Abstract
The cortactin oncoprotein is frequently overexpressed in head and neck squamous cell carcinoma (HNSCC), often due to amplification of the encoding gene (CTTN). While cortactin overexpression enhances invasive potential, recent research indicates that it also promotes cell proliferation, but how cortactin regulates the cell cycle machinery is unclear. In this article we report that stable short hairpin RNA-mediated cortactin knockdown in the 11q13-amplified cell line FaDu led to increased expression of the Cip/Kip cyclin-dependent kinase inhibitors (CDKIs) p21WAF1/Cip1, p27Kip1, and p57Kip2 and inhibition of S-phase entry. These effects were associated with increased binding of p21WAF1/Cip1 and p27Kip1 to cyclin D1- and E1-containing complexes and decreased retinoblastoma protein phosphorylation. Cortactin regulated expression of p21WAF1/Cip1 and p27Kip1 at the transcriptional and posttranscriptional levels, respectively. The direct roles of p21WAF1/Cip1, p27Kip1, and p57Kip2 downstream of cortactin were confirmed by the transient knockdown of each CDKI by specific small interfering RNAs, which led to partial rescue of cell cycle progression. Interestingly, FaDu cells with reduced cortactin levels also exhibited a significant diminution in RhoA expression and activity, together with decreased expression of Skp2, a critical component of the SCF ubiquitin ligase that targets p27Kip1 and p57Kip2 for degradation. Transient knockdown of RhoA in FaDu cells decreased expression of Skp2, enhanced the level of Cip/Kip CDKIs, and attenuated S-phase entry. These findings identify a novel mechanism for regulation of proliferation in 11q13-amplified HNSCC cells, in which overexpressed cortactin acts via RhoA to decrease expression of Cip/Kip CDKIs, and highlight Skp2 as a downstream effector for RhoA in this process.
Cortactin is an F-actin binding protein involved in a variety of cellular processes, including endocytosis, vesicle trafficking, and the formation of cellular protrusions such as lamellipodia (14). In order to mediate these functions, cortactin interacts with a variety of proteins depending on the cell type and subcellular compartment, and this is achieved via distinct binding domains located within the cortactin molecule. The N terminus of cortactin harbors an acidic region that represents the interaction site for the actin-polymerizing Arp2/Arp3 complex, and this is followed by a repeat region containing the F-actin binding site. A Src homology 3 (SH3) domain located at the C terminus of cortactin recruits diverse proteins, including components of the endocytosis machinery (e.g., CD2AP and dynamin) and regulators of Rho family GTPases (e.g., BPGAP1 and Fgd1) and actin polymerization (e.g., N-WASP), while an adjacent proline-rich region contains phosphorylation sites for Src family kinases (14).
The cortactin gene (CTTN) is located at chromosome band 11q13, a region that is commonly amplified in many human malignancies, particularly breast, ovarian, and bladder cancers and head and neck squamous cell carcinoma (HNSCC) (43). Several genes lie within this chromosomal region and are overexpressed upon its amplification. However, of these genes, the amplification of cyclin D1 (CCND1) and CTTN is most frequently associated with poor clinical outcomes such as decreased patient survival and increased metastasis (34, 43). Chromosomal mapping of the 11q13 locus has revealed four distinct regions that can be individually or coordinately amplified (12, 20, 34). Within this locus, CCND1 and CTTN are located on different amplicons, and independent amplification of these genes has been demonstrated (34). In HNSCC, a tumor type in which 11q13 amplification occurs at the relatively high frequency of ∼30% (43), CTTN amplification has been identified as an independent predictor of reduced disease-specific survival while CCND1 amplification is not prognostic in this tumor type (16, 18, 38, 39). This strongly suggests that cortactin overexpression can act independently to promote tumor progression in cases of HNSCC.
Due to the ability of cortactin to promote actin polymerization, many previous studies on cancer cells have focused on the role of cortactin in promoting cell motility and invasion (35, 40, 54), effects mediated by increased lamellipodial persistence (5), invadopodia formation (4), and protease secretion (10, 11). In agreement with this, cortactin overexpression has been correlated with enhanced lymph node metastasis in clinical studies (28, 30, 40) and increased metastasis in experimental models (30).
While the ability of cortactin overexpression to increase migratory capacity is well established, this does not account for the presence of CTTN amplification in primary tumors nor for the positive effect of cortactin on tumor growth in xenograft models (9, 30), indicating a proliferative or survival advantage for cortactin-overexpressing cells. The mechanisms behind this selective advantage have not been completely elucidated although we recently demonstrated that cortactin overexpression attenuates ligand-induced epidermal growth factor receptor (EGFR) degradation, leading to increased mitogenic signaling (48, 49). Additionally, a recent study involving the modulation of cortactin in HNSCC cell lines suggested that cortactin may influence proliferation by increasing autocrine growth factor secretion (9).
Deregulation of cell cycle control mechanisms leading to unrestrained proliferation is a hallmark of cancer. Progression through different stages of the mammalian cell cycle is controlled by specific cyclin/cyclin-dependent kinase (Cdk) complexes, which in turn are regulated by a variety of processes including changes in cyclin abundance, posttranslational modification including phosphorylation, and association with Cdk inhibitors (CDKIs) (6). During G1 phase the major cyclin/Cdk complexes are cyclin D1/Cdk4 and cyclin E/Cdk2, and these phosphorylate the retinoblastoma gene product, Rb, to promote progression from G1 to S phase. Two families of CDKIs regulate the assembly and/or activity of cyclin D1/Cdk4 and cyclin E/Cdk2 complexes: the Cip/Kip family (p21WAF1/Cip1, p27Kip1, and p57Kip2), which act on both complexes, and the INK4 family, which exhibits selectivity for Cdk4 over Cdk2. Cip/Kip CDKIs are potent inhibitors of cyclin E/Cdk2 complexes but have a dual function toward cyclin D1/Cdk4 complexes, acting as assembly factors or inhibitors at low and high concentrations, respectively (8, 25). The activity of G1 cyclin/Cdk complexes is regulated by a variety of signaling pathways, including those emanating from activated growth factor receptors and Rho family GTPases. For example, Ras/Erk signaling positively regulates cyclin D1 transcription, while RhoA activation increases expression of the F-box protein Skp2 that functions in combination with the Skp1-Cullin-F-box protein (SCF) E3 ubiquitin protein ligase to promote proteasomal degradation of p27Kip1 (56).
Surprisingly, despite several studies demonstrating that high cortactin levels promote mitogenic signaling and/or cancer cell proliferation (9, 30, 48, 49), how cortactin overexpression affects the cell cycle machinery has not been characterized. We have now addressed this question and, in doing so, have identified a novel mechanism linking cortactin overexpression to deregulation of Cip/Kip family CDKIs. This mechanism provides new insights into how cortactin promotes proliferation in 11q13-amplified HNSCC cells.
MATERIALS AND METHODS
Plasmids.
The pSIREN-RetroQ-ZsGreen (Clontech) constructs containing short hairpin RNA (shRNA) targeting cortactin and green fluorescent protein ([GFP] negative control) were constructed by the ligation of synthesized oligonucleotides into the BamHI and EcoRI sites of pSIREN. The DNA sequences used for construction of the oligonucleotides to create cortactin-targeting shRNA were based on small interfering RNA (siRNA) previously used to knock down cortactin expression in HNSCC cell lines (49). The following oligonucleotides were used: shRNA 1, GATCCAAGCTGAGGGAGAATGTCTTTTCAAGAGAAAGACATTCTCCCTCAGCTTTTTTTTACGCGTG; shRNA 2, GATCCGACTGGTTTTGGAGGCAAATTTTCAAGAGAAATTTGCCTCCAAAACCAGTCTTTTTTACGCGTG; and negative-control sequence targeting GFP (Ambion 4626).
The wild-type and 3YF mutant myc-tagged murine cortactin genes were PCR amplified from plasmids kindly donated by X. Zhan (19) using the following primers: a, AATTCCGCGGATGGAACAAAAGCTTATTTCTGAAGAAGA; b, TAGGATCCCTACTGCCGCAGCTCCACATAGTT. The resulting PCR products were purified using the Wizard PCR cleanup system (Promega) and cloned into the SacII and BamHI restriction sites of pRetroX-IRES-DsRed Express vector (Clontech). Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. A stop codon was introduced into the wild-type murine cortactin gene at residue 496 to produce the ΔSH3 mutant (19), using the following primers: a, GCATCACAGCCATCGCCTAGTATGACTACCAGGCTG; b, CAGCCTGGTAGTCATACTAGGCGATGGCTGTGATGC.
Antibodies and reagents.
The cortactin monoclonal 4F11 antibody was purchased from Upstate Biotechnologies, Inc. Antibodies against phosphorylated Akt (T308 and S473), Erk (T202/Y204), Rb (S780), and myosin light chain 2 ([MLC2] T18/S19), as well as antibodies against total Akt, Erk, RhoA, Skp2, and MLC2 were from Cell Signaling Technology. p57Kip2 (C-20), cyclin E1 monoclonal (HE12), and cyclin E1 (C-19) polyclonal antibodies were purchased from Santa Cruz Biotechnology. The cyclin D1 (AB-3) polyclonal antibody was from Neomarkers while the cyclin D1 monoclonal antibody (DCS6) was from Novacastra. Monoclonal antibodies against p21WAF1/Cip1 (610233), p27Kip1 (610241), and total Rb were from BD Transduction Laboratories. The β-actin monoclonal antibody (AC-15) was purchased from Sigma.
Aphidicolin, hydroxyurea, and phleomycin were purchased from Calbiochem. The Akt inhibitor Akti-1/2 was a gift from Peter Shepherd (Auckland, New Zealand). ROCK inhibitor Y27632 was purchased from Merck.
Tissue culture and generation of stable cell lines.
The FaDu HNSCC line was maintained as previously described (48, 49). HEK293 cells were maintained in minimal essential medium (Gibco) supplemented with 10% fetal calf serum (FCS), penicillin (50 units/ml), and streptomycin (50 μg/ml). For the production of stable cell lines, Phoenix cells were transfected with pSIREN or pRetroX constructs using Polyfect (Qiagen Pty., Ltd), according to the manufacturer's instructions. Viral supernatants were collected 48 h later and filtered through 20-μm-pore-size filter caps (Millipore), and 8 μg/ml of Polybrene was added. FaDu cells were transiently transfected with pQCXIN_EcoR, and 48 h later the medium was removed and replaced with viral supernatants. Positive cells were selected by cell sorting on a fluorescence-activated cell sorter (FACS) Vantage instrument (Becton Dickinson), based on the bicistronic expression of ZsGreen or DsRed.
Cell lysis, immunoprecipitation, and immunoblotting.
Cell lysates were prepared using 1% Triton-X lysis buffer containing 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM sodium orthovanadate, and 1 mM phenylmethylsulfonyl fluoride. Immunoprecipitations were performed by overnight incubation with 25 μl of protein G-Sepharose, 2 to 5 μg of primary antibody, and 200 to 500 μg of lysate. For GTPase activity assays, cell lysates were collected in a high-magnesium lysis buffer (25 mM HEPES, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 10% glycerol, 25 mM NaF, 10 mM MgCl2, 1 mM EDTA). GTP-bound RhoA was isolated from 100 to 200 μg of lysate using Rhotekin receptor binding domain-glutathione S-transferase (RBD-GST) beads (Cytoskeleton, Denver, CO), according to the manufacturer's instructions. GTP-bound Rac and Cdc42 were isolated from 200 μg of lysate using GST-p21-activated kinase (PAK) Sepharose, as previously described (15).
For immunoblotting, all antibodies were used at a 1:1,000 dilution in Tris-buffered saline (TBS)-1% bovine serum albumin (BSA)-0.02% sodium azide, except for the 4F11 cortactin monoclonal antibody (1:3,000) and the AC-15 actin monoclonal antibody (1:50,000). Relevant anti-mouse and anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies (Amersham) were used at 1:5,000 in TBS-0.1% Tween 20-5% skim milk powder.
Colony-forming assays.
These were undertaken essentially as previously described (49). Briefly, 1,000 cells were seeded into the wells of a six-well plate and allowed to grow for 10 days. Resulting colonies were fixed and stained using Diff-Quick (Lab Aids), and colony size and number were determined using the particle analysis function of ImageJ software (version 1.37).
Cell cycle analysis.
For synchronization of cells in G1 phase, FaDu pSIREN cells were seeded into six-well plates at 2 × 105 cells/well and incubated for 24 h. The cells were then serum starved for 48 h, followed by the addition of hydroxyurea (2 mM) in serum-free medium for a further 24 h. The cells were released from this block by washing with phosphate-buffered saline (PBS) and the addition of complete medium containing 10% FCS for the time periods indicated in the relevant figures. Cell cycle distribution analysis was undertaken by flow cytometry on a FACS Canto I instrument (Becton Dickinson), using either propidium iodide or bromodeoxyuridine (BrdU)-propidium iodide (PI) staining, as previously described (32, 46). Data analysis was performed using WinMDI (version 2.9).
siRNA treatment.
Small interfering RNA (siRNA) targeting p21WAF1/Cip1 (siRNA s415; Ambion Silencer Select), p27Kip1 (siRNA s2838; Ambion Silencer Select), p57Kip2 (siRNA s230039; Ambion Silencer Select,), and RhoA (siRNA 1 s11099 and siRNA 2 s11100; Ambion Silencer Select) were used at the commencement of the serum starvation step of the synchronization procedure. Negative control for Ambion Silencer Select siRNAs was negative-control 2 siRNA (4390846). Briefly, 2.5 μl of Lipofectamine 2000 was added to 250 μl of OptiMem and incubated at room temperature for 5 min. A separate tube was prepared containing 250 μl OptiMem and the necessary amount of siRNA for a final concentration of 10 nM, and this was also incubated at room temperature for 5 min. The two solutions were then combined and incubated at room temperature for a further 20 min. Following this, the 500-μl siRNA-Lipofectamine 2000 mixture was added, in a dropwise fashion, to cells undergoing serum starvation in 1.5 ml of serum-free medium.
qPCR analysis.
Quantitative PCR (qPCR) analysis was performed on mRNA extracted from synchronized and released FaDu pSIREN cells to determine the mRNA levels of p21WAF1/Cip1, p27Kip1, or RhoA genes (CDKN1A, CDKN1B, and RHOA, respectively). mRNA was extracted using an AnalytikJena innuPREP RNA Mini Kit according to the manufacturer's instructions. Reverse transcription was performed with a Reverse Transcription System (Promega) according to the manufacturer's instructions, and the resulting cDNA was diluted 1:8. Quantitative reverse transcription-PCR (qRT-PCR) was performed on an Applied Biosystems ABI 7900 qPCR machine (absolute quantification setting) using TaqMan Gene Expression Assays, also from Applied Biosystems. Each reaction mixture contained 4.5 μl of cDNA, 5 μl of TaqMan Universal PCR Master Mix, and 0.5 μl of the Gene Expression Assay (for p21WAF1/Cip1, Hs00355782_m1; p27Kip1, Hs00153277_m1; RhoA, Hs00236938_m1; ribosomal large protein [RPLPO], 4326314E; glyceraldehyde-3-phosphate dehydrogenase [GAPDH], 4326317E). Reaction mixtures were prepared in triplicate using an EpMotion 5070 system (Eppendorf). Standard curves were constructed for each TaqMan probe using a serial dilution of 1 in 10, and data were analyzed through the 2−ΔΔCT (where CT is threshold cycle) method, corrected for the efficiency of each TaqMan probe and normalized to the RPLPO or GAPDH housekeeping gene.
Cortactin overexpression in HEK293 cells.
The pRetroX vector containing human cortactin cDNA was transfected into HEK293 cells using Polyfect (Qiagen Pty Ltd), according to the manufacturer's instructions. After 48 h, four populations of increasing cortactin expression were selected by cell sorting on a FACS Vantage instrument (Becton Dickinson), based on the bicistronic expression of DsRed. A control population was obtained by transfection with the empty pRetroX vector and collection across all four sorting gates. Following sorting, the cells were replated at a density of 5 × 105 cells/6-cm dish for a further 48 h before harvesting.
RESULTS
Cortactin promotes G1-to-S-phase cell cycle progression in 11q13-amplified HNSCC cells.
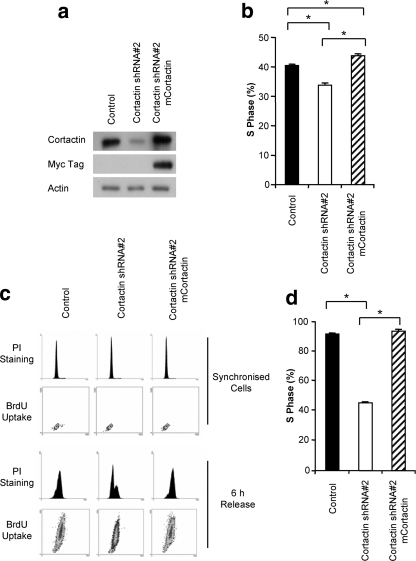
We have previously demonstrated that the transient knockdown of cortactin in HNSCC cell lines overexpressing this protein results in a decreased S-phase population and a reduction in cell proliferation (49). In order to interrogate the mechanism underpinning this effect, we established a model system in which cortactin was stably knocked down in the 11q13-amplified cell line FaDu by retrovirus-mediated expression of shRNA targeted to human cortactin. This resulted in an ∼80% reduction in cortactin expression (Fig. 1 a), producing expression levels comparable to the expression level of the HNSCC cell line Scc9, which lacks 11q13 amplification (49). Additionally, we reexpressed murine cortactin, which is resistant to the human-specific cortactin shRNA, in this knockdown cell line to provide a robust control (Fig. 1a). In accordance with our previous findings, cortactin knockdown induced a significant decrease in the S-phase population of asynchronous cells, which was rescued upon the reexpression of murine cortactin (Fig. 1b).
FIG. 1.
Stable cortactin knockdown in 11q13-amplified HNSCC cells decreases S-phase population. (a) Western blot analysis of cortactin expression in cortactin knockdown cells and in cells reconstituted with shRNA-resistant myc-tagged murine cortactin (mCortactin). (b) The effect of cortactin knockdown on the S-phase population in asynchronous cells. Following a 2-h pulse with BrdU, cell cycle analysis was performed using anti-BrdU and propidium iodide staining and flow cytometry. Data represent mean ± standard error from three independent experiments. *, P < 0.05. (c) The effect of cortactin knockdown on cell cycle progression following a G0/G1 phase block. FaDu cells were synchronized into G1 phase by serum starvation and hydroxyurea treatment and then released into complete medium for 6 h, with the addition of BrdU for the final 2 h. Representative data for BrdU uptake and PI staining are shown. (d) Effect of cortactin knockdown on S-phase entry for synchronized cells. The histogram indicates the S-phase population from experiments outlined in panel C. Data represent the mean ± standard error from three independent experiments. *, P < 0.05.
To facilitate characterization of the mechanism whereby cortactin regulates cell cycle progression, we synchronized cells in G1 phase. In FaDu cells serum starvation alone led to only partial synchrony, and therefore cells were synchronized in G1 phase by a combination of serum starvation and hydroxyurea treatment (Fig. 1c) (53). Upon release from this G1 block into medium containing serum but lacking hydroxyurea, bromodeoxyuridine (BrdU) incorporation was used to quantify the percentage of cells that had entered S phase after 6 h (Fig. 1c). Under these conditions, a ∼50% reduction in S-phase population was observed for the cortactin knockdown cells, which was completely rescued by the reexpression of murine cortactin (Fig. 1c and d).
Cortactin knockdown modulates expression of key regulators of G1-to-S-phase progression.
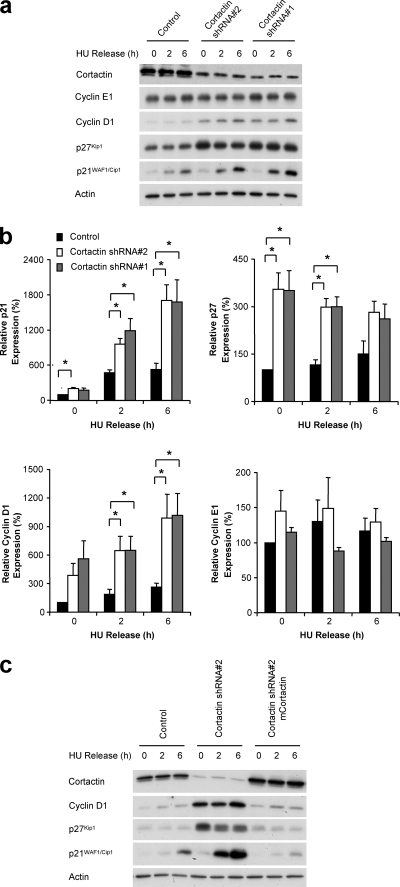
Using this synchronization model, we sought to analyze the levels of regulatory proteins known to be involved in the G1/S-phase progression, namely, cyclins D1 and E1 and the CDKIs p21WAF1/Cip1 and p27Kip1 (Fig. 2). Also, to further reinforce the specificity of our cortactin-targeting shRNA, we utilized an additional FaDu cell line expressing a second shRNA which displayed a similar level of cortactin knockdown (Fig. 2a). Western blotting of lysates revealed that p21WAF1/Cip1 expression was low in synchronized control cells and increased ∼5-fold at both 2 and 6 h following release. However, while p21WAF1/Cip1 levels in synchronized cortactin knockdown cells were slightly higher than those in the control line, the fold induction of p21WAF1/Cip1 upon release was approximately two times higher than that of the control at the 2-h time point and ∼3-fold higher at 6 h (Fig. 2a and b). The levels of p27Kip1 were elevated ∼3.5-fold in synchronized cortactin knockdown cells over control cells and decreased slightly upon release (Fig. 2a and b). Cyclin D1 expression was ∼3- to 4-fold higher in cortactin knockdown cells at each time point, which was statistically significant at the 2- and 6-h time points, while the levels of cyclin E1 were not significantly different at any time point (Fig. 2a and b).
FIG. 2.
The effect of cortactin knockdown on key cell cycle regulators. (A) Western blot analysis of specific cyclins and CDKIs. FaDu control and cortactin knockdown cells were synchronized into G1 phase by serum starvation and hydroxyurea treatment and then released into complete medium for the time periods indicated. The expression of the indicated proteins was then analyzed by Western blotting. (B) Quantification of Western blotting results. Data were normalized using actin as a loading control and then expressed relative to the value for control cells, which was arbitrarily set at 100%. Data represent the mean ± standard error from three independent experiments. *, P < 0.05. (C) Expression of murine cortactin reverses the effect of human cortactin knockdown. Cells were treated and analyzed as in panel A. Data shown are representative of at least three independent experiments. HU, hydroxyurea.
Confirmation of the role of cortactin in modulating the levels of these regulatory proteins was provided by their complete reversion to control levels upon the reexpression of murine cortactin within cortactin knockdown cells (Fig. 2c). Furthermore, the same pattern of increased p21WAF1/Cip1, p27Kip1, and cyclin D1 expression and of an inhibition of S-phase entry from G1 synchronization was observed in FaDu cells treated with cortactin siRNA in a transient fashion (see Fig. S1 in the supplemental material), confirming that this effect was not an artifact arising from the construction of a stable cortactin knockdown cell line.
A potential caveat to the approach of cell synchronization using agents such as hydroxyurea is the induction of double-stranded DNA breaks (52) and the associated activation of DNA repair mechanisms, which can result in the upregulation of p21WAF1/Cip1 via p53 transactivation (37). Within this model system, an increase in ATM/ATR substrate phosphorylation and p53 phosphorylation was observed upon hydroxyurea treatment; however, this did not result in an increase in p21WAF1/Cip1 expression (see Fig. S2 in the supplemental material), presumably due to the inactive version of p53 expressed by FaDu cells (23). In agreement with this, treatment with the DNA damage-inducing agent phleomycin induced p53 phosphorylation, also without any subsequent increase in p21WAF1/Cip1 expression (see Fig. S2).
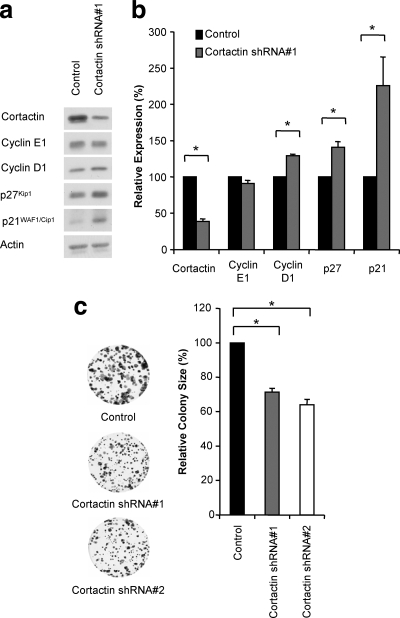
To confirm that the effects of cortactin knockdown were not restricted to this model system, we synchronized cells using the DNA polymerase inhibitor, aphidicolin. Again, cortactin knockdown resulted in a significant reduction in S-phase entry following release (data not shown). Furthermore, asynchronous cells also displayed significantly increased levels of p21WAF1/Cip1, p27Kip1, and cyclin D1 expression upon cortactin knockdown (Fig. 3 a and b), confirming that these are responses to cortactin knockdown rather than an artifact of cell synchronization using hydroxyurea. Of note, cortactin knockdown with either shRNA construct resulted in decreased proliferation in colony-forming assays, with both lines displaying a 30 to 40% reduction in colony size (Fig. 3C), while no variation in colony number was observed (data not shown).
FIG. 3.
Cortactin knockdown affects cell cycle regulators in asynchronous cells, and cell proliferation. (A) Western blot analysis of the indicated cyclins and CDKIs in asynchronous FaDu control and cortactin knockdown cells. (B) Quantification of Western blotting results. Data were analyzed as described in the legend to Fig. 2 and are derived from three independent experiments. *, P < 0.05. (C) The effect of cortactin knockdown on colony formation. Stained colonies were scanned and analyzed for colony number and size. Data shown represent the mean ± standard error from six independent experiments, with 100% representing the corresponding value for control cells.*, P < 0.05.
Determination of the functional roles of p21WAF1/Cip1 and p27Kip1 in cortactin-regulated cell cycle progression.
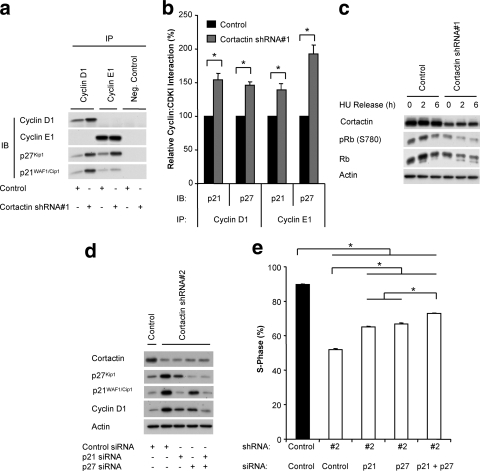
To establish a direct link between the increased levels of p21WAF1/Cip1/p27Kip1 and the inhibition of S-phase progression in cortactin knockdown cells, we first sought to confirm that the enhanced expression of these CDKIs resulted in an increased interaction with particular cyclin/Cdk complexes. Coimmunoprecipitation of p21WAF1/Cip1 and p27Kip1 with both cyclin D1 and cyclin E1 confirmed that the observed elevation in expression of these CDKIs resulted in a significant increase in their complex formation with these cyclins (Fig. 4 a and b). This was observed even when the interaction with cyclin D1 was normalized for its increased expression in cortactin knockdown cells (Fig. 4b). As a further measure of G1 cyclin/Cdk activity, we measured the phosphorylation state of the retinoblastoma protein (Rb), a critical regulator of G1/S-phase progression. Accordingly, in cortactin knockdown cells there was an increase in the hypophosphorylated form of Rb, evident as a higher mobility band in Western blots of total Rb protein, and decreased phosphorylation at a Rb residue (Ser780) targeted by specific Cdk complexes.
FIG. 4.
Determination of the functional role of p21WAF1/Cip1 and p27Kip1 downstream of cortactin. (A) Coimmunoprecipitation analysis. Cyclin D1 and cyclin E1 were immunoprecipitated from lysates of control and cortactin knockdown cells that had been synchronized and released for 4 h. Western blotting was undertaken to detect the coimmunoprecipitation of p21WAF1/Cip1 and p27Kip1. (B) Quantification of coimmunoprecipitation data. The levels of associating p21WAF1/Cip1 or p27Kip1 were normalized for the amount of cyclin immunoprecipitated and then expressed relative to the value for control cells, which was arbitrarily set at 100%. Data represent the mean ± standard error of three independent experiments. *, P < 0.05. (C) Rb phosphorylation. Control and cortactin knockdown cells were synchronized into G1 phase and released for the time points indicated. Cell lysates were subjected to Western blotting with the indicated antibodies. Data shown are representative of at least three independent experiments. HU, hydroxyurea. (D) Reduction in p21WAF1/Cip1 and p27Kip1 expression levels using specific siRNAs. These CDKIs were transiently depleted by the addition of specific siRNAs during the serum starvation step of the synchronization procedure. Following a 6-h release, Western blotting was undertaken as indicated. Data shown are representative of at least three independent experiments. (E) Reduction of p21WAF1/Cip1 and p27Kip1 expression enhances cell cycle progression in cortactin knockdown cells. Entry into S-phase after a 6-h release was determined by uptake of BrdU during a 2 h pulse. The incorporation of BrdU was measured by flow cytometry. Data represent the mean ± standard error of three independent experiments. *, P < 0.05).
To determine whether the increased expression of p21WAF1/Cip1 and p27Kip1 in cortactin knockdown cells contributed to the observed decrease in S-phase progression, transient knockdown of p21WAF1/Cip1 and p27Kip1, alone and in combination, was undertaken. This reduced expression of these CDKIs to levels similar to those in control cells (Fig. 4d) and partially rescued S-phase entry (Fig. 4d and e), demonstrating that increased expression of these CDKIs contributes to the inhibition of G1/S-phase progression. These results obtained for the transient knockdown of p21WAF1/Cip1 and p27Kip1 were also replicated with a separate set of control and specific siRNA pools from a separate manufacturer (data not shown).
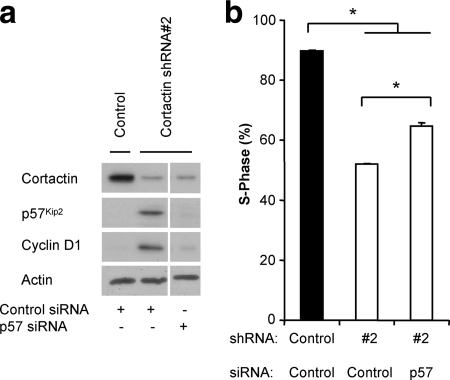
The combined knockdown of both p21WAF1/Cip1 and p27Kip1 was more effective than knockdown of either alone but did not completely rescue the defect induced by cortactin knockdown (Fig. 4d and e), and therefore the role of p57Kip2 was also investigated. p57Kip2 is a relative of p27Kip1, and their expression levels are often regulated in the same manner (21, 36). Accordingly, p57Kip2 was also upregulated in cortactin knockdown cells (Fig. 5 a). Furthermore, transient knockdown of p57Kip2 partially rescued S-phase entry (Fig. 5).
FIG. 5.
Upregulation of p57Kip2 contributes to decreased cell cycle progression in cortactin knockdown cells. (A) Western blot analysis of p57Kip2 and cyclin D1 expression. Control and cortactin knockdown cells were transiently transfected with the indicated siRNAs during the serum starvation step of the synchronization procedure. Cell lysates were subjected to Western blotting as indicated. Data presented are derived from an individual experiment although intervening bands have been removed for the sake of clarity. Data are also representative of at least three independent experiments. (B) Effect of p57Kip2 knockdown on cell cycle progression. Entry into S-phase after a 6-h release was determined by uptake of BrdU during a 2-h pulse. The incorporation of BrdU was measured by flow cytometry. Data shown represent the mean ± standard error of three independent experiments. *, P < 0.05.
Interestingly, the knockdown of each individual CDKI also reduced the expression of cyclin D1 back to levels approaching those observed in control cells (Fig. 4d and 5a), suggesting that the increase in cyclin D1 levels is secondary to the increase in CDKI expression. Taken together, these findings indicate that the elevated CDKI expression induced by cortactin knockdown in HNSCC cells is responsible for the decrease in S-phase entry, due to their increased inhibition of the cell cycle machinery.
p21WAF1/Cip1 and p27Kip1 protein levels can be regulated by both transcriptional and posttranscriptional mechanisms (1). In order to determine which mechanism was responsible for the elevated levels of these proteins following cortactin knockdown, we assayed levels of their encoding mRNA transcripts via quantitative RT-PCR (Fig. 6). Analysis of p21WAF1/Cip1 mRNA levels during release from G1-phase synchronization revealed a progressive increase in p21WAF1/Cip1 mRNA, which was further elevated in the cortactin knockdown cells and became significantly different from the control at the 6-h time point (Fig. 6a). This enhancement was comparable in magnitude to the increase in p21WAF1/Cip1 protein levels, suggesting that the modulation of p21WAF1/Cip1 expression by cortactin was occurring at a transcriptional level. Conversely, the levels of p27Kip1 mRNA remained relatively constant following release and were not significantly different between cortactin knockdown and control cells at any of the time points analyzed (Fig. 6b), suggesting that regulation of p27Kip1 by cortactin was at the posttranscriptional level.
FIG. 6.
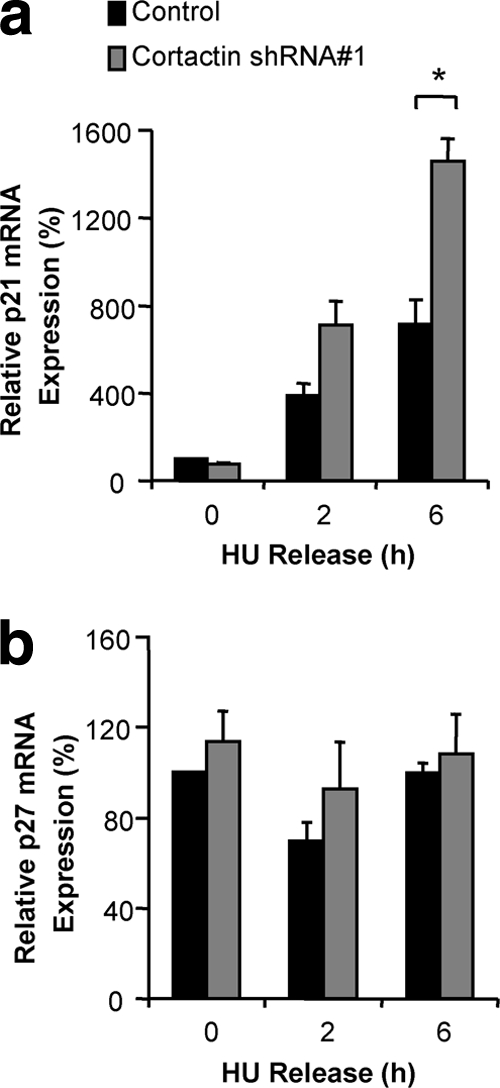
Effect of cortactin knockdown on p21WAF1/Cip1 and p27Kip1 mRNA levels. FaDu control and cortactin knockdown cells were synchronized into G1 phase by serum starvation and hydroxyurea (HU) treatment and then released into complete medium for the time periods indicated. Following this, mRNA was extracted from the cells, and qRT-PCR was performed to determine the relative amounts of p21WAF1/Cip1 (A) and p27Kip1 (B) mRNA in each sample, as described in Materials and Methods. Normalized data are expressed relative to the value for control cells at 0 h, which is arbitrarily set at 100%. Data represent the mean ± standard error of three independent experiments. *, P < 0.05.
Cortactin regulates CDKI expression via modulation of RhoA signaling.
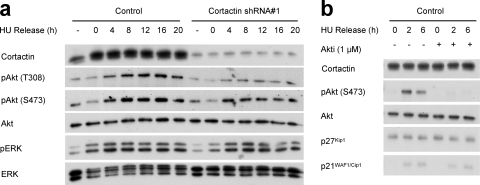
In order to determine the signaling pathway(s) responsible for mediating the effects of cortactin knockdown on expression of the Cip/Kip family of inhibitors, we analyzed pathways known to be important in either the transcriptional or posttranscriptional control of these CDKIs. Previously, we reported that transient knockdown of cortactin in serum-supplemented FaDu cells led to decreased activation of Erk and Akt (49). However, in our synchronization model, stable cortactin knockdown affected only Akt activity, as evident by lower levels of phosphorylation on S473 and T308 (Fig. 7 a). In order to determine the contribution of Akt signaling to regulation of p21WAF1/Cip1 and p27Kip1, we treated control cells with a selective pharmacological inhibitor of Akt, Akti-1/2 (3). Administration of the inhibitor markedly decreased Akt activation but did not alter expression of p21WAF1/Cip1 or p27Kip1 (Fig. 7b). Consequently, the observed modulation of Akt activity by cortactin cannot, by itself, explain the effects on p21WAF1/Cip1 and p27Kip1 levels.
FIG. 7.
Determination of the role of Erk and Akt signaling in regulation of p21WAF1/Cip1 and p27Kip1 expression. (A) Western blot analysis of pathway activation. FaDu control and cortactin knockdown cells were synchronized into G1 phase by serum starvation and hydroxyurea (HU) treatment and then released into complete medium for the time periods indicated. Cell lysates were subjected to Western blotting with the indicated antibodies. Data shown are representative of at least three independent experiments. (B) Inhibition of Akt does not affect p21WAF1/Cip1 and p27Kip1 levels. The Akt inhibitor Akti-1/2 (1 μM) was administered during synchronization and release of FaDu control cells. Western blotting was undertaken as indicated. Data shown are representative of at least three independent experiments.
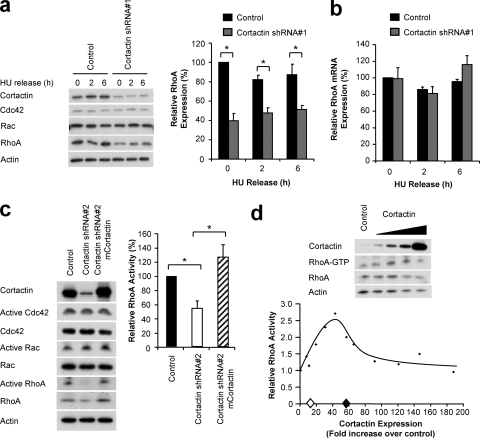
Several studies have implicated RhoA in regulation of G1/S progression via modulation of p21WAF1/Cip1 or p27Kip1 expression (56). Interestingly, in our model, the stable knockdown of cortactin in FaDu cells resulted in a 40 to 60% reduction in RhoA expression while there was no alteration in the expression of the other Rho family members Cdc42 or Rac (Fig. 8 a). This decrease in RhoA protein level in cortactin knockdown cells was not accompanied by changes in the levels of the RhoA mRNA transcript, which did not differ significantly between control or cortactin knockdown cells at any of the time points measured (Fig. 8b). The activity of RhoA, when normalized to its total levels, was also decreased by ∼40% in synchronized cortactin knockdown cells while there was no alteration in the activity of Cdc42 or Rac (Fig. 8c). Both the expression levels of RhoA and its specific activity were restored to control levels upon the reexpression of shRNA-resistant murine cortactin (Fig. 8c).
FIG. 8.
RhoA expression and activity are decreased upon cortactin knockdown. (A) Effect of cortactin knockdown on expression of Rho family GTPases. FaDu control and cortactin knockdown cells were synchronized into G1 phase by serum starvation and hydroxyurea (HU) treatment and then released into complete medium for the time periods indicated. The expression of RhoA, Cdc42, and Rac was determined by Western blotting. Protein levels were normalized using actin as a loading control and then expressed relative to the level for control cells at 0 h, which was arbitrarily set at 100%. Data shown represent the mean ± standard error of three independent experiments. *, P < 0.05. (B) Effect of cortactin knockdown on RhoA mRNA levels. mRNA was extracted from FaDu cells treated as described for panel A, and qRT-PCR was performed to determine the relative amounts of RhoA mRNA in each sample, as described in Materials and Methods. Normalized data are expressed relative to the value for control cells at 0 h, which is arbitrarily set at 100%. (C) Effect of cortactin knockdown on Rho family GTPase activation. Synchronized FaDu control, cortactin knockdown, and murine cortactin-reexpressing cells were lysed, and active GTPases were isolated as described in Materials and Methods. The active and total levels of each GTPase were determined by Western blotting. Activity was normalized relative to the level of total protein in each cell type and is expressed relative to the level of activity in control cells, which was arbitrarily set at 100%. Data shown represent the mean ± standard error of at least three independent experiments. *, P < 0.05. (D) Effect of increasing cortactin expression on RhoA activation. GTP-bound RhoA was isolated from cortactin-overexpressing HEK293 cells. Cell lysates were also subjected to Western blotting with the indicated antibodies. RhoA activation and cortactin expression are expressed relative to empty vector-transfected cells, which are assigned an arbitrary value of 1.0. Western blotting is representative of three independent experiments, while the graph contains the individual data points from three experiments due to variations in the cortactin expression obtained between repeat experiments. The comparative levels of cortactin expression in control (⧫) and cortactin knockdown (⋄) FaDu cells are indicated on the x axis.
Cortactin gene knockout fibroblasts do not exhibit altered RhoA activation (26), indicating that, at normal expression levels, cortactin does not regulate RhoA. To determine whether cortactin overexpression affects RhoA activation, we generated pools of HEK293 cells expressing increasing amounts of cortactin. Strikingly, a 10- to 50-fold overexpression of cortactin led to a progressive increase in RhoA activation, up to a maximum of approximately 2.5-fold (Fig. 8d). However, this response of RhoA was biphasic, such that increasing cortactin overexpression further led to a gradual decrease in RhoA activation back to control levels. Importantly, direct comparison of the cortactin expression levels in this experiment with those in control and cortactin knockdown FaDu cells revealed that the range of expression in the latter largely corresponds to the region of the dose-response curve where cortactin overexpression enhances RhoA activation and is consistent with the decrease in RhoA activation observed in the cortactin knockdown cells. These data represent the first demonstration that cortactin overexpression modulates RhoA activation. However, we note that in the HEK293 system, RhoA expression levels were not significantly affected upon cortactin overexpression (Fig. 8d and data not shown). Consequently, this effect of cortactin (Fig. 8a) appears to be more specific to HNSCC cells.
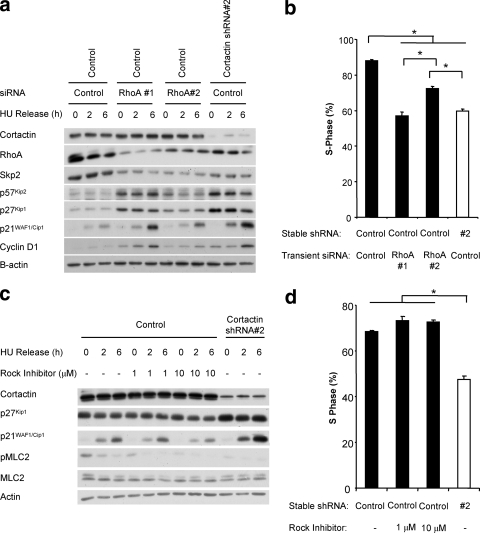
In order to interrogate the functional role of RhoA in the FaDu system, we used two different siRNAs (RhoA siRNA 1 and siRNA 2) to transiently reduce expression of RhoA in synchronized control FaDu cells. Strikingly, transfection of either siRNA resulted in increased expression of p21WAF1/Cip1, p27Kip1, p57Kip2, and cyclin D1 (Fig. 9 a) and inhibited the ability of the cells to enter S phase (Fig. 9b), mirroring the effects of cortactin knockdown. Furthermore, a reduction in either RhoA or cortactin levels resulted in decreased expression of the F-box protein Skp2 (Fig. 9a), which is known to function as a vital component of the SCF complex that promotes proteasomal degradation of p27Kip1 and p57Kip2 (7, 21, 45, 50). Consequently, decreased SCF-Skp2-mediated degradation of p27Kip1 and p57Kip2 likely contributes to the increased expression of these CDKIs in cortactin knockdown cells, consistent with our observation that cortactin regulates p27Kip1 at the posttranscriptional level (Fig. 6). However, effects of cortactin/RhoA knockdown on Skp2 expression did not accurately correlate with changes in p27Kip1 or p57Kip2 levels. For example, RhoA siRNA 1 exerted a greater effect on these CDKIs than siRNA 2, despite a similar suppression of Skp2, and the enhancement of p27Kip1 and p57Kip2 expression reflected the degree of RhoA knockdown. Consequently, RhoA must utilize additional mechanisms for regulating p27Kip1 and p57Kip2 levels in addition to regulating Skp2 expression. In combination, these data indicate that cortactin overexpression leads to an enhancement of RhoA signaling that suppresses expression of Cip/Kip CDKIs and suggest that a cortactin/RhoA/Skp2 pathway contributes to the effect of cortactin on p27Kip1 and p57Kip2.
FIG. 9.
Determination of the role of RhoA/ROCK signaling in regulation of Cip/Kip CDKIs. (A) Effect of RhoA knockdown on expression of cell cycle regulators. siRNA against RhoA (10 nM) was applied to control cells during the serum starvation step of the synchronization procedure. Following synchronization, cells were released for the time periods indicated. Western blotting was undertaken as indicated. Data shown are representative of at least three independent experiments. (B) Effect of RhoA knockdown on cell cycle progression. Entry into S phase after a 6-h release was determined by uptake of BrdU during a 2-h pulse. The incorporation of BrdU was measured by flow cytometry. Data shown represent the mean ± standard error of three independent experiments. *, P < 0.05. (C) Effect of ROCK inhibition on expression of p21WAF1/Cip1 and p27Kip1. The ROCK inhibitor Y27632 was included during the hydroxyurea (HU) treatment and during release in growth medium at the concentrations indicated. Cell lysates were subjected to Western blotting as indicated. Data shown are representative of at least three independent experiments. (D) Effect of ROCK inhibition on cell cycle progression. Cells were treated as described for panel C, and entry into S phase after a 6-h release was determined by uptake of BrdU during a 2-h pulse. The incorporation of BrdU was measured by flow cytometry. Data shown represent the mean ± standard error of three independent experiments. *, P < 0.05.
In certain cellular contexts, RhoA can signal through its effector ROCK to decrease expression of p21WAF1/Cip1 and p27Kip1 (13, 27). We therefore sought to determine the role of ROCK in our system by treating synchronized control cells with Y27632, a pharmacological inhibitor of this enzyme. Administration of Y27632 decreased the phosphorylation of the ROCK target MLC2, indicating effective inhibition of ROCK signaling. However, it did not affect the expression of either p21WAF1/Cip1 or p27Kip1 (Fig. 9c), nor did it decrease S-phase entry (Fig. 9d), indicating that the effects of RhoA on cell cycle progression in HNSCC do not require ROCK activation and must involve an alternative RhoA effector pathway.
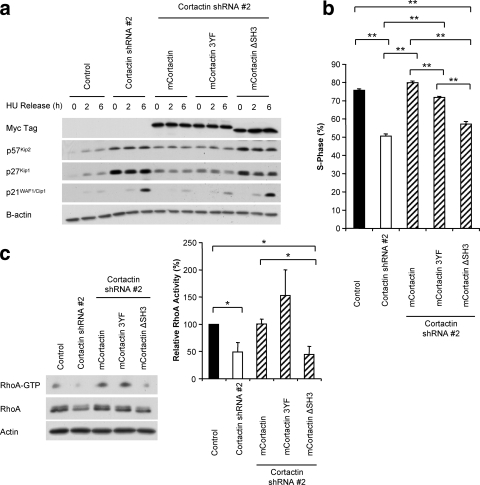
To further refine the functional link between cortactin and RhoA activation, we undertook complementation analysis using two key cortactin mutants (Fig. 10 a). In the first mutant (3YF), the three major sites of tyrosine phosphorylation are mutated to phenylalanine, which blocks phosphotyrosine-dependent interactions with Src family kinases and Nck (14, 47). The second mutant (ΔSH3) lacks the SH3 domain so that it does not bind a subset of cortactin interactors (e.g., BPGAP1 and Fgd1) but retains binding of F-actin, Arp2/3, and p120 catenin (14). Murine cDNAs encoding these mutants but resistant to the human cortactin-selective shRNA were reintroduced into cortactin knockdown cells. Expression of 3YF restored CDKI expression (Fig. 10a), S-phase entry (Fig. 10b), and RhoA activity (Fig. 10c) to levels similar to those of control FaDu cells. However, cortactin ΔSH3 did not affect these parameters. These data demonstrate a critical role for the cortactin SH3 domain in promotion of RhoA activation and cell cycle progression and indicate that recruitment of N-terminal binding partners is not sufficient for these effects. In addition, they demonstrate that tyrosine phosphorylation of cortactin is not required. While not formally defining the exact mechanism for cortactin's effects on RhoA, these data rule out certain candidates (e.g., p120 catenin) and implicate one or more targets of the cortactin SH3 domain.
FIG. 10.
The cortactin SH3 domain is required for cortactin to promote S-phase entry and RhoA activation. (A) FaDu control and cortactin knockdown cells, along with cortactin knockdown cells expressing shRNA-resistant myc-tagged wild-type murine cortactin, 3YF or ΔSH3 mutant versions, were synchronized into G1 phase by serum starvation and hydroxyurea (HU) treatment and then released into complete medium for the time periods indicated. Expression of the indicated proteins was then analyzed by Western blotting. Data shown are representative of three independent experiments. (B) Effect of cortactin mutants on cell cycle progression. Entry into S phase after a 6-h release was determined by uptake of BrdU during a 2-h pulse. The incorporation of BrdU was measured by flow cytometry. Data shown represent the mean ± standard error of three independent experiments. **, P < 0.01. (C) Cortactin lacking the SH3 domain does not rescue RhoA activity. Active RhoA was isolated from synchronized cells using Rhotekin RBD-GST beads. Active and total levels of RhoA were then determined by Western blotting. Activity was normalized relative to the level of total protein in each cell type and is expressed relative to the level of activity in control cells, which was arbitrarily set at 100%. Data shown represent the mean ± standard error of at least three independent experiments. *, P < 0.05.
DISCUSSION
Recent evidence indicates that cortactin overexpression promotes the proliferation of HNSCC cells (9, 30, 49) although the underlying mechanisms require further clarification. In particular, how cortactin overexpression impacts upon cell cycle regulation has not been addressed. In the manuscript we demonstrate that in 11q13-amplified HNSCC cells, cortactin promotes cell cycle progression by downregulating expression of p21WAF1/Cip1, p27Kip1, and p57Kip2. In addition, we report that in these cells, enhancement of RhoA signaling represents a key mechanism whereby cortactin modulates the levels of Cip/Kip CDKIs. These findings provide important insights into the oncogenic role of cortactin and lend further support to our hypothesis that the selective pressure for CTTN amplification in primary tumors is its ability to enhance cancer cell proliferation.
FaDu cells with stable cortactin knockdown exhibited marked increases in the expression of p21WAF1/Cip1, p27Kip1, and p57Kip2. This represents the first demonstration that cortactin can modulate the expression of Cip/Kip CDKIs. Consistent with a functional role for these CDKIs in mediating the effects of cortactin knockdown, enhanced CDKI expression was associated with increased coimmunoprecipitation of p21WAF1/Cip1 and p27Kip1 with cyclin D1 and cyclin E1 and Rb hypophosphorylation. Direct evidence that these CDKIs attenuate cell cycle progression in cortactin knockdown cells was obtained by manipulating their expression. Individual knockdown of p21WAF1/Cip1, p27Kip1, and p57Kip2 resulted in a partial rescue of cell cycle progression in cortactin knockdown cells, and although we were unable to achieve effective knockdown of all three CDKIs simultaneously, reducing the expression of p21WAF1/Cip1 and p27Kip1 in combination resulted in a further enhancement of S-phase entry. While the degree of cell cycle progression obtained with this dual knockdown was still less than that of control cells, the residual defect in S-phase entry was comparable to that reversible by p57Kip2 knockdown. These data indicate that p21WAF1/Cip1 and p27Kip1 do not play redundant roles and strongly suggest that the effect of cortactin knockdown on cell cycle progression requires upregulation of all three CDKIs.
In contrast to our data regarding CDKI expression, the increase in cyclin D1 expression observed upon cortactin knockdown initially seems counter-intuitive as the role of cyclin D1 in promoting the G1/S transition is well characterized (6). Additionally, the possibility of Cip/Kip CDKIs acting as assembly factors for cyclin D1/Cdk4 complexes at low concentrations (25) further complicates the interpretation of this data. However, both total Rb phosphorylation and specific phosphorylation at Ser780, a residue targeted by cyclin D1-dependent CDKs (24), were reduced by cortactin knockdown, arguing that the increased levels of cyclin D1 did not result in an increase in associated Cdk activity. This suggests that within our model of cortactin knockdown, the levels of p21WAF1/Cip1, p27Kip1, and p57Kip2 are high enough to act as inhibitors of cyclin D1/Cdk activity. It is therefore likely that this increase in cyclin D1 levels reflects a nuclear accumulation of inactive cyclin D1/Cdk/CDKI complexes that have escaped proteasomal degradation. This theory is supported by previous studies demonstrating the ability of p21WAF1/Cip1, p27Kip1, and p57Kip2 to direct cyclin D1 accumulation in the nucleus (25); the increased stability of cyclin D1 in the presence of p21WAF1/Cip1 and p27Kip1 (8); and the refractory nature of nuclear cyclin D1/p21WAF1/Cip1 complexes toward proteasomal degradation (2). Furthermore, the reversion of cyclin D1 expression toward control levels upon transient p21WAF1/Cip1, p27Kip1, or p57Kip2 knockdown in cells with reduced cortactin expression (Fig. 6) highlights their ability to stabilize cyclin D1 in our model.
Several lines of evidence support a role for RhoA in mediating the effect of cortactin on cell cycle progression. First, both RhoA activity and expression were reduced upon cortactin knockdown, and lowering the expression of either RhoA or cortactin led to decreased Skp2 levels and enhanced expression of Cip/Kip CDKIs and cyclin D1. Second, cortactin overexpression reduced p21WAF1/Cip1 and p27Kip1 expression at the transcriptional and posttranscriptional levels, respectively, consistent with previous studies demonstrating that RhoA signaling can repress p21WAF1/Cip1 transcription (33) and promote p27Kip1 degradation (55). Third, the ability of cortactin and RhoA to enhance Skp2 expression provides one mechanism for posttranscriptional regulation of p27Kip1 and, presumably, p57Kip2 as Skp2 plays a critical role in the proteasomal degradation of Kip family CDKIs (7, 21, 36, 45, 50). However, we note that for a given change in RhoA levels, cortactin knockdown exerted a more potent effect on CDKI and cyclin D1 expression and on cell cycle progression than siRNA-mediated manipulation of RhoA alone. A potential explanation is provided by our observation that cortactin knockdown affects RhoA activity as well as expression since reducing RhoA using siRNA may not achieve comparable effects on downstream signaling. Alternatively, cortactin may use additional pathways to signal to these cell cycle regulators. With regard to the latter hypothesis, Akt signaling is unlikely to make a major contribution since pharmacological inhibition of this kinase did not affect p21WAF1/Cip1 or p27Kip1 expression. Furthermore, cortactin cannot signal to p21WAF1/Cip1 via p53 since the latter protein is mutated and inactive in FaDu cells (23).
An additional insight provided by the effects of RhoA knockdown on cell cycle regulators (Fig. 9) is that RhoA must utilize additional mechanisms for regulating p27Kip1 and p57Kip2 levels in addition to enhancing Skp2 expression. A potential mechanism is via regulation of protein synthesis since RhoA negatively regulates p27Kip1 mRNA translation via a Rho-responsive element in the 3′ untranslated region of the p27Kip1 transcript (51). In addition, p27Kip1 is degraded in G1 by the Kip ubiquitin-promoting complex (22) although effects of RhoA signaling on this process have not been reported.
Depending on the cellular context, RhoA can suppress p21WAF1/Cip1 transcription and p27Kip1 expression by ROCK-dependent and -independent mechanisms (13, 27, 31, 41, 42, 57). In control FaDu cells, RhoA knockdown inhibited cell cycle progression and enhanced p21WAF1/Cip1 and p27Kip1 levels, whereas these parameters were unaffected by the ROCK inhibitor Y27632, indicating that RhoA-mediated regulation of these CDKIs must utilize one or more alternative effector pathways. A strong candidate involves the formin mammalian Diaphanous (mDia), since mDia activity is required for RhoA to enhance Skp2 expression and promote p27Kip1 degradation during cell spreading (31). In addition, a recent paper on gastric cancer cells reported that RhoA required mDia to suppress p21WAF1/Cip1 and p27Kip1 while RhoA/ROCK signaling downregulated INK4 family tumor suppressors (57). This raises the interesting possibility that two proteins intimately involved in regulation of actin polymerization, cortactin and mDia, function in a signaling pathway that promotes cell cycle progression in HNSCC. Since both proteins also promote cell migration (14, 17), targeting these proteins therapeutically could impact both cancer growth and metastasis.
Regulation of RhoA signaling does not appear to be a normal physiological function of cortactin (26). However, our data indicate that marked elevation of cortactin modulates RhoA activation in a biphasic manner, explaining why cortactin knockdown in 11q13-amplified FaDu cells results in a decrease in RhoA-GTP loading. There are several possible explanations for this effect, which are not mutually exclusive. Previously, we reported that cortactin overexpression in HNSCC was associated with impaired ligand-induced downregulation of the EGFR and MET and more sustained mitogenic signaling (48, 49). In addition, high cortactin expression promotes recycling of the G protein-coupled receptor CXCR4 (29). Consequently, the increased RhoA activation observed upon cortactin overexpression may reflect altered endocytic trafficking by particular growth factor receptors. However, it should be noted that while certain G protein-coupled receptors can stimulate RhoA directly, activation of RhoA by receptor tyrosine kinases often occurs secondary to effects on Rac, and we did not detect changes in Rac-GTP levels in our model system. Also, since the effect on RhoA was observed in synchronized cells in the absence of serum, stimulation of such receptors must result from autocrine production of growth factors by the cancer cells.
In addition, cortactin may modulate RhoA activation independently of receptor activation, for example, via interaction with one or more regulators of this GTPase. Since the effect on RhoA requires marked overexpression of cortactin, it might reflect a weak interaction that becomes functionally significant only at high cortactin levels and/or sequestration of a negative regulator. Importantly, our structure-function analysis rules out certain candidates (e.g., p120 catenin) and highlights binding partners of the cortactin SH3 domain for further investigation. One such protein is BPGAP1, which exhibits GTPase-activating protein (GAP) activity toward RhoA both in vitro and in vivo (44, 54). Whether the regulation of RhoA expression observed in FaDu cells reflects posttranslational modification of RhoA, for example, by ubiquitylation, is currently under investigation.
In conclusion, we have identified a novel RhoA-mediated mechanism contributing to cortactin-stimulated cell proliferation in HNSCC, which is of particular relevance in this cancer type due to the relatively high frequency of cortactin overexpression, and the association of this clinicopathological parameter with decreased patient survival. This finding is a significant step forward in understanding the biological role of the cortactin oncoprotein and may assist in the development of therapeutic strategies aimed at inhibiting cortactin signaling in HNSCC.
Supplementary Material
Acknowledgments
Grants were received from the Cancer Institute of New South Wales (CINSW), the Cancer Council of New South Wales, and the National Health and Medical Research Council of Australia. D.R.C. and E.A.M. hold CINSW Early Career Development and Career Development and Support fellowships, respectively.
Footnotes
Published ahead of print on 30 August 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abukhdeir, A. M., and B. H. Park. 2008. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev. Mol. Med. 10:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alt, J. R., A. B. Gladden, and J. A. Diehl. 2002. p21Cip1 promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export. J. Biol. Chem. 277:8517-8523. [DOI] [PubMed] [Google Scholar]
- 3.Bain, J., L. Plater, M. Elliott, N. Shpiro, C. J. Hastie, H. McLauchlan, I. Klevernic, J. S. Arthur, D. R. Alessi, and P. Cohen. 2007. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408:297-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden, E. T., M. Barth, D. Thomas, R. I. Glazer, and S. C. Mueller. 1999. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene 18:4440-4449. [DOI] [PubMed] [Google Scholar]
- 5.Bryce, N. S., E. S. Clark, J. L. Leysath, J. D. Currie, D. J. Webb, and A. M. Weaver. 2005. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 15:1276-1285. [DOI] [PubMed] [Google Scholar]
- 6.Caldon, C. E., R. J. Daly, R. L. Sutherland, and E. A. Musgrove. 2006. Cell cycle control in breast cancer cells. J. Cell Biochem. 97:261-274. [DOI] [PubMed] [Google Scholar]
- 7.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, M., P. Olivier, J. A. Diehl, M. Fero, M. F. Roussel, J. M. Roberts, and C. J. Sherr. 1999. The p21(Cip1) and p27(Kip1) CDK “inhibitors” are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18:1571-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, E. S., B. Brown, A. S. Whigham, A. Kochaishvili, W. G. Yarbrough, and A. M. Weaver. 2009. Aggressiveness of HNSCC tumors depends on expression levels of cortactin, a gene in the 11q13 amplicon. Oncogene 28:431-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, E. S., and A. M. Weaver. 2008. A new role for cortactin in invadopodia: regulation of protease secretion. Eur. J. Cell Biol. 87:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, E. S., A. S. Whigham, W. G. Yarbrough, and A. M. Weaver. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67:4227-4235. [DOI] [PubMed] [Google Scholar]
- 12.Courjal, F., M. Cuny, J. Simony-Lafontaine, G. Louason, P. Speiser, R. Zeillinger, C. Rodriguez, and C. Theillet. 1997. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 57:4360-4367. [PubMed] [Google Scholar]
- 13.Croft, D. R., and M. F. Olson. 2006. The Rho GTPase effector ROCK regulates cyclin A, cyclin D1, and p27Kip1 levels by distinct mechanisms. Mol. Cell. Biol. 26:4612-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly, R. J. 2004. Cortactin signalling and dynamic actin networks. Biochem. J. 382:13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edlund, S., M. Landstrom, C. H. Heldin, and P. Aspenstrom. 2002. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol. Biol. Cell 13:902-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibcus, J. H., M. F. Mastik, L. Menkema, G. H. de Bock, P. M. Kluin, E. Schuuring, and J. E. van der Wal. 2008. Cortactin expression predicts poor survival in laryngeal carcinoma. Br. J. Cancer 98:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupton, S. L., K. Eisenmann, A. S. Alberts, and C. M. Waterman-Storer. 2007. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J. Cell Sci. 120:3475-3487. [DOI] [PubMed] [Google Scholar]
- 18.Hofman, P., C. Butori, K. Havet, V. Hofman, E. Selva, N. Guevara, J. Santini, and E. Van Obberghen-Schilling. 2008. Prognostic significance of cortactin levels in head and neck squamous cell carcinoma: comparison with epidermal growth factor receptor status. Br. J. Cancer. 98:956-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, C., J. Liu, C. C. Haudenschild, and X. Zhan. 1998. The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J. Biol. Chem. 273:25770-25776. [DOI] [PubMed] [Google Scholar]
- 20.Hui, R., D. H. Campbell, C. S. Lee, K. McCaul, D. J. Horsfall, E. A. Musgrove, R. J. Daly, R. Seshadri, and R. L. Sutherland. 1997. EMS1 amplification can occur independently of CCND1 or INT-2 amplification at 11q13 and may identify different phenotypes in primary breast cancer. Oncogene 15:1617-1623. [DOI] [PubMed] [Google Scholar]
- 21.Kamura, T., T. Hara, S. Kotoshiba, M. Yada, N. Ishida, H. Imaki, S. Hatakeyama, K. Nakayama, and K. I. Nakayama. 2003. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc. Natl. Acad. Sci. U. S. A. 100:10231-10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamura, T., T. Hara, M. Matsumoto, N. Ishida, F. Okumura, S. Hatakeyama, M. Yoshida, K. Nakayama, and K. I. Nakayama. 2004. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 6:1229-1235. [DOI] [PubMed] [Google Scholar]
- 23.Kim, M. S., S. L. Li, C. N. Bertolami, H. M. Cherrick, and N. H. Park. 1993. State of p53, Rb and DCC tumor suppressor genes in human oral cancer cell lines. Anticancer Res. 13:1405-1413. [PubMed] [Google Scholar]
- 24.Kitagawa, M., H. Higashi, H. K. Jung, I. Suzuki-Takahashi, M. Ikeda, K. Tamai, J. Kato, K. Segawa, E. Yoshida, S. Nishimura, and Y. Taya. 1996. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15:7060-7069. [PMC free article] [PubMed] [Google Scholar]
- 25.LaBaer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11:847-862. [DOI] [PubMed] [Google Scholar]
- 26.Lai, F. P., M. Szczodrak, J. M. Oelkers, M. Ladwein, F. Acconcia, S. Benesch, S. Auinger, J. Faix, J. V. Small, S. Polo, T. E. Stradal, and K. Rottner. 2009. Cortactin promotes migration and platelet-derived growth factor-induced actin reorganization by signaling to Rho-GTPases. Mol. Biol. Cell 20:3209-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, J. M., S. Wu, D. Y. Huang, and Z. F. Chang. 2002. Cytosolic retention of phosphorylated extracellular signal-regulated kinase and a Rho-associated kinase-mediated signal impair expression of p21Cip1/Waf1 in phorbol 12-myristate-13-acetate-induced apoptotic cells. Mol. Cell. Biol. 22:7581-7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, H. S., H. H. Lu, M. T. Lui, E. H. Yu, W. Shen, Y. P. Chen, K. W. Chang, and H. F. Tu. 2009. Detection of copy number amplification of cyclin D1 (CCND1) and cortactin (CTTN) in oral carcinoma and oral brushed samples from areca chewers. Oral Oncol. 45:1032-1036. [DOI] [PubMed] [Google Scholar]
- 29.Luo, C., H. Pan, M. Mines, K. Watson, J. Zhang, and G. H. Fan. 2006. CXCL12 induces tyrosine phosphorylation of cortactin, which plays a role in CXC chemokine receptor 4-mediated extracellular signal-regulated kinase activation and chemotaxis. J. Biol. Chem. 281:30081-30093. [DOI] [PubMed] [Google Scholar]
- 30.Luo, M. L., X. M. Shen, Y. Zhang, F. Wei, X. Xu, Y. Cai, X. Zhang, Y. T. Sun, Q. M. Zhan, M. Wu, and M. R. Wang. 2006. Amplification and overexpression of CTTN (EMS1) contribute to the metastasis of esophageal squamous cell carcinoma by promoting cell migration and anoikis resistance. Cancer Res. 66:11690-11699. [DOI] [PubMed] [Google Scholar]
- 31.Mammoto, A., S. Huang, K. Moore, P. Oh, and D. E. Ingber. 2004. Role of RhoA, mDia, and ROCK in cell shape-dependent control of the Skp2-p27kip1 pathway and the G1/S transition. J. Biol. Chem. 279:26323-26330. [DOI] [PubMed] [Google Scholar]
- 32.Musgrove, E. A., L. J. Hunter, C. S. Lee, A. Swarbrick, R. Hui, and R. L. Sutherland. 2001. Cyclin D1 overexpression induces progestin resistance in T-47D breast cancer cells despite p27Kip1 association with cyclin E-Cdk2. J. Biol. Chem. 276:47675-47683. [DOI] [PubMed] [Google Scholar]
- 33.Olson, M. F., H. F. Paterson, and C. J. Marshall. 1998. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature 394:295-299. [DOI] [PubMed] [Google Scholar]
- 34.Ormandy, C. J., E. A. Musgrove, R. Hui, R. J. Daly, and R. L. Sutherland. 2003. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res. Treat. 78:323-335. [DOI] [PubMed] [Google Scholar]
- 35.Patel, A. S., G. L. Schechter, W. J. Wasilenko, and K. D. Somers. 1998. Overexpression of EMS1/cortactin in NIH3T3 fibroblasts causes increased cell motility and invasion in vitro. Oncogene 16:3227-3232. [DOI] [PubMed] [Google Scholar]
- 36.Pateras, I. S., K. Apostolopoulou, K. Niforou, A. Kotsinas, and V. G. Gorgoulis. 2009. p57KIP2: “Kip”ing the cell under control. Mol. Cancer Res. 7:1902-1919. [DOI] [PubMed] [Google Scholar]
- 37.Paulsen, R. D., and K. A. Cimprich. 2007. The ATR pathway: fine-tuning the fork. DNA Repair (Amst.) 6:953-966. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigo, J. P., D. Garcia-Carracedo, L. A. Garcia, S. Menendez, E. Allonca, M. V. Gonzalez, M. F. Fresno, C. Suarez, and J. M. Garcia-Pedrero. 2009. Distinctive clinicopathological associations of amplification of the cortactin gene at 11q13 in head and neck squamous cell carcinomas. J. Pathol. 217:516-523. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigo, J. P., L. A. Garcia, S. Ramos, P. S. Lazo, and C. Suarez. 2000. EMS1 gene amplification correlates with poor prognosis in squamous cell carcinomas of the head and neck. Clin. Cancer Res. 6:3177-3182. [PubMed] [Google Scholar]
- 40.Rothschild, B. L., A. H. Shim, A. G. Ammer, L. C. Kelley, K. B. Irby, J. A. Head, L. Chen, M. Varella-Garcia, P. G. Sacks, B. Frederick, D. Raben, and S. A. Weed. 2006. Cortactin overexpression regulates actin-related protein 2/3 complex activity, motility, and invasion in carcinomas with chromosome 11q13 amplification. Cancer Res. 66:8017-8025. [DOI] [PubMed] [Google Scholar]
- 41.Sahai, E., T. Ishizaki, S. Narumiya, and R. Treisman. 1999. Transformation mediated by RhoA requires activity of ROCK kinases. Curr. Biol. 9:136-145. [DOI] [PubMed] [Google Scholar]
- 42.Sahai, E., M. F. Olson, and C. J. Marshall. 2001. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 20:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuuring, E. 1995. The involvement of the chromosome 11q13 region in human malignancies: cyclin D1 and EMS1 are two new candidate oncogenes—a review. Gene 159:83-96. [DOI] [PubMed] [Google Scholar]
- 44.Shang, X., Y. T. Zhou, and B. C. Low. 2003. Concerted regulation of cell dynamics by BNIP-2 and Cdc42GAP homology/Sec14p-like, proline-rich, and GTPase-activating protein domains of a novel Rho GTPase-activating protein, BPGAP1. J. Biol. Chem. 278:45903-45914. [DOI] [PubMed] [Google Scholar]
- 45.Sutterluty, H., E. Chatelain, A. Marti, C. Wirbelauer, M. Senften, U. Muller, and W. Krek. 1999. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1:207-214. [DOI] [PubMed] [Google Scholar]
- 46.Swarbrick, A., C. S. Lee, R. L. Sutherland, and E. A. Musgrove. 2000. Cooperation of p27Kip1 and p18INK4c in progestin-mediated cell cycle arrest in T-47D breast cancer cells. Mol. Cell. Biol. 20:2581-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tehrani, S., N. Tomasevic, S. Weed, R. Sakowicz, and J. A. Cooper. 2007. Src phosphorylation of cortactin enhances actin assembly. Proc. Natl. Acad. Sci. U. S. A. 104:11933-11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timpson, P., D. K. Lynch, D. Schramek, F. Walker, and R. J. Daly. 2005. Cortactin overexpression inhibits ligand-induced down-regulation of the epidermal growth factor receptor. Cancer Res. 65:3273-3280. [DOI] [PubMed] [Google Scholar]
- 49.Timpson, P., A. S. Wilson, G. M. Lehrbach, R. L. Sutherland, E. A. Musgrove, and R. J. Daly. 2007. Aberrant expression of cortactin in head and neck squamous cell carcinoma cells is associated with enhanced cell proliferation and resistance to the epidermal growth factor receptor inhibitor gefitinib. Cancer Res. 67:9304-9314. [DOI] [PubMed] [Google Scholar]
- 50.Tsvetkov, L. M., K. H. Yeh, S. J. Lee, H. Sun, and H. Zhang. 1999. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9:661-664. [DOI] [PubMed] [Google Scholar]
- 51.Vidal, C., B. Geny, J. Melle, M. Jandrot-Perrus, and M. Fontenay-Roupie. 2002. Cdc42/Rac1-dependent activation of the p21-activated kinase (PAK) regulates human platelet lamellipodia spreading: implication of the cortical-actin binding protein cortactin. Blood 100:4462-4469. [DOI] [PubMed] [Google Scholar]
- 52.Walker, I. G., R. W. Yatscoff, and R. Sridhar. 1977. Hydroxyurea: induction of breaks in template strands of replicating DNA. Biochem. Biophys. Res. Commun. 77:403-408. [DOI] [PubMed] [Google Scholar]
- 53.Wang, W., L. Chen, Y. Ding, J. Jin, and K. Liao. 2008. Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J. Cell Sci. 121:1334-1343. [DOI] [PubMed] [Google Scholar]
- 54.Weaver, A. M. 2008. Cortactin in tumor invasiveness. Cancer Lett. 265:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber, J. D., W. Hu, S. C. Jefcoat, Jr., D. M. Raben, and J. J. Baldassare. 1997. Ras-stimulated extracellular signal-related kinase 1 and RhoA activities coordinate platelet-derived growth factor-induced G1 progression through the independent regulation of cyclin D1 and p27. J. Biol. Chem. 272:32966-32971. [DOI] [PubMed] [Google Scholar]
- 56.Welsh, C. F. 2004. Rho GTPases as key transducers of proliferative signals in g1 cell cycle regulation. Breast Cancer Res. Treat 84:33-42. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, S., Q. Tang, F. Xu, Y. Xue, Z. Zhen, Y. Deng, M. Liu, J. Chen, S. Liu, M. Qiu, Z. Liao, Z. Li, D. Luo, F. Shi, Y. Zheng, and F. Bi. 2009. RhoA regulates G1-S progression of gastric cancer cells by modulation of multiple INK4 family tumor suppressors. Mol. Cancer Res. 7:570-580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.