Abstract
Polyamines critically regulate all mammalian cell growth and proliferation by mechanisms such as the repression of growth-inhibitory proteins, including JunD. Decreasing the levels of cellular polyamines stabilizes JunD mRNA without affecting its transcription, but the exact mechanism whereby polyamines regulate JunD mRNA degradation has not been elucidated. RNA-binding proteins HuR and AUF1 associate with labile mRNAs bearing AU-rich elements located in the 3′ untranslated regions (3′-UTRs) and modulate their stability. Here, we show that JunD mRNA is a target of HuR and AUF1 and that polyamines modulate JunD mRNA degradation by altering the competitive binding of HuR and AUF1 to the JunD 3′-UTR. The depletion of cellular polyamines enhanced HuR binding to JunD mRNA and decreased the levels of JunD transcript associated with AUF1, thus stabilizing JunD mRNA. The silencing of HuR increased AUF1 binding to the JunD mRNA, decreased the abundance of HuR-JunD mRNA complexes, rendered the JunD mRNA unstable, and prevented increases in JunD mRNA and protein in polyamine-deficient cells. Conversely, increasing the cellular polyamines repressed JunD mRNA interaction with HuR and enhanced its association with AUF1, resulting in an inhibition of JunD expression. These results indicate that polyamines modulate the stability of JunD mRNA in intestinal epithelial cells through HuR and AUF1 and provide new insight into the molecular functions of cellular polyamines.
JunD is a basic region leucine zipper DNA-binding protein belonging to the family of Jun proteins that function as primary components of the activating protein 1 (AP-1) transcription factors (14). Jun proteins can form AP-1 homodimers or heterodimers among themselves or with members of the related Fos or ATF (activating transcription factor) protein families and regulate the transcription of target genes by binding to specific promoter DNA elements such as TGAGTCA and TGACGTCA (17, 41, 58, 59). All three Jun proteins (c-Jun, JunB, and JunD) are similar in DNA-binding affinity, but their patterns of expression vary in response to stress and during cell proliferation and transformation (6, 10, 17, 48, 56, 59). Although c-Jun and JunB behave as immediate-early response genes and enhance the G1-to-S-phase transition upon mitogenic stimulation, the overexpression of JunD inhibits cell proliferation (14, 29, 38). JunD also regulates the expression of genes involved in antioxidant defense and hydrogen peroxide production (10, 26, 37) and reduces tumor angiogenesis by repressing vascular endothelial growth factor transcription (3, 10). Mice lacking JunD exhibit multiple defects in their reproductive system (47), enhanced cardiomyocyte apoptosis and hypertrophic growth (15), chronic kidney disease (42), and increased bone formation (20). Our previous studies have shown that JunD plays an important role in the maintenance of normal intestinal epithelial integrity by modulating the transcription of cyclin-dependent kinase 4 (CDK4) (59) and zonula occludens-1 genes (9) through dimerization with ATF2 (58, 59).
The natural polyamines spermidine and spermine and their precursor putrescine (Put) are organic cations found in all eukaryotic cells. They have been long recognized as key molecules that control multiple signaling pathways and distinct cellular functions (8, 11). The levels of cellular polyamines are tightly regulated and depend on the dynamic balance among polyamine biosynthesis, degradation, and transport (11, 50, 52). Cellular polyamine content increases rapidly in cells stimulated to grow and divide (7, 49), whereas decreasing cellular polyamines stops cell cycle progression and causes growth arrest in the G1 phase (27, 40). Studies from our laboratory (27, 28, 40, 49-51, 60, 62) and other laboratories (36, 45) show that in normal intestinal mucosa, growth and repair after injury require the supply of polyamines to the dividing cells in the crypts. These studies also have shown that reducing cellular polyamines by inhibiting ornithine decarboxylase (ODC), the rate-limiting enzyme in polyamine biosynthesis (11), represses intestinal epithelial cell (IEC) renewal and delays wound healing in vivo and in vitro. Although the exact molecular processes governed by polyamines remain largely unknown, polyamines are shown to regulate IEC proliferation by controlling the expression of growth-related genes (11, 32, 52). In this regard, polyamines negatively regulate the posttranscription of the JunD gene, and the depletion of cellular polyamines stabilizes JunD mRNA without effect on its transcription (29). However, the exact mechanisms whereby polyamines modulate the stability of JunD mRNA at the molecular level remain to be investigated.
The mRNAs in mammalian cells typically are targeted for rapid degradation through a process involving the interaction of specific mRNA sequences (cis elements) with specific trans-acting factors such as RNA-binding proteins (RBPs) and microRNAs (18, 21). U- and UA-rich elements (AREs) are the best-characterized cis-acting sequences located in the 3′ untranslated regions (3′-UTRs) of many labile mRNAs (5, 55). Many RBPs selectively recognize and bind to mRNAs bearing AREs, and such ribonucleoprotein (RNP) associations modulate mRNA stability and/or translation (12, 19, 21, 22, 39, 55). HuR (Hu-antigen R) and AUF1 (AU-binding factor 1) are the most extensively studied RBPs, and they are shown to exert opposing influence on the stability and translation of several target mRNAs (1, 2, 16, 23, 35, 44). HuR binds with high affinity and specificity to target mRNAs and increases their stability and/or translation (16, 32-34). In contrast, AUF1 originally was identified as an RBP that promoted mRNA decay (23, 35, 44). AUF1 is expressed as four isoforms (p37, p40, p42, and p45) arising through the alternative splicing of a common pre-mRNA and causes the destabilization of some target transcripts (4, 44), although in some instances AUF1 enhanced mRNA stability (45, 46) and translation (30). Little is known about the roles of HuR and AUF1 and their functional interactions in the regulation of JunD mRNA stability.
Our previous studies showed that polyamines modulate HuR subcellular localization in IECs (63, 64), as decreasing cellular polyamines increased cytoplasmic HuR levels leading to the stabilization of several mRNAs, such as those encoding p53, nucleophosmin (NPM), ATF2, XIAP, and MEK-1 (53, 58, 61, 63, 64). Given that HuR and AUF1 can bind target transcripts on both distinct, nonoverlapping sites and on common sites in a competitive fashion (23, 35), and that en masse analyses revealed that both HuR and AUF1 could associate with JunD mRNA (33, 35), we set out to assess the association of endogenous HuR and AUF1 with JunD mRNA after modulating the cellular polyamine levels and to investigate their influence upon JunD mRNA stability. Our results indicate that HuR and AUF1 jointly bind JunD mRNA and control its stability. HuR was found to stabilize JunD mRNA, whereas AUF1 destabilized it. Moreover, polyamines regulated JunD mRNA stability by modulating the competitive binding of HuR and AUF1 with the JunD 3′-UTR.
MATERIALS AND METHODS
Chemicals and cell culture.
Tissue culture medium and dialyzed fetal bovine serum were from Invitrogen (Carlsbad, CA), and biochemicals were from Sigma (St. Louis, MO). The antibodies recognizing JunD, HuR, AUF1, TIAR T-cell-restricted intracellular antigen 1-related protein), and β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and BD Bioscience, and the secondary antibody conjugated to horseradish peroxidase was from Sigma. α-Difluoromethylornithine (DMFO) was purchased from Genzyme (Cambridge, MA).
The IEC-6 cell line, derived from normal rat intestinal crypt cells (43), was used at passages 15 to 20; cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% heat-inactivated fetal bovine serum. ODC-overexpressing IEC-6 (ODC-IEC) cells were developed as described in our previous studies (31, 63) and expressed a more stable ODC variant with full enzyme activity. Caco-2 cells (a human colon carcinoma cell line) were obtained from the American Type Culture Collection (ATCC) at passage 16. They were maintained in Eagle's minimum essential medium with 10% heat-inactivated fetal bovine serum and 50 μg/ml gentamicin, and passages 18 to 23 were used for the experiments as described previously (29, 40).
Plasmid construction.
Recombinant adenoviral plasmids containing human HuR were constructed by using the Adeno-X expression system according to the protocol provided by the manufacturer (Clontech) (55, 61). pAdeno-X, which was the recombinant replication-incompetent adenovirus carrying no HuR cDNA insert (Adnull), served as the control adenovirus. The AUF1 expression vector was described previously (24).
The chimeric firefly luciferase reporter construct of the JunD 3′-UTR was generated as described previously (32, 61). The full-length 558-bp JunD 3′-UTR and its different ARE fragments were amplified and subcloned into the pGL3-Luc plasmid (Promega) at the XbaI site to generate various chimeric pGL3-Luc-JunD-3′UTR reporter constructs. The sequence and orientation of the fragment in the luciferase reporter were confirmed by DNA sequencing and enzyme digestion. Transient transfections were performed using the Lipofectamine reagent and performed as recommended by the manufacturer (Invitrogen). The luciferase reporter constructs were transfected into cells along with phRL-null, a Renilla luciferase control reporter vector from Promega, to monitor transfection efficiencies as described previously (53). Luciferase activity was measured using the dual luciferase assay system, and the levels of the pGL3-Luc-JunD-3′UTR luciferase activity were normalized to Renilla luciferase activity in every experiment.
RNA interference.
The silencing RNA duplexes that were designed to specifically cleave HuR mRNA were synthesized and transfected into cells as described previously (53, 61). The sequence of small interfering RNA (siRNA) that specifically targets HuR mRNA (siHuR) was AACACGCTGAACGGCTTGAGG, while the sequence of the control siRNA (C-siRNA) was AAGTGTAGTAGATCACCAGGC. The siRNA specifically targeting AUF1 mRNA (siAUF1, CACUCUGAAGUUAGAUCCUAUCACA) was obtained from Invitrogen. Cells were harvested for various assays 48 h after transfection.
Western blot analysis.
Whole-cell lysates were prepared using 2% SDS, sonicated, and centrifuged (12,000 rpm) at 4°C for 15 min. The supernatants were boiled for 5 min and size-fractionated by SDS-PAGE (7.5% acrylamide). After transferring proteins onto nitrocellulose filters, the blots were incubated with primary antibodies recognizing HuR, AUF1, or JunD. Following incubations with secondary antibodies, immunocomplexes were developed by using chemiluminescence.
RT-PCR and real-time PCR analysis.
Total RNA was isolated by using an RNeasy mini kit (Qiagen, Valencia, CA) and was used in reverse transcription-PCR (RT-PCR) amplification reactions as described previously (57). PCR primers for detecting JunD mRNA were ATGGACACGCAGGAGCGCAT and AGCAGCTGGCAGCCGCTGTT (sense and antisense, respectively). The levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PCR product were assessed to monitor the evenness in RNA input in RT-PCR samples. Real-time quantitative PCR (Q-PCR) analysis was performed using 7500-fast real-time PCR systems with specific primers, probes, and software (Applied Biosystems, Foster City, CA).
Biotin pulldown assays.
The synthesis of biotinylated transcripts and analysis of RBPs bound to biotinylated RNA were done as previously described (63). cDNA from IEC-6 cells was used as a template for the PCR amplification of the coding region (CR) and 3′-UTR of JunD. The 5′ primers contained the T7 RNA polymerase promoter sequence [(T7) CCAAGCTTCTAATACGACTCACTATAGGGAGA]. To prepare the JunD CR template (spanning positions 264 to 1289), primers (T7)ATGGAAACGCCCTTCTATGGC and TCCCGCGCTCGGGCTCAATAC were used. To prepare the JunD 3′-UTR template (spanning position 1289 to 1847), primers (T7)GTATTGAGCCCGAGCGCGGGA and AGCAGCTGGCAGCCGCTGTT were used. All sequences of oligonucleotides for the preparation of various short RNA probes for mapping the JunD 3′-UTR are described in the supplemental material (see Table S1 in the supplemental material). PCR-amplified products were used as templates to transcribe biotinylated RNAs by using T7 RNA polymerase in the presence of biotin-CTP as described previously (61). Biotinylated transcripts (6 μg) were incubated with 120 μg of cytoplasmic lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway) and analyzed by Western blot analysis.
RNP IP assays.
To assess the association of endogenous HuR and AUF1 with endogenous JunD mRNA, the immunoprecipitation (IP) of ribonucleoprotein (RNP) complexes was performed as described previously (53, 61). Twenty million cells were collected per sample, and lysates were used for IP for 4 h at room temperature in the presence of excess (30 μg) IP antibody (IgG, anti-HuR, anti-AUF1, or anti-TIAR). RNA in IP materials was used in RT followed by PCR and Q-PCR analysis to detect the presence of JunD and GAPDH mRNAs.
Immunofluorescence staining.
Immunofluorescence was performed as described previously (64). Cells were fixed using 3.7% formaldehyde, and the rehydrated samples were incubated overnight at 4°C with primary antibody anti-HuR or anti-AUF1 diluted 1:300 in blocking buffer and then incubated with secondary antibody conjugated with Alexa Fluor-594 (Molecular Probes, Eugene, OR) for 2 h at room temperature. After being rinsed, slides were incubated with 1 μM TO-PRO3 (Molecular Probes) for 10 min to stain nuclei, rinsed again, mounted, and viewed through a Zeiss confocal microscope (model LSM410). Images were processed using Photoshop software (Adobe, San Jose, CA).
Assays for ODC enzyme activity and cellular polyamine content.
ODC activity was determined by a radiometric technique in which the amount of 14CO2 liberated from l-[1-14C]ornithine was estimated (50), and enzymatic activity was expressed as picomoles of CO2 per milligram of protein per hour. The cellular polyamine content was analyzed by high-performance liquid chromatography (HPLC) analysis as described previously (31). After 0.5 M perchloric acid was added, the cells were frozen at −80°C until ready for extraction, dansylation, and HPLC analysis. The standard curve encompassed 0.31 to 10 μM. Values that fell >25% below the curve were considered undetectable. The results are expressed as nanomoles of polyamines per milligram of protein.
Statistics.
Values are shown as the means ± standard errors (SE) from three to six samples. Immunoblotting and RNP IP assays were repeated three times. The significance of the difference between means was determined by analysis of variance. The level of significance was determined using Duncan's multiple range test (13).
RESULTS
Polyamine depletion increases the association of JunD mRNA with HuR and reduces its binding to AUF1.
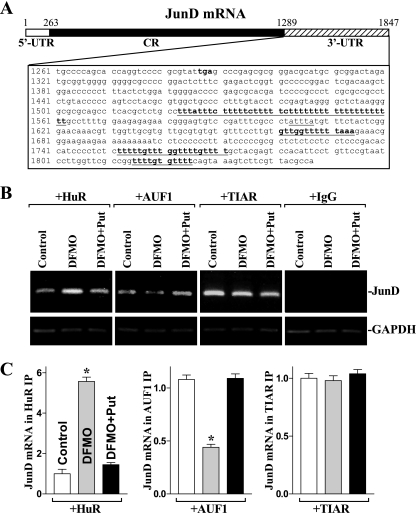
Decreased levels of cellular polyamines are shown to stabilize JunD mRNA (29), but the exact mechanism underlying this process remains unknown. There are three computationally predicted hits of the HuR or AUF1 motif in the JunD mRNA 3′-UTR (Fig. 1A), suggesting that JunD mRNA is a direct target of HuR and/or AUF1. To define the roles of HuR and AUF1 in the regulation of JunD mRNA stability after polyamine depletion, the following four sets of experiments were performed. First, we examined if JunD mRNA associated with HuR and/or AUF1 by performing RNP IP assays using specific antibodies that recognized either HuR or AUF1 under conditions that preserved RNP integrity (19, 23). The interactions of JunD mRNA with HuR and AUF1 were examined by isolating RNA from the IP materials and subjecting them to RT, followed by either conventional PCR or real-time Q-PCR analyses. As shown in Fig. 1B and C (control lanes), the JunD PCR products were highly enriched in samples immunoprecipitated with antibodies that recognized HuR, AUF1, or TIAR compared to levels for control IgG samples in IEC-6 cells (as well as in Caco-2 cells; data not shown). The enrichment of c-Myc PCR product also was examined and served as a positive control (data not shown), since c-Myc mRNA is a target of both HuR (32) and AUF1 (30). In addition, the amplification of GAPDH PCR products, found in all samples as a low-level contaminating housekeeping transcript (not HuR, AUF1, or TIAR targets), served to monitor the evenness of sample input, as reported previously (1, 61).
FIG. 1.
Polyamine depletion increases HuR binding to the JunD mRNA but decreases AUF1 association with the JunD transcripts in IEC-6 cells. (A) Schematic representation of the JunD mRNA and the predicted hits of the HuR or AUF1 signature motif in its 3′-UTR, indicated by underlining. (B) Association of endogenous HuR, AUF1, or TIAR with endogenous JunD mRNA in cells exposed to DFMO (5 mM) alone or DFMO plus putrescine (Put; 10 μM) for 6 days as measured by RNP immunoprecipitation (IP) analysis. After IP of RNA-protein complexes from cell lysates using antibodies (Ab) recognizing HuR, AUF1, or TIAR or with control IgG, RNA was isolated and used in RT reactions. The top panel shows representative RT-PCR products of JunD mRNA visualized in ethidium bromide-stained agarose gels; the low-level amplification of GAPDH (a housekeeping mRNA that is not a target of HuR or AUF1) served as a loading control. (C) Fold differences in JunD mRNA abundance in HuR, AUF1, or TIAR RNP IP compared to that of IgG IP, as measured by Q-PCR analysis. Values were the means ± SE from six samples. *, P < 0.05 compared to results for controls and cells treated with DFMO plus Put.
Second, we determined if polyamine depletion altered JunD mRNA interaction with HuR, AUF1, and TIAR. Consistently with our previous studies (27, 29), the inhibition of ODC activity by treatment with 5 mM DFMO almost completely depleted cellular polyamines in IEC-6 cells. The levels of putrescine and spermidine were undetectable at 6 days after treatment with DFMO, and spermine was decreased by ∼60% (data not shown). Results presented in Fig. 1B and C (left) showed that polyamine depletion increased the amount of HuR-JunD mRNA complex, as indicated by an increase in the levels of JunD mRNA in the HuR-IP materials from DFMO-treated cells when measured by RT followed by conventional PCR and Q-PCR analyses, but this induction was prevented by putrescine given together with DFMO. In contrast, decreased levels of cellular polyamines repressed the formation of AUF1-JunD mRNA complexes (Fig. 1B and C, +AUF). The levels of JunD mRNA were decreased in the AUF1 IP materials in DFMO-treated cells, which were overcome by exogenous putrescine. Polyamine depletion failed to alter TIAR binding to the JunD mRNA, because there was no significant change in the levels of TIAR-JunD mRNA complex in control relative to DFMO-treated cells with or without putrescine (Fig. 1B and C, +TIAR). In addition, the JunD mRNA was undetectable in nonspecific IgG IPs in all treatment groups (Fig. 1B, +IgG).
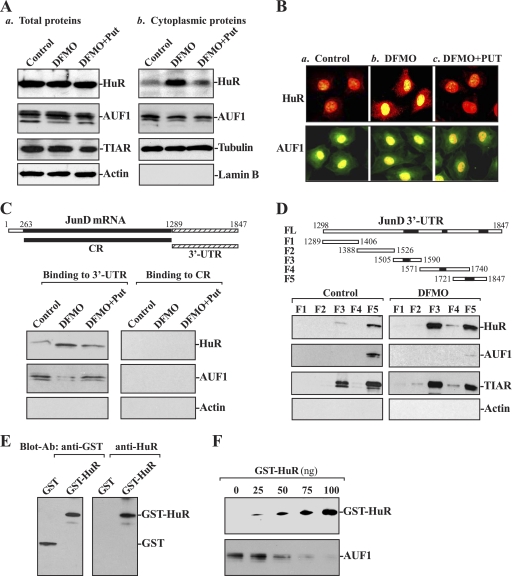
Third, we determined whether polyamine depletion alters whole-cell HuR and AUF1 levels and/or their subcellular distribution, and we further examined if polyamines regulate HuR and/or AUF1 association with the JunD mRNA through their interactions with the JunD 3′-UTR by using biotinylated transcripts that spanned the JunD mRNA regions. As shown in Fig. 2A, polyamine depletion by DFMO did not alter whole-cell levels of HuR, AUF1, and TIAR proteins, but it increased cytoplasmic HuR levels without significantly changing the subcellular distribution of AUF1. Supplementation with putrescine reversed the DFMO-triggered changes in HuR subcellular distribution, as did spermidine supplementation (data not shown). To monitor the quality and abundance of the cytoplasmic fractions, we examined the levels of β-tubulin (a cytoplasmic protein) and lamin B (a nuclear protein). The assessment of these markers revealed that there was no contamination between cytoplasmic and nuclear fractions. Immunofluorescence staining study further showed that HuR immunostaining was low in the cytoplasm of control cells (Fig. 2B, image a), but it increased significantly in DFMO-treated cells (Fig. 2B, image b), associated with a decrease in its nuclear levels. Combined treatment with DFMO and putrescine prevented the increased cytoplasmic HuR signal, rendering the subcellular HuR staining pattern similar to that of control cells (Fig. 2B, image c). Consistently with the Western blotting results, there were no changes in AUF1 immunostaining between controls and cells exposed to DFMO alone or DFMO plus putrescine (Fig. 2B, bottom). The JunD 3′-UTR transcript readily associated with cytoplasmic HuR and AUF1, as detected by the Western blot analysis of the pulldown material (Fig. 2C, control lanes). The binding intensity of HuR increased significantly when using lysates prepared from cells that were treated with DFMO, but it was reduced when cells had been treated with DFMO plus putrescine. This increase in the levels of HuR binding to JunD 3′-UTR is specific, because transcripts corresponding to the JunD CR did not bind to HuR in controls and cells exposed to DFMO alone or DFMO plus putrescine (Fig. 2C, right). On the other hand, the association of AUF1 with the JunD 3′-UTR was decreased in DFMO-treated cells, and this reduction was completely prevented by putrescine. AUF1 did not interact with the JunD CR regardless of the presence or absence of cellular polyamines.
FIG. 2.
Changes in the binding of HuR and AUF1 to the 3′-UTR and coding region (CR) of JunD mRNA in cells described in Fig. 1. (A) Representative immunoblots of HuR, AUF1, and TIAR after polyamine depletion. Shown are total protein levels (a) and cytoplasmic proteins (b). Whole-cell lysates and cytoplasmic proteins were prepared and subjected to SDS-PAGE. Equal loading was monitored by the immunoblotting of β-actin in whole-cell lysates and β-tubulin in cytoplasmic proteins. (B) Cellular distributions of HuR and AUF1 in cells described for panel A. Shown are the control (a), DFMO (b), and DFMO plus putrescine (c). After HuR (red) or AUF1 (green) immunostaining (see Materials and Methods), nuclei (yellow) were visualized with TO-PRO3. Original magnification, ×1,000. (C) Representative HuR and AUF1 immunoblots using the pulldown materials by different fractions of JunD mRNA. The top panel shows a schematic representation of the JunD-biotinylated transcripts (CR and 3′-UTR) used in this study. The left panel shows the binding of HuR or AUF1 to the 3′-UTR. The right panel shows HuR or AUF1 binding to CR. Cytoplasmic lysates prepared from control cells, and cells exposed to DFMO alone or DFMO plus Put were incubated with 6 μg of biotinylated JunD 3′-UTR or CR for 30 min at 25°C, and the resulting RNP complexes were pulled down by using streptavidin-coated beads. The presence of HuR and AUF1 in the pulldown material was assayed by Western blotting. β-Actin in the pulldown material also was examined and served as a negative control. (D) Representative HuR, AUF1, and TIAR immunoblots in the material pulled down by different biotinylated fractions of the JunD mRNA 3′-UTR. The top panel shows the schematic representation of the JunD 3′-UTR-biotinylated transcripts used in this study. The lower panel shows representative HuR, AUF1, and TIAR immunoblots in the material pulled down in control cells (left) and DFMO-treated cells (right) by using different biotinylated fractions of the JunD mRNA 3′-UTR (F1 to F5). (E) GST-HuR fusion protein identified by anti-GST antibody (left) or recognized by anti-HuR antibody (right). (F) Effect of GST-HuR added to the binding reaction on interactions of HuR and AUF1 with F5 of the JunD 3′-UTR. Various concentrations of GST-HuR were used; the levels of binding complexes were detected by pulldown assays. Three independent experiments were performed showing similar results.
To further examine whether the binding of HuR and AUF1 to the JunD 3′-UTR is mediated through the specific sites containing predicted hits of the HuR and/or AUF1 motif, partial biotinylated transcripts spanning the JunD 3′-UTR were prepared (Fig. 2D, schematic), and their associations with HuR or AUF1 in controls (Fig. 2D, left) and polyamine-deficient cells (Fig. 2D, right) were tested by pulldown assays. HuR was found to bind to F3 and F5, two transcripts that contained HuR motif hits, but it did not bind to F1, F2, and F4 (although F4 also contained a potential HuR or AUF1 hit). In contrast, AUF1 only bound to F5 in control cells, and this interaction was almost undetectable in polyamine-deficient cells. In addition, TIAR also bound to F3 and F5, but its binding activity was not altered by polyamine depletion, which served as a control in this study. Consistently with the sequences of previously reported HuR and AUF1 targets (33, 35), F3 and F5 contained higher percentages of AU-rich elements than those identified in F1, F2, and F4 (see Table S2 in the supplemental material).
Fourth, we examined the effect of GST-HuR added to the binding reaction on AUF1 association with F5 of the JunD 3′-UTR. The construct expressing the glutathione S-transferase (GST)-HuR fusion protein was generated, and its product GST-HuR protein was characterized (Fig. 2E). As shown in Fig. 2F, F5 association with HuR was progressively increased when increasing concentrations of GST-HuR were added to the binding reaction, but its interaction with AUF1 was reduced with increasing GST-HuR levels. Neither HuR binding nor AUF1 binding to F5 of JunD 3′-UTR was affected by GST added to the binding reaction (data not shown). Taken together, these findings indicate that HuR and AUF1 competitively bind to the JunD mRNA and that polyamine depletion enhances HuR levels in the cytoplasm and HuR interaction with the JunD mRNA via its 3′-UTR, while AUF1 binding to the JunD transcript is reduced.
Increasing cellular polyamines reduces HuR-JunD mRNA complex and enhances AUF1 binding to the JunD mRNA.
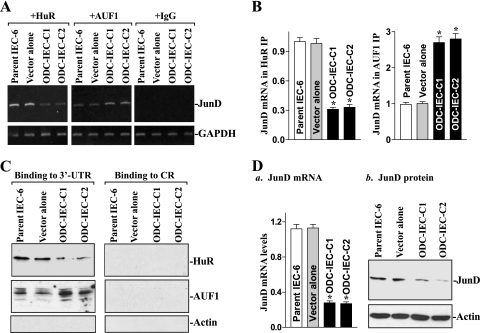
To determine the effect of increasing the levels of cellular polyamines on JunD mRNA interaction with HuR and AUF1, two clonal populations of IECs stably expressing ODC (ODC-IEC) (31) were used in this study. Consistently with our previous studies (31, 63), ODC-IECs exhibited very high levels of ODC protein and greater-than 50-fold increases in ODC enzyme activity. The levels of putrescine, spermidine, and spermine in stable ODC-IECs were increased by ∼12-fold, ∼2-fold, and ∼25%, respectively, compared to those of cells transfected with the control vector lacking ODC cDNA (see Table S3 in the supplemental material). Increasing cellular polyamines by ODC overexpression repressed the formation of HuR-JunD mRNA complexes (Fig. 3A and B, left), but it enhanced AUF1 association with the JunD mRNA (Fig. 3A, middle, and B, left). The levels of JunD mRNA associated with HuR were decreased in stable ODC-IECs compared to those observed in parent IEC-6 cells and in cells transfected with the control vector, whereas the levels of JunD mRNA associated with AUF1 were increased in cells overexpressing ODC. These changes in JunD mRNA interactions with HuR and AUF1 following increased levels of cellular polyamines were mediated through the JunD 3′-UTR, because neither HuR nor AUF1 bound to the transcript corresponding to the JunD CR in stable ODC-IECs (Fig. 3C, right). Results presented in Fig. 3D further show that increasing cellular polyamines by ODC overexpression also repressed JunD expression, as shown by a decrease in the levels of JunD mRNA and protein. These results indicate that increased levels of cellular polyamines repress JunD expression by inhibiting HuR binding to the JunD mRNA and by inducing AUF1 association with JunD mRNA.
FIG. 3.
Increasing cellular polyamines repress JunD mRNA association with HuR and enhance its binding to AUF1. (A) Changes in the association of endogenous HuR and AUF1 with endogenous JunD mRNA in parent IEC-6 cells, controls (vector alone), and stable ODC-IEC cells (clones C1 and C2). IEC-6 cells were infected with either the retroviral vector containing the sequence encoding ODC cDNA or a control retroviral vector lacking the ODC cDNA. Clones resistant to the selection medium containing 0.6 mg/ml G418 were isolated and screened for ODC expression. Whole-cell lysates were tested in IP reactions with antibodies against HuR or AUF1 or with nonspecific IgG. RNA in the IP material was used in RT-PCRs to detect the presence of JunD mRNA; the resulting PCR products of JunD and GAPDH mRNAs were visualized in agarose gels. (B) Quantitative analysis derived from densitometric scans of PCR-amplified products of JunD mRNA described for panel A. Values were normalized to the amounts of GAPDH and expressed as means ± SE from three samples. *, P < 0.05 compared to results for parent IEC-6 cells and cells infected with control vector. (C) Representative immunoblots of HuR and AUF1 after pulldown using JunD 3′-UTR (left) or CR (right) in cells described for panel A. Cytoplasmic lysates were incubated with the biotinylated JunD 3′-UTR or CR, and the resulting RNP complexes were pulled down by using streptavidin-coated beads. Levels of HuR and AUF1 proteins in the pulldown materials were measured by Western blot analysis. (D) Levels of JunD expression in cells described for panel A: JunD mRNA (a) and JunD protein (b). Total cellular RNA was isolated, and the levels of JunD and GAPDH mRNAs were measured by Q-PCR analysis. Values are the means ± SE from triplicate samples. *, P < 0.05 compared to parent IEC-6 cells and controls. Levels of JunD protein were examined by Western blot analysis, and equal loading was monitored by β-actin immunoblotting.
HuR silencing prevents the increased stability of JunD mRNA in polyamine-deficient cells.
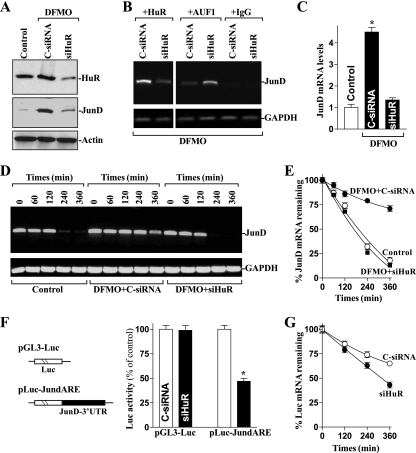
To directly examine the putative role of HuR in the increased JunD mRNA stability in polyamine-deficient cells, small interfering RNA (siRNA) targeting the HuR mRNA (siHuR) was used to reduce HuR levels. With >95% of cells transfected (data not shown), siHuR potently and specifically silenced HuR expression in polyamine-deficient cells (Fig. 4A). As shown in Fig. 4B, silencing HuR in polyamine-deficient cells decreased HuR-JunD mRNA complex by ∼70%, but it increased the amount of JunD mRNA associated with AUF1 (by ∼2.6-fold). HuR silencing also prevented the increased levels of total JunD mRNA (Fig. 4C). This reduction in JunD mRNA by HuR silencing resulted primarily from the destabilization of JunD transcript, because the half-life of JunD mRNA in DFMO-treated cells transfected with siHuR was similar to that measured in control cells as determined by RT-PCR (Fig. 4D) and Q-PCR analyses (Fig. 4E). Furthermore, in HuR-silenced populations, the increase in JunD protein levels after polyamine depletion also was prevented (Fig. 4A, middle) and was reduced to levels similar to those seen in control cells. Transfection with C-siRNA had no effect on the JunD mRNA stability and the levels of JunD protein in polyamine-deficient cells.
FIG. 4.
Effect of HuR silencing on JunD mRNA stability in polyamine-deficient cells. (A) Representative HuR and JunD immunoblots. After cells were cultured in the presence of DFMO for 4 days, they were transfected with either siRNA targeting the HuR mRNA coding region (siHuR) or control siRNA (C-siRNA), and whole-cell lysates were harvested 48 h thereafter. The levels of HuR and JunD proteins were measured by Western blot analysis, and equal loading was monitored by β-actin immunoblotting. (B) Changes in the association of endogenous HuR and AUF1 with endogenous JunD mRNA in cells described for panel A. Whole-cell lysates were used for RNP IP, and the levels of JunD mRNA in the IP materials were measured by RT-PCR analysis. (C) Levels of JunD mRNA in cells treated as described for panel A. Total RNA from each group was harvested, and JunD mRNA levels were measured by Q-PCR analysis. The data were normalized to the amount of GAPDH mRNA, and the values are represented as the means ± SE of data from six samples. *, P < 0.05 compared to results for controls and DFMO-treated cells transfected with C-siRNA. (D) Half-life of the JunD mRNA in cells that were transfected and treated as described for panel A. Total cellular RNA was isolated at the indicated times after the administration of actinomycin D, and the remaining levels of JunD and GAPDH mRNAs were measured by RT-PCR analysis. (E) Percentages of JunD mRNA remaining in cells that were treated as described for panel D as measured by Q-PCR analysis. Values are means ± SE from triplicate samples. (F) Changes in JunD 3′-UTR activity as measured by using pGL3-Luc-JundARE reporter assays in cells that were processed as described for panel A. On the left is a schematic of plasmids, namely, control (pGL3-Luc) and chimeric firefly luciferase (Luc)-JunD 3′-UTR (pLuc-JundARE). On the right are levels of luciferase reporter activity of the JunD-3′UTR. The pLuc-JundARE or pGL3-Luc (negative control) plasmid was cotransfected with a Renilla luciferase reporter, and firefly and Renilla luciferase activities were assayed 24 h thereafter. Luciferase values were normalized by transfection efficiency and expressed as the means ± SE of data from six samples. *, P < 0.05 compared to results for cells transfected with C-siRNA. (G) The half-life of the Luc mRNA expressed from pLuc-JundARE in cells described for panel F. After the administration of actinomycin D, total RNA was isolated at the indicated times and the remaining levels of Luc mRNA were measured by Q-PCR analysis. Data were normalized to GAPDH mRNA levels.
To further determine if HuR regulates the stability of JunD mRNA by interacting with its 3′-UTR, we used a firefly luciferase reporter gene construct containing the JunD 3′-UTR (pLuc-JundARE) and the negative-control vector pGL3-Luc (Fig. 4F, schematic). A plasmid expressing Renilla luciferase was cotransfected as an internal normalization control. HuR silencing reduced the levels of luciferase activity of pLuc-JundARE, and this decrease was due to the destabilization of luciferase transcript, because the half-life of luciferase mRNA expressed from pLuc-JundARE decreased in HuR-silenced cells compared to that observed in cells transfected with C-siRNA (Fig. 4G). These findings strongly suggest that HuR stabilizes JunD mRNA by interacting with its 3′-UTR and that the stabilization of JunD mRNA by polyamine depletion results from both the enhancement of its interaction with HuR and the reduction of its interaction with AUF1.
HuR overexpression induces JunD expression by stabilizing its mRNA.
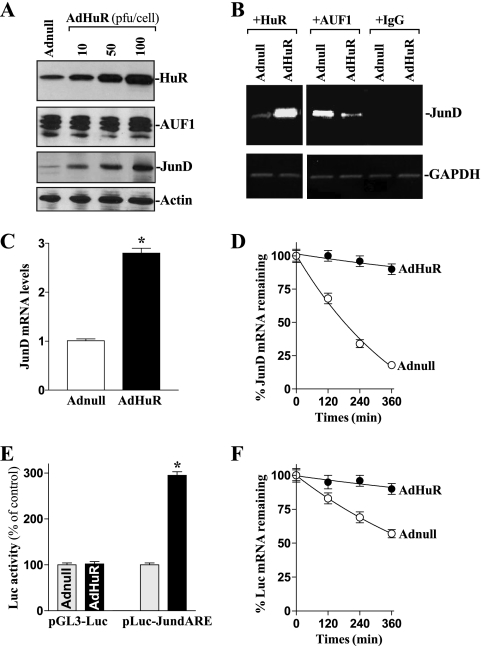
To investigate the exact role of HuR in the regulation of JunD mRNA stability, we examined the effect of overexpressing wild-type HuR upon the half-life of JunD mRNA. The adenoviral vector containing the corresponding HuR cDNA under the control of the human cytomegalovirus immediate-early gene promoter (AdHuR) was generated and used as described previously (32). As shown, infection with AdHuR dramatically increased total HuR protein and also enhanced the expression of JunD, although it had no effect on AUF1 levels (Fig. 5A). HuR overexpression also induced the levels of HuR-JunD mRNA complex but decreased the amount of JunD mRNA with AUF1 (Fig. 5B), which was accompanied by an increase in total JunD mRNA levels (Fig. 5C) as a result of an induction in its stability (Fig. 5D). This stimulatory effect also was mediated by HuR interacting with the JunD 3′-UTR, because HuR overexpression increased reporter luciferase mRNA levels expressed from pLuc-JundARE (Fig. 5E), which is associated with enhanced luciferase mRNA stability (Fig. 5F), compared to levels for control adenovirus (Adnull) infections. These results indicate that HuR overexpression enhances JunD expression by stabilizing the JunD mRNA as a result of both increased HuR-JunD mRNA association and reduced AUF1 binding.
FIG. 5.
Changes in JunD mRNA stability after ectopic HuR overexpression. (A) Representative immunoblots of HuR, AUF1, and JunD proteins after HuR overexpression. Cells were infected with the recombinant adenoviral vector encoding HuR cDNA (AdHuR) or adenoviral vector lacking HuR cDNA (Adnull) at a multiplicity of infection of 10 to 100 PFU/cell; the levels of HuR, AUF1, and JunD proteins were analyzed 48 h after the infection. (B) Association of endogenous HuR and AUF1 with endogenous JunD mRNA in cells infected with AdHuR or Adnull at a concentration of 100 PFU/cell for 48 h. Using whole-cell lysates, the levels of JunD mRNA in HuR or AUF1 RNP complexes were measured by RT-PCR analysis. (C) Levels of JunD mRNA as measured by Q-PCR analysis in cells that were treated as described for panel B. Data were normalized to GAPDH mRNA levels, and values are shown as the means ± SE from six samples. *, P < 0.05 compared to results for cells infected with Adnull. (D) Half-life of the JunD mRNA as measured by Q-PCR analysis by using actinomycin D in the cells described for panel B. Values are the means ± SE from triplicate samples. (E) Changes in luciferase reporter activity as measured by using the pLuc-JundARE construct in cells that were processed as described for panel B. Twenty-four hours after cells were transfected with pLuc-JundARE or pGL3-Luc (negative control), the levels of luciferase activity were examined and the values expressed as the means ± SE of data from six samples. *, P < 0.05 compared with cells transfected with Adnull. (F) The half-life of the Luc mRNA expressed from pLuc-JundARE in cells that were treated as described for panel B and measured by using actinomycin D. The remaining levels of Luc mRNA at the indicated times were measured by Q-PCR analysis, and results were normalized to GAPDH mRNA levels.
AUF1-JunD mRNA interactions decrease the stability of JunD mRNA.
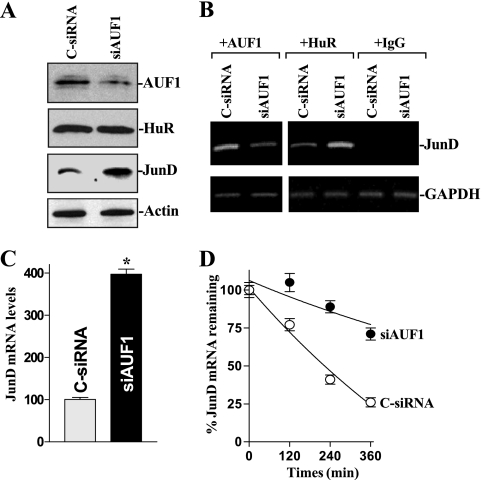
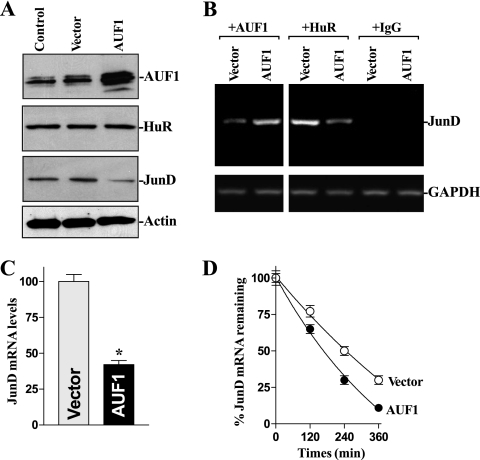
Given that AUF1 and HuR bind the JunD ARE and that modulating cellular polyamine levels has opposite effects on their binding to the JunD 3-UTR, we investigated the effect of the AUF1-JunD mRNA association upon JunD mRNA stability. To reduce AUF1 levels, cells were transfected with siRNA targeting the AUF1 mRNA (siAUF1). As shown in Fig. 6A, AUF1 silencing by transfection with siAUF1 induced JunD expression without altering HuR levels. AUF1 silencing decreased the amount of JunD mRNA associated with AUF1 and increased the levels of JunD mRNA associated with HuR (Fig. 6B). AUF1 silencing also increased levels of total JunD mRNA (Fig. 6C), and this induction in JunD mRNA was linked to an increase in its stability. The half-life of JunD mRNA was increased significantly in AUF1-silenced cells (Fig. 6D). In contrast, as shown in Fig. 7, ectopic AUF1 overexpression increased the levels of JunD mRNA associated with AUF1, reduced the amount of JunD mRNA associated with HuR, and reduced JunD abundance. This inhibition of JunD expression by AUF1 results in part from the destabilization of JunD mRNA, because the degradation of JunD mRNA increased in cells overexpressing AUF1 (Fig. 7D). Taken together, these results indicate that AUF1 inhibits JunD expression by destabilizing its mRNA, further supporting the notion that polyamine depletion stabilizes JunD mRNA both by decreasing the association of JunD mRNA with AUF1 and by increasing the abundance of the HuR-JunD mRNA complex.
FIG. 6.
Changes in JunD mRNA stability after AUF1 silencing. (A) Representative immunoblots of AUF1, HuR, and JunD proteins in AUF1-silenced cells. Forty-eight hours after transfecting cells with either siRNA targeting the AUF1 mRNA coding region (siAUF1) or control siRNA (C-siRNA), whole-cell lysates were harvested for Western blot analysis to monitor the expression of AUF1, HuR, and JunD and the loading control β-actin. (B) Association of endogenous AUF1 and HuR with endogenous JunD mRNA in cells described for panel A. JunD mRNA levels in the RNP complexes immunoprecipitated by using antibodies that recognized AUF1 or HuR were measured by RT-PCR analysis. (C) Levels of JunD mRNA in cells that were processed as described for panel A. Total RNA from each group was harvested, and the levels of JunD mRNA were measured by Q-PCR analysis. Values are means ± SE of data from six samples. *, P < 0.05 compared to results for cells transfected with C-siRNA. (D) Half-life of the JunD mRNA in cells described for panel A. Total cellular RNA was isolated at the indicated times after the administration of actinomycin D, and the remaining levels of JunD and GAPDH mRNAs were measured by Q-PCR analysis. Values were means ± SE from triplicate experiments.
FIG. 7.
Effect of ectopic AUF1 overexpression on JunD mRNA stability. (A) Representative immunoblots of AUF1, HuR, and JunD proteins after AUF1 overexpression. Cells were transfected with the AUF1 expression vectors (collectively indicated as AUF1) or control vector lacking AUF1 (vector); the levels of AUF1, HuR, and JunD proteins was analyzed by Western blot analysis 48 h after the transfection. (B) Association of endogenous AUF1 and HuR with endogenous JunD mRNA in cells described for panel A. Levels of JunD mRNA in the RNP complexes that were immunoprecipitated using anti-AUF1 or anti-HuR antibodies were measured by RT-PCR analysis. (C) Levels of JunD mRNA as measured by Q-PCR analysis in cells that were treated as described for panel A. Data were normalized to GAPDH mRNA levels, and the values are shown as the means ± SE of data from six samples. *, P < 0.05 compared with cells transfected with the vector alone. (D) Half-life of the JunD mRNA as measured by Q-PCR analysis by using actinomycin D in cells treated as described for panel A. Values are the means ± SE from triplicate samples.
Deletion of ARE-binding sites from the JunD 3′-UTR abolishes its regulatory effect.
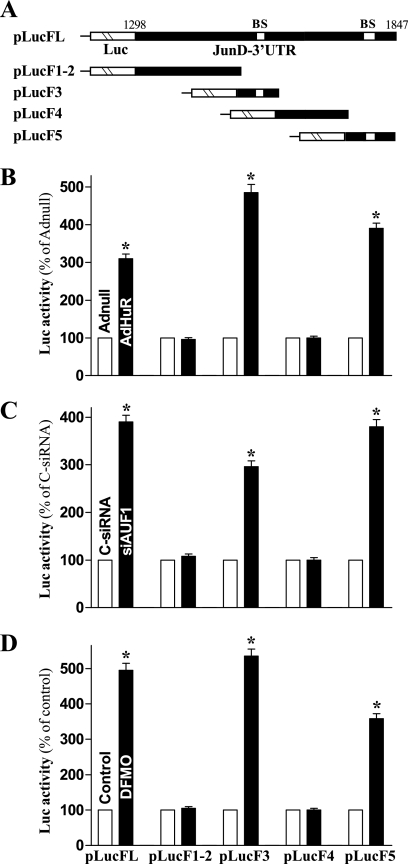
Based on observations from mapping the JunD 3′-UTR (Fig. 2C), HuR binds F3 and F5 of JunD transcripts, which contained HuR motif hits, whereas AUF1 only associates with F5. To determine if HuR and AUF1 regulate the stability of JunD mRNA by directly interacting with these specific ARE-binding sites, we prepared reporter constructs that expressed chimeric RNA containing the luciferase and partial transcripts spanning the JunD 3′-UTR with or without the ARE-binding sites, as shown in Fig. 8A. The results presented in Fig. 8B show that ectopic HuR overexpression increased the levels of luciferase reporter gene activity when cells were transfected with pLucFL (containing full-length JunD 3′-UTR), pLucF3, or pLucF5 (containing the ARE-binding sites), but it failed to induce the reporter activity of pLuc1-2 and pLucF4 in which the ARE-binding sites were deleted. On the other hand, the activation of the JunD 3′-UTR activity in AUF1-silenced cells also required these specific ARE-binding sites. AUF1 silencing increased the levels of reporter luciferase activity of pLucFL, pLucF3, and pLucF5 (Fig. 8C), although there were no significant differences in reporter activity between AUF1-silenced cells and cells transfected with C-siRNA when pLucF1-2 and pLucF4 were used. To further define the role of these ARE-binding sites in an induction of JunD mRNA stability following polyamine depletion, changes in JunD 3′-UTR-driven luciferase reporter activity were measured in polyamine-deficient cells by using various pLuc-JunD 3′-UTR reporter constructs. Consistently with results from the full-length 3′-UTR (pLucFL), the reporter activity also was substantially increased when DFMO-treated cells were transfected with pLucF3 and pLucF5 (Fig. 8C). However, this stimulatory effect disappeared when DFMO-treated cells were transfected with pLucF1-2 and pLucF4. Taken together, these results indicate that HuR and AUF1 regulate JunD mRNA stability by competitively binding to specific ARE sites within the JunD 3′-UTR.
FIG. 8.
Changes in the levels of JunD 3′-UTR luciferase reporter activity after deletion of HuR/AUF1 binding sites. (A) Schematic of plasmids of different chimeric firefly luciferase (Luc)-JunD 3′-UTR reporters. BS, HuR- and/or AUF1-binding site. (B) Activity of the pLuc-JundARE luciferase reporter with (pLucF3 and pLucF5) or without (pLucF1-2 and pLucF4) BS after HuR overexpression. Twenty- four hours after infection with AdHuR or Adnull, cells were cotransfected with pLuc-JundARE constructs or pGL3-Luc (negative control) and a Renilla luciferase reporter. Levels of firefly and Renilla luciferase activities were assayed 24 h later. Results were normalized to the Renilla luciferase activity and expressed as the means ± SE of data from triplicate experiments. *, P < 0.05 compared with cells infected with Adnull. (C) Activity of various pLuc-JundARE luciferase reporters after AUF1 silencing. Cells initially were transfected with either siAUF1 or C-siRNA for 24 h, and then pLuc-JundARE was cotransfected with the Renilla luciferase reporter. *, P < 0.05 compared to results for cells transfected with C-siRNA. (D) Activity of various pLuc-JundARE luciferase reporters in cells exposed to DFMO for 6 days. *, P < 0.05 compared to results for control cells.
DISCUSSION
Polyamines are essential for all mammalian cell proliferation and growth (8, 11, 52), but the exact roles of polyamines at the molecular level are not well understood. Our previous studies have shown that polyamines regulate JunD expression posttranscriptionally and that the depletion of cellular polyamines stabilizes JunD mRNA without affecting JunD transcription (29). In the present study, we highlight the novel functions of the RBPs HuR and AUF1 in the regulation of JunD mRNA stability and provide new evidence to show that polyamines modulate JunD mRNA turnover by altering the association of HuR and AUF1 with JunD mRNA. We identified JunD mRNA as a target of HuR and AUF1 and found that JunD mRNA stability increased with HuR association but decreased with AUF1 binding. Interestingly, altering the association of HuR with JunD mRNA affected the levels of mRNA associated with AUF1, which was regulated by cellular polyamines. Increasing polyamines destabilized JunD mRNA by repressing JunD mRNA association with HuR and concomitantly enhancing its interaction with AUF1. Conversely, increased JunD stabilization following polyamine depletion resulted from both the increased HuR binding and decreased AUF1 binding to the JunD 3′-UTR.
Our results indicate that JunD mRNA directly interacts with both HuR and AUF1 via its 3′-UTR and that the dynamic balance of their binding regulates JunD mRNA stability. Based on the present findings, we propose that HuR and AUF1 compete for binding to the JunD 3′-UTR in the cytoplasm. JunD mRNA association with HuR stabilizes JunD mRNA, as the JunD mRNA half-life decreased in HuR-silenced cells but increased in HuR-overexpressing cells. Our observations are consistent with studies from others who demonstrated that HuR binds to specific ARE-bearing target mRNAs and stabilizes them, as shown for mRNAs encoding p53, p21, ATF2, NPM, XIAP, c-Fos, and MEK-1 (2, 28, 53, 57, 61, 63). On the other hand, the association of AUF1 with JunD mRNA promoted its decay. Thus, JunD mRNA joins a few mRNAs whose stability is determined by competitive interaction between the stability-promoting HuR and the decay-promoting AUF1, notably the cyclin D1 and p21 mRNAs (23).
The most significant findings from the present study are that the competitive binding of HuR and AUF1 to the JunD mRNA in IECs is tightly regulated by polyamines. Decreasing the levels of cellular polyamines by inhibiting ODC with DFMO led to an increase in the levels of HuR in the cytoplasm and consequently elevated the abundance of HuR-JunD mRNA complexes and decreased the levels of JunD mRNA associated with AUF1 (Fig. 1 and 2), which caused JunD stabilization (29). Since increased HuR binding and decreased AUF1 binding to the JunD 3′-UTR in DFMO-treated cells were completely prevented by the addition of exogenous putrescine, the observed changes in JunD mRNA association with HuR and AUF1 more likely are related to polyamine depletion rather than to the nonspecific effect of DFMO. In addition, increasing cellular polyamines by ectopic ODC overexpression repressed the formation of HuR-JunD mRNA complex but promoted AUF1 association with the JunD mRNA, thus inhibiting JunD expression (Fig. 3).
The precise mechanisms by which cellular polyamines modulate HuR and AUF1 association with JunD mRNA are likely to be complex and multiple. The availability of HuR and AUF1 and their relative cytoplasmic abundances are important factors affecting the formation of either HuR-JunD mRNA or AUF1-JunD mRNA complex after changes in the levels of cellular polyamines. While neither whole-cell levels of HuR and AUF1 nor cytoplasmic AUF1 levels were altered by polyamine depletion, the cytoplasmic abundance of HuR increased significantly in polyamine-deficient cells (Fig. 2A and B), associated with an induction in HuR-JunD mRNA complex and a reduction in AUF1-bound JunD mRNA. At the same time, increasing cellular polyamines by ODC overexpression increased the AMP-activated protein kinase (AMPK) activity, which in turn lowered the cytoplasmic levels of HuR through importin α1-mediated events (64), allowing AUF1 to bind to the JunD mRNA (Fig. 3). Moreover, HuR silencing increased AUF1 binding to JunD mRNA and triggered the degradation of JunD mRNA and reduced the expression of JunD protein (Fig. 4), whereas HuR overexpression decreased the JunD mRNA association with AUF1, increased its interaction with HuR, and induced the stability of JunD transcript (Fig. 5). Conversely, the downregulation of AUF1 by siAUF1 increased HuR binding to JunD mRNA and stabilized it (Fig. 6). Consistently with these findings, AUF1 silencing also induced the binding of HuR to p21 and cyclin D1 mRNAs and increased their stability and protein levels (23). In addition, the function of AUF1 also is modulated by signal-induced phosphorylation and ubiquitination (25, 54), but their involvement in the regulation of JunD mRNA stability after altering the levels of cellular polyamines remains unknown and is the focus of our ongoing studies.
The results reported here also show that the JunD 3′-UTR contains a nonoverlapping binding site for HuR and a common site for both HuR and AUF1, because HuR interacted with the JunD transcript at segments F3 (spanning positions 1505 to 1590) and F5 (spanning positions 1721 to 1847), whereas AUF1 associated with the JunD mRNA at F5 only (Fig. 2D). Both F3 and F5 contained predicted hits of the HuR and/or AUF1 signature motif and were highly AU rich (see Table S2 in the supplemental material), which is consistent with the sequences of previously reported HuR and AUF1 targets (35). In this study, we did not identify the specific JunD 3′-UTR nucleotides with which HuR and AUF1 interact, as this would require more specialized biochemical, crystallographic, and molecular methods than those used here. The results presented in Fig. 8 show that the deletion of these binding sites abolished the JunD 3′-UTR-driven luciferase reporter activity, suggesting that HuR and AUF1 regulate the stability of JunD mRNA through direct interactions with these specific AREs. The AREs also were necessary for causing JunD stabilization following polyamine depletion, because increased reporter activity in DFMO-treated cells was prevented by deleting these binding sites (Fig. 8D).
In summary, these results indicate that HuR and AUF1 bind to the JunD 3′-UTR competitively and regulate JunD mRNA stability in IECs. The depletion of cellular polyamines increased HuR translocation to the cytoplasm but had no effect on the subcellular distribution of AUF1. HuR binding to the JunD mRNA increased in polyamine-deficient cells, but the levels of JunD transcript associated with AUF1 decreased. The silencing of HuR in polyamine-deficient cells not only increased AUF1 binding to the JunD mRNA but also rendered the JunD mRNA unstable, thus preventing the accumulation of JunD mRNA and protein. In contrast, increasing polyamines repressed JunD mRNA association with HuR and enhanced its interaction with AUF1, resulting in JunD mRNA decay. Since polyamines are required for maintaining intestinal epithelial integrity and their cellular levels are tightly regulated by the status of cell growth and stress stimulation, these findings suggest that the stabilization of JunD mRNA by preferential association with HuR contributes to the inhibition of IEC proliferation seen following polyamine depletion and plays an important role in intestinal epithelial homeostasis.
Supplementary Material
Acknowledgments
This work was supported by a merit review grant (to J.-Y.W.) from the Department of Veterans Affairs and by NIH grants DK-57819, DK-61972, and DK-68491 (to J.-Y.W.). J.-Y.W. is a Research Career Scientist, Medical Research Service, U.S. Department of Veterans Affairs. M.G. is supported by the NIA-IRP, NIH.
Footnotes
Published ahead of print on 30 August 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdelmohsen, K., R. Pullmann, Jr., A. Lal, H. H. Kim, S. Galban, X. Yang, J. D. Blethrow, M. Walker, J. Shubert, D. A. Gillespie, H. Furneaux, and M. Gorospe. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25:543-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelmohsen, K., Y. Kuwano, H. H. Kim, and M. Gorospe. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 389:243-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdul-Hafez, A., R. Shu, and B. D. Uhal. 2009. JunD and HIF-1α mediate transcriptional activation of angiotensinogen by TGF-β1 in human lung fibroblasts. FASEB J. 23:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arao, Y., A. Kikuchi, M. Kishida, M. Yonekura, A. Inoue, S. Yasuda, S. Wada, K. Ikeda, and F. Kayama. 2004. Stability of A+U-rich element binding factor 1 (AUF1)-binding messenger ribonucleic acid correlates with the subcellular relocalization of AUF1 in the rat uterus upon estrogen treatment. Mol. Endocrinol. 18:2255-2267. [DOI] [PubMed] [Google Scholar]
- 5.Bakheet, T., M. Frevel, B. R. Williams, W. Greer, and K. S. Khabar. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behmoaras, J., G. Bhangal, J. Smith, K. McDonald, B. Mutch, P. C. Lai, J. Domin, L. Game, A. Salama, B. M. Foxwell, C. D. Pusey, H. T. Cook, and T. J. Aitman. 2008. JunD is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nat. Genet. 40:553-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belting, M., L. Borsig, M. M. Fuster, J. R. Brown, L. Persson, L. A. Fransson, and J. D. Esko. 2002. Tumor attenuation by combined heparan sulfate and polyamine depletion. Proc. Natl. Acad. Sci. U. S. A. 99:371-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casero, R. A., Jr., and L. J. Marton. 2007. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 6:373-390. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., L. Xiao, J. N. Rao, T. Zou, L. Liu, E. Bellavance, M. Gorospe, and J. Y. Wang. 2008. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol. Biol. Cell 19:3701-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerald, D., E. Berra, Y. M. Frapart, D. A. Chan, A. J. Giaccia, D. Mansuy, J. Pouysségur, M. Yaniv, and F. Mechta-Grigoriou. 2004. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell 118:781-794. [DOI] [PubMed] [Google Scholar]
- 11.Gerner, E. W., and F. L. Meyskens, Jr. 2004. Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer 4:781-792. [DOI] [PubMed] [Google Scholar]
- 12.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmüller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14:571-583. [DOI] [PubMed] [Google Scholar]
- 13.Harter, J. L. 1960. Critical values for Duncan's new multiple range test. Biometrics 16:671-685. [Google Scholar]
- 14.Hernandez, J. M., D. H. Floyd, K. N. Weilbaecher, P. L. Green, and K. Boris-Lawrie. 2008. Multiple facets of junD gene expression are atypical among AP-1 family members. Oncogene 27:4757-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilfiker-Kleiner, D., A. Hilfiker, K. Kaminski, A. Schaefer, J. K. Park, K. Michel, A. Quint, M. Yaniv, J. B. Weitzman, and H. Drexler. 2005. Lack of JunD promotes pressure overload-induced apoptosis, hypertrophic growth, and angiogenesis in the heart. Circulation 112:1470-1477. [DOI] [PubMed] [Google Scholar]
- 16.Hinman, M. N., and H. Lou. 2008. Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 65:3168-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hock, T. D., K. Liby, M. M. Wright, S. McConnell, M. Schorpp-Kistner, T. M. Ryan, and A. Agarwal. 2007. JunB and JunD regulate human heme oxygenase-1 gene expression in renal epithelial cells. J. Biol. Chem. 282:6875-6886. [DOI] [PubMed] [Google Scholar]
- 18.Houseley, J., and D. Tollervey. 2009. The many pathways of RNA degradation. Cell 136:763-776. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, T., A. Lal, X. Yang, S. Galban, K. Mazan-Mamczarz, and M. Gorospe. 2006. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 26:3295-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawamata, A., Y. Izu, H. Yokoyama, T. Amagasa, E. F. Wagner, K. Nakashima, Y. Ezura, T. Hayata, and M. Noda. 2008. JunD suppresses bone formation and contributes to low bone mass induced by estrogen depletion. J. Cell Biochem. 103:1037-1045. [DOI] [PubMed] [Google Scholar]
- 21.Keene, J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8:533-543. [DOI] [PubMed] [Google Scholar]
- 22.Kim, H. H., Y. Kuwano, S. Srikantan, E. K. Lee, J. L. Martindale, and M. Gorospe. 2009. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23:1743-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J. L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23:3092-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal, A., K. Abdelmohsen, R. Pullmann, T. Kawai, S. Galban, X. Yang, G. Brewer, and M. Gorospe. 2006. Posttranscriptional derepression of GADD45α by genotoxic stress. Mol. Cell 22:117-128. [DOI] [PubMed] [Google Scholar]
- 25.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 26.Laurent, G., F. Solari, B. Mateescu, M. Karaca, J. Castel, B. Bourachot, C. Magnan, M. Billaud, and F. Mechta-Grigoriou. 2008. Oxidative stress contributes to aging by enhancing pancreatic angiogenesis and insulin signaling. Cell Metab. 7:113-124. [DOI] [PubMed] [Google Scholar]
- 27.Li, L., J. Li, J. N. Rao, M. Li, B. L. Bass, and J. Y. Wang. 1999. Inhibition of polyamine synthesis induces p53 gene expression but not apoptosis. Am. J. Physiol. Cell Physiol. 276:C946-C954. [DOI] [PubMed] [Google Scholar]
- 28.Li, L., J. N. Rao, X. Guo, L. Liu, R. Santora, B. L. Bass, and J. Y. Wang. 2001. Polyamine depletion stabilizes p53 resulting in inhibition of normal intestinal epithelial cell proliferation. Am. J. Physiol. Cell Physiol. 281:C941-C953. [DOI] [PubMed] [Google Scholar]
- 29.Li, L., L. Liu, J. N. Rao, A. Esmaili, E. D. Strauch, B. L. Bass, and J. Y. Wang. 2002. JunD stabilization results in inhibition of normal intestinal epithelial cell growth through P21 after polyamine depletion. Gastroenterology 123:764-779. [DOI] [PubMed] [Google Scholar]
- 30.Liao, B., Y. Hu, and G. Brewer. 2007. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 14:511-518. [DOI] [PubMed] [Google Scholar]
- 31.Liu, L., X. Guo, J. N. Rao, T. Zou, B. S. Marasa, J. Chen, J. Greenspon, R. A. Casero, Jr., and J. Y. Wang. 2006. Polyamine-modulated c-Myc expression in normal intestinal epithelial cells regulates p21Cip1 transcription through a proximal promoter region. Biochem. J. 398:257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, L., J. N. Rao, T. Zou, L. Xiao, P. Y. Wang, D. J. Turner, M. Gorospe, and J. Y. Wang. 2009. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol. Biol. Cell 20:4885-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López de Silanes, I., M. Zhan, A. Lal, X. Yang, and M. Gorospe. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U. S. A. 101:2987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazan-Mamczarz, K., S. Galbán, I. López de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U. S. A. 100:8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazan-Mamczarz, K., Y. Kuwano, M. Zhan, E. J. White, J. L. Martindale, A. Lal, and M. Gorospe. 2009. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 37:204-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCormack, S. A., and L. R. Johnson. 1991. Role of polyamines in gastrointestinal mucosal growth. Am. J. Physiol. Gastrointest. Liver Physiol. 260:G795-G806. [DOI] [PubMed] [Google Scholar]
- 37.Mehraein-Ghomi, F., E. Lee, D. R. Church, T. A. Thompson, H. S. Basu, and G. Wilding. 2008. JunD mediates antidrogen-induced oxidative stress in androgen dependent LNCaP human prostate cancer cells. Prostate 68:924-934. [DOI] [PubMed] [Google Scholar]
- 38.Meixner, A., F. Karreth, L. Kenner, J. M. Penninger, and E. F. Wagner. 2010. Jun and JunD-dependent functions in cell proliferation and stress response. Cell Death Differ. 17:1409-1419. [DOI] [PubMed] [Google Scholar]
- 39.Moore, M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514-1518. [DOI] [PubMed] [Google Scholar]
- 40.Patel, A. R., J. Li, B. L. Bass, and J. Y. Wang. 1998. Expression of the transforming growth factor-β gene during growth inhibition following polyamine depletion. Am. J. Physiol. Cell Physiol. 275:C590-C598. [DOI] [PubMed] [Google Scholar]
- 41.Patel, A. R., and J. Y. Wang. 1999. Polyamine depletion is associated with and increase in JunD/AP-1 activity in small intestinal crypt cells. Am. J. Physiol. Gastrointest. Liver Physiol. 276:G441-G450. [DOI] [PubMed] [Google Scholar]
- 42.Pillebout, E., J. B. Weitzman, M. Burtin, C. Martino, P. Federici, M. Yaniv, G. Friedlander, and F. Terzi. 2003. JunD protects against chronic kidney disease by regulating paracrine mitogens. J. Clin. Investig. 112:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quaroni, A., J. Wands, R. L. Trelstad, and K. J. Isselbacher. 1979. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J. Cell Biol. 80:248-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray, R. M., B. J. Zimmerman, S. A. McCormack, T. B. Patel, and L. R. Johnson. 1999. Polyamine depletion arrests cell cycle and induces inhibitors p21(Waf1/Cip1), p27(Kip1), and p53 in IEC-6 cells. Am. J. Physiol. Cell Physiol. 276:C684-C691. [DOI] [PubMed] [Google Scholar]
- 46.Sela-Brown, A., J. Silver, G. Brewer, and T. Naveh-Many. 2000. Identification of AUF1 as a parathyroid hormone mRNA 3′-untranslated region-binding protein that determines parathyroid hormone mRNA stability. J. Biol. Chem. 275:7424-7429. [DOI] [PubMed] [Google Scholar]
- 47.Thépot, D., J. B. Weitzman, J. Barra, D. Segretain, M. G. Stinnakre, C. Babinet, and M. Yaniv. 2000. Targeted disruption of the murine junD gene results in multiple defects in male reproductive function. Development 127:143-153. [DOI] [PubMed] [Google Scholar]
- 48.Tsuji, Y. 2005. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene 24:7567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, J. Y., S. A. McCormack, M. J. Viar, and L. R. Johnson. 1991. Stimulation of proximal small intestinal mucosal growth by luminal polyamines. Am. J. Physiol. Gastrointest. Liver Physiol. 261:G504-G511. [DOI] [PubMed] [Google Scholar]
- 50.Wang, J. Y., and L. R. Johnson. 1991. Polyamines and ornithine decarboxylase during repair of duodenal mucosa after stress in rats. Gastroenterology 100:333-343. [DOI] [PubMed] [Google Scholar]
- 51.Wang, J. Y., and L. R. Johnson. 1994. Expression of protooncogenes c-fos and c-myc in healing of gastric mucosal stress ulcers. Am. J. Physiol. Gastrointest. Liver Physiol. 266:G878-G886. [DOI] [PubMed] [Google Scholar]
- 52.Wang, J. Y. 2007. Polyamines and mRNA stability in regulation of intestinal mucosal growth. Amino Acids 33:241-252. [DOI] [PubMed] [Google Scholar]
- 53.Wang, P. Y., J. N. Rao, T. Zou, L. Liu, L. Xiao, T. X. Yu, D. J. Turner, M. Gorospe, and J. Y. Wang. 2010. Post-transcriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem. J. 426:293-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, G. M., J. Lu, K. Sutphen, Y. Suarez, S. Sinha, B. Brewer, E. C. Villanueva-Feliciano, R. M. Ysla, S. Charles, and G. Brewer. 2003. Phosphorylation of p40AUF1 regulates binding to A+U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J. Biol. Chem. 278:33039-33048. [DOI] [PubMed] [Google Scholar]
- 55.Wilusz, C. J., and J. Wilusz. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20:491-497. [DOI] [PubMed] [Google Scholar]
- 56.Wisdom, R., R. S. Johnson, and C. Moore. 1999. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18:188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao, L., J. N. Rao, T. Zou, L. Liu, B. S. Marasa, J. Chen, D. J. Turner, A. Passaniti, and J. Y. Wang. 2007. Induced JunD in intestinal epithelial cells represses CDK4 transcription through its proximal promoter region following polyamine depletion. Biochem. J. 403:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao, L., J. N. Rao, T. Zou, L. Liu, B. S. Marasa, J. Chen, D. J. Turner, H. Zhou, M. Gorospe, and J. Y. Wang. 2007. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol. Biol. Cell 18:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao, L., J. N. Rao, T. Zou, L. Liu, T. X. Yu, X. Y. Zhu, J. M. Donahue, and J. Y. Wang. 2010. Induced ATF-2 represses CDK4 transcription through dimerization with JunD inhibiting intestinal epithelial cell growth after polyamine depletion. Am. J. Physiol. Cell Physiol. 298:C1226-C1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, A. H., J. N. Rao, T. Zou, L. liu, B. S. Marasa, L. Xiao, J. Chen, D. J. Turner, and J. Y. Wang. 2007. p53-dependent NDRG1 expression induces inhibition of intestinal epithelial cell proliferation but not apoptosis after polyamine depletion. Am. J. Physiol. Cell Physiol. 293:C379-C389. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, X., T. Zou, J. N. Rao, L. Liu, L. Xiao, P. Y. Wang, Y. H. Cui, M. Gorospe, and J. Y. Wang. 2009. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 37:7623-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou, T., J. N. Rao, L. Liu, B. S. Marasa, K. M. Keledjian, A. H. Zhang, L. Xiao, B. L. Bass, and J. Y. Wang. 2005. Polyamine depletion induces nucleophosmin modulating stability and transcriptional activity of p53 in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 289:C686-C696. [DOI] [PubMed] [Google Scholar]
- 63.Zou, T., K. Mazan-Mamczarz, J. N. Rao, L. Liu, B. S. Marasa, A. H. Zhang, L. Xiao, R. Pullmann, M. Gorospe, and J. Y. Wang. 2006. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J. Biol. Chem. 281:19387-19394. [DOI] [PubMed] [Google Scholar]
- 64.Zou, T., L. Liu, J. N. Rao, B. S. Marasa, J. Chen, L. Xiao, H. Zhou, M. Gorospe, and J. Y. Wang. 2008. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1. Biochem. J. 409:389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.