan Mueller for providing expertise and the Chrono-log luminometer. We also acknowledge support from the MUSC Sequencing facility and the Flow Cytometry & Cell Sorting Shared Resource of the Hollings Cancer Center. We also thank Clayton Polite and other members of the DLAR staff.
Footnotes
Published ahead of print on 7 September 2010.
REFERENCES
- 1.Arvand, A., S. M. Welford, M. A. Teitell, and C. T. Denny. 2001. The COOH-terminal domain of FLI-1 is necessary for full tumorigenesis and transcriptional modulation by EWS/FLI-1. Cancer Res. 61:5311-5317. [PubMed] [Google Scholar]
- 2.Asano, Y., M. Markiewicz, M. Kubo, G. Szalai, D. K. Watson, and M. Trojanowska. 2009. Transcription factor Fli1 regulates collagen fibrillogenesis in mouse skin. Mol. Cell. Biol. 29:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athanasiou, M., P. A. Clausen, G. J. Mavrothalassitis, X. K. Zhang, D. K. Watson, and D. G. Blair. 1996. Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell Growth Differ. 7:1525-1534. [PubMed] [Google Scholar]
- 4.Ben-David, Y., E. B. Giddens, K. Letwin, and A. Bernstein. 1991. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 5:908-918. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron, D., B. Barbeau, C. Leger, and E. Rassart. 1995. Experimental bias in the evaluation of the cellular transient expression in DNA co-transfection experiments. Cell Mol. Biol. Res. 41:155-159. [PubMed] [Google Scholar]
- 6.Bergeron, D., J. Houde, L. Poliquin, B. Barbeau, and E. Rassart. 1993. Expression and DNA rearrangement of proto-oncogenes in Cas-Br-E-induced non-T-, non-B-cell leukemias. Leukemia 7:954-962. [PubMed] [Google Scholar]
- 7.Bouillet, P., D. Metcalf, D. C. Huang, D. M. Tarlinton, T. W. Kay, F. Kontgen, J. M. Adams, and A. Strasser. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286:1735-1738. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, K. E., and P. J. Farnham. 1999. Identification of target genes of oncogenic transcription factors. Proc. Soc. Exp. Biol. Med. 222:9-28. [DOI] [PubMed] [Google Scholar]
- 9.Carrell, N. A., L. A. Fitzgerald, B. Steiner, H. P. Erickson, and D. R. Phillips. 1985. Structure of human platelet membrane glycoproteins IIb and IIIa as determined by electron microscopy. J. Biol. Chem. 260:1743-1749. [PubMed] [Google Scholar]
- 10.Dalgleish, P., and A. D. Sharrocks. 2000. The mechanism of complex formation between Fli-1 and SRF transcription factors. Nucleic Acids Res. 28:560-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darby, T. G., J. D. Meissner, A. Ruhlmann, W. H. Mueller, and R. J. Scheibe. 1997. Functional interference between retinoic acid or steroid hormone receptors and the oncoprotein Fli-1. Oncogene 15:3067-3082. [DOI] [PubMed] [Google Scholar]
- 12.Denicourt, C., E. Edouard, and E. Rassart. 1999. Oncogene activation in myeloid leukemias by Graffi murine leukemia virus proviral integration. J. Virol. 73:4439-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deveaux, S., A. Filipe, V. Lemarchandel, J. Ghysdael, P. H. Romeo, and V. Mignotte. 1996. Analysis of the thrombopoietin receptor (MPL) promoter implicates GATA and Ets proteins in the coregulation of megakaryocyte-specific genes. Blood 87:4678-4685. [PubMed] [Google Scholar]
- 14.Eisbacher, M., M. L. Holmes, A. Newton, P. J. Hogg, L. M. Khachigian, M. Crossley, and B. H. Chong. 2003. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol. Cell. Biol. 23:3427-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzsimmons, D., W. Hodsdon, W. Wheat, S. M. Maira, B. Wasylyk, and J. Hagman. 1996. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 10:2198-2211. [DOI] [PubMed] [Google Scholar]
- 16.Fitzsimmons, D., R. Lutz, W. Wheat, H. M. Chamberlin, and J. Hagman. 2001. Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucleic Acids Res. 29:4154-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara, Y., C. P. Browne, K. Cunniff, S. C. Goff, and S. H. Orkin. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. U. S. A. 93:12355-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart, A., F. Melet, P. Grossfeld, K. Chien, C. Jones, A. Tunnacliffe, R. Favier, and A. Bernstein. 2000. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity 13:167-177. [DOI] [PubMed] [Google Scholar]
- 19.Holmes, M. L., N. Bartle, M. Eisbacher, and B. H. Chong. 2002. Cloning and analysis of the thrombopoietin-induced megakaryocyte-specific glycoprotein VI promoter and its regulation by GATA-1, Fli-1, and Sp1. J. Biol. Chem. 277:48333-48341. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, T., M. Trojanowska, and D. K. Watson. 2004. Ets proteins in biological control and cancer. J. Cell Biochem. 91:896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, H., M. Yu, T. E. Akie, T. B. Moran, A. J. Woo, N. Tu, Z. Waldon, Y. Y. Lin, H. Steen, and A. B. Cantor. 2009. Differentiation-dependent interactions between RUNX-1 and FLI-1 during megakaryocyte development. Mol. Cell. Biol. 29:4103-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichikawa, M., T. Asai, T. Saito, S. Seo, I. Yamazaki, T. Yamagata, K. Mitani, S. Chiba, S. Ogawa, M. Kurokawa, and H. Hirai. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10:299-304. [DOI] [PubMed] [Google Scholar]
- 23.Jackers, P., G. Szalai, O. Moussa, and D. K. Watson. 2004. Ets-dependent regulation of target gene expression during megakaryopoiesis. J. Biol. Chem. 279:52183-52190. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, C. W. 1973. Cholinesterase as a possible marker for early cells of the megakaryocytic series. Blood 42:413-421. [PubMed] [Google Scholar]
- 25.Kawada, H., T. Ito, P. N. Pharr, D. D. Spyropoulos, D. K. Watson, and M. Ogawa. 2001. Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. Int. J. Hematol. 73:463-468. [DOI] [PubMed] [Google Scholar]
- 26.Kruse, E. A., S. J. Loughran, T. M. Baldwin, E. C. Josefsson, S. Ellis, D. K. Watson, P. Nurden, D. Metcalf, D. J. Hilton, W. S. Alexander, and B. T. Kile. 2009. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc. Natl. Acad. Sci. U. S. A. 106:13814-13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwiatkowski, B. A., A. G. Zielinska-Kwiatkowska, T. R. Bauer, Jr., and D. D. Hickstein. 2000. The ETS family member Tel antagonizes the Fli-1 phenotype in hematopoietic cells. Blood Cells Mol. Dis. 26:84-90. [DOI] [PubMed] [Google Scholar]
- 28.Lecine, P., J. L. Villeval, P. Vyas, B. Swencki, Y. Xu, and R. A. Shivdasani. 1998. Mice lacking transcription factor NF-E2 provide in vivo validation of the proplatelet model of thrombocytopoiesis and show a platelet production defect that is intrinsic to megakaryocytes. Blood 92:1608-1616. [PubMed] [Google Scholar]
- 29.Lemarchandel, V., J. Ghysdael, V. Mignotte, C. Rahuel, and P. H. Romeo. 1993. GATA and Ets cis-acting sequences mediate megakaryocyte-specific expression. Mol. Cell. Biol. 13:668-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leon, C., B. Hechler, M. Freund, A. Eckly, C. Vial, P. Ohlmann, A. Dierich, M. LeMeur, J. P. Cazenave, and C. Gachet. 1999. Defective platelet aggregation and increased resistance to thrombosis in purinergic P2Y(1) receptor-null mice. J. Clin. Invest. 104:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levene, R. B., J. M. Lamaziere, H. E. Broxmeyer, L. Lu, and E. M. Rabellino. 1985. Human megakaryocytes. V. Changes in the phenotypic profile of differentiating megakaryocytes. J. Exp. Med. 161:457-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin, J., J. P. Peng, G. R. Baker, J. L. Villeval, P. Lecine, S. A. Burstein, and R. A. Shivdasani. 1999. Pathophysiology of thrombocytopenia and anemia in mice lacking transcription factor NF-E2. Blood 94:3037-3047. [PubMed] [Google Scholar]
- 33.Loughran, S. J., E. A. Kruse, D. F. Hacking, C. A. de Graaf, C. D. Hyland, T. A. Willson, K. J. Henley, S. Ellis, A. K. Voss, D. Metcalf, D. J. Hilton, W. S. Alexander, and B. T. Kile. 2008. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat. Immunol. 9:810-819. [DOI] [PubMed] [Google Scholar]
- 34.Maier, H., R. Ostraat, S. Parenti, D. Fitzsimmons, L. J. Abraham, C. W. Garvie, and J. Hagman. 2003. Requirements for selective recruitment of Ets proteins and activation of mb-1/Ig-alpha gene transcription by Pax-5 (BSAP). Nucleic Acids Res. 31:5483-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao, X., S. Miesfeldt, H. Yang, J. M. Leiden, and C. B. Thompson. 1994. The FLI-1 and chimeric EWS-FLI-1 oncoproteins display similar DNA binding specificities. J. Biol. Chem. 269:18216-18222. [PubMed] [Google Scholar]
- 36.Masuya, M., O. Moussa, T. Abe, T. Deguchi, T. Higuchi, Y. Ebihara, D. D. Spyropoulos, D. K. Watson, and M. Ogawa. 2005. Dysregulation of granulocyte, erythrocyte, and NK cell lineages in Fli-1 gene-targeted mice. Blood 105:95-102. [DOI] [PubMed] [Google Scholar]
- 37.Mathur, A., Y. Hong, G. Wang, and J. D. Erusalimsky. 2004. Assays of megakaryocyte development: surface antigen expression, ploidy, and size. Methods Mol. Biol. 272:309-322. [DOI] [PubMed] [Google Scholar]
- 38.Mignotte, V., I. Vigon, E. Boucher de Crevecoeur, P. H. Romeo, V. Lemarchandel, and S. Chretien. 1994. Structure and transcription of the human c-mpl gene (MPL). Genomics 20:5-12. [DOI] [PubMed] [Google Scholar]
- 39.Niitsu, N., Y. Yamamoto-Yamaguchi, H. Miyoshi, K. Shimizu, M. Ohki, M. Umeda, and Y. Honma. 1997. AML1a but not AML1b inhibits erythroid differentiation induced by sodium butyrate and enhances the megakaryocytic differentiation of K562 leukemia cells. Cell Growth Differ. 8:319-326. [PubMed] [Google Scholar]
- 40.Oettgen, P., E. Finger, Z. Sun, Y. Akbarali, U. Thamrongsak, J. Boltax, F. Grall, A. Dube, A. Weiss, L. Brown, G. Quinn, K. Kas, G. Endress, C. Kunsch, and T. A. Libermann. 2000. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J. Biol. Chem. 275:1216-1225. [DOI] [PubMed] [Google Scholar]
- 41.Ohno, T., V. N. Rao, and E. S. Reddy. 1993. EWS/Fli-1 chimeric protein is a transcriptional activator. Cancer Res. 53:5859-5863. [PubMed] [Google Scholar]
- 42.Ott, D. E., J. Keller, and A. Rein. 1994. 10A1 MuLV induces a murine leukemia that expresses hematopoietic stem cell markers by a mechanism that includes fli-1 integration. Virology 205:563-568. [DOI] [PubMed] [Google Scholar]
- 43.Pang, L., H. H. Xue, G. Szalai, X. Wang, Y. Wang, D. K. Watson, W. J. Leonard, G. A. Blobel, and M. Poncz. 2006. Maturation stage-specific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood 108:2198-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips, D. R., I. F. Charo, L. V. Parise, and L. A. Fitzgerald. 1988. The platelet membrane glycoprotein IIb-IIIa complex. Blood 71:831-843. [PubMed] [Google Scholar]
- 45.Putz, G., A. Rosner, I. Nuesslein, N. Schmitz, and F. Buchholz. 2006. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene 25:929-939. [DOI] [PubMed] [Google Scholar]
- 46.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabellino, E. M., R. B. Levene, L. L. Leung, and R. L. Nachman. 1981. Human megakaryocytes. II. Expression of platelet proteins in early marrow megakaryocytes. J. Exp. Med. 154:88-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramakers, C., J. M. Ruijter, R. H. Deprez, and A. F. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 49.Rao, V. N., T. Ohno, D. D. Prasad, G. Bhattacharya, and E. S. Reddy. 1993. Analysis of the DNA-binding and transcriptional activation functions of human Fli-1 protein. Oncogene 8:2167-2173. [PubMed] [Google Scholar]
- 50.Raslova, H., E. Komura, J. P. Le Couedic, F. Larbret, N. Debili, J. Feunteun, O. Danos, O. Albagli, W. Vainchenker, and R. Favier. 2004. FLI1 monoallelic expression combined with its hemizygous loss underlies Paris-Trousseau/Jacobsen thrombopenia. J. Clin. Invest. 114:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulze, H., and R. A. Shivdasani. 2005. Mechanisms of thrombopoiesis. J. Thromb. Haemost. 3:1717-1724. [DOI] [PubMed] [Google Scholar]
- 52.Schwer, H. D., P. Lecine, S. Tiwari, J. E. Italiano, Jr., J. H. Hartwig, and R. A. Shivdasani. 2001. A lineage-restricted and divergent beta-tubulin isoform is essential for the biogenesis, structure and function of blood platelets. Curr. Biol. 11:579-586. [DOI] [PubMed] [Google Scholar]
- 53.Seth, A., L. Robinson, D. M. Thompson, D. K. Watson, and T. S. Papas. 1993. Transactivation of GATA-1 promoter with ETS1, ETS2 and ERGB/Hu-FLI-1 proteins: stabilization of the ETS1 protein binding on GATA-1 promoter sequences by monoclonal antibody. Oncogene 8:1783-1790. [PubMed] [Google Scholar]
- 54.Seth, A., and D. K. Watson. 2005. ETS transcription factors and their emerging roles in human cancer. Eur. J. Cancer. 41:2462-2478. [DOI] [PubMed] [Google Scholar]
- 55.Shavit, J. A., H. Motohashi, K. Onodera, J. Akasaka, M. Yamamoto, and J. D. Engel. 1998. Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev. 12:2164-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shivdasani, R. A., Y. Fujiwara, M. A. McDevitt, and S. H. Orkin. 1997. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 16:3965-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shivdasani, R. A., and S. H. Orkin. 1995. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc. Natl. Acad. Sci. U. S. A. 92:8690-8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spyropoulos, D. D., P. N. Pharr, K. R. Lavenburg, P. Jackers, T. S. Papas, M. Ogawa, and D. K. Watson. 2000. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 20:5643-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stankiewicz, M. J., and J. D. Crispino. 2009. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood 113:3337-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starck, J., N. Cohet, C. Gonnet, S. Sarrazin, Z. Doubeikovskaia, A. Doubeikovski, A. Verger, M. Duterque-Coquillaud, and F. Morle. 2003. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol. Cell. Biol. 23:1390-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szalai, G., A. C. LaRue, and D. K. Watson. 2006. Molecular mechanisms of megakaryopoiesis. Cell. Mol. Life Sci. 63:2460-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waga, K., Y. Nakamura, K. Maki, H. Arai, T. Yamagata, K. Sasaki, M. Kurokawa, H. Hirai, and K. Mitani. 2003. Leukemia-related transcription factor TEL accelerates differentiation of Friend erythroleukemia cells. Oncogene 22:59-68. [DOI] [PubMed] [Google Scholar]
- 63.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U. S. A. 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, X., J. D. Crispino, D. L. Letting, M. Nakazawa, M. Poncz, and G. A. Blobel. 2002. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 21:5225-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watson, D. K., L. Robinson, D. R. Hodge, I. Kola, T. S. Papas, and A. Seth. 1997. FLI1 and EWS-FLI1 function as ternary complex factors and ELK1 and SAP1a function as ternary and quaternary complex factors on the Egr1 promoter serum response elements. Oncogene 14:213-221. [DOI] [PubMed] [Google Scholar]
- 66.Watson, D. K., F. E. Smyth, D. M. Thompson, J. Q. Cheng, J. R. Testa, T. S. Papas, and A. Seth. 1992. The ERGB/Fli-1 gene: isolation and characterization of a new member of the family of human ETS transcription factors. Cell Growth Differ. 3:705-713. [PubMed] [Google Scholar]
- 67.Yordy, J. S., R. Li, V. I. Sementchenko, H. Pei, R. C. Muise-Helmericks, and D. K. Watson. 2004. SP100 expression modulates ETS1 transcriptional activity and inhibits cell invasion. Oncogene 23:6654-6665. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, L., A. Eddy, Y.-T. Teng, M. Fritzler, M. Kluppel, F. Melet, and A. Bernstein. 1995. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol. Cell. Biol. 15:6961-6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, L., V. Lemarchandel, P. H. Romeo, Y. Ben-David, P. Greer, and A. Bernstein. 1993. The Fli-1 proto-oncogene, involved in erythroleukemia and Ewing's sarcoma, encodes a transcriptional activator with DNA-binding specificities distinct from other Ets family members. Oncogene 8:1621-1630. [PubMed] [Google Scholar]
- 70.Zhang, X. K., O. Moussa, A. LaRue, S. Bradshaw, I. Molano, D. D. Spyropoulos, G. S. Gilkeson, and D. K. Watson. 2008. The transcription factor Fli-1 modulates marginal zone and follicular B cell development in mice. J. Immunol. 181:1644-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng, C., R. Yang, Z. Han, B. Zhou, L. Liang, and M. Lu. 2008. TPO-independent megakaryocytopoiesis. Crit. Rev. Oncol. Hematol. 65:212-222. [DOI] [PubMed] [Google Scholar]