Abstract
Concomitantly with an outbreak of fatal anaplasmosis in a cattle herd in Switzerland in 2002, we detected two bovine hemoplasma species in diseased animals: Mycoplasma wenyonii (formerly Eperythrozoon wenyonii) and a second, novel bovine hemoplasma species later designated “Candidatus Mycoplasma haemobos” (synonym, “Candidatus Mycoplasma haemobovis”). The second species was characterized by a shorter 16S rRNA gene. The aims of the present study were to provide a detailed molecular characterization of this species, to develop specific quantitative real-time PCR assays for the two bovine hemoplasma species, and to apply these assays in order to evaluate the prevalence and clinical significance of the hemoplasmas. Sequencing of the near-complete 16S rRNA gene of the second hemoplasma revealed that it was 94% identical to that of Mycoplasma haemofelis, an anemia-inducing feline hemoplasma species, but less than 85% identical to that of the bovine hemoplasma M. wenyonii. Using the newly developed assays, a total of 159 animals from the anaplasmosis outbreak were reexamined. In addition, we tested 57 clinically ill and 61 healthy Swiss cattle, as well as 47 calves. Both hemoplasmas were highly prevalent in adult cattle but occurred rarely in calves. Animals from the herd with the fatal anemia outbreak were more frequently infected with M. wenyonii and exhibited higher M. wenyonii blood loads than animals with unrelated diseases and healthy animals. Coinfections may increase the pathogenicity and clinical significance of bovine hemoplasmosis.
In connection with an outbreak of anaplasmosis in a cattle herd in eastern Switzerland in 2002, more than 300 animals were culled. Most of these cattle exhibited pronounced anemia. The anemia was statistically associated with the detection of Anaplasma marginale, Babesia spp., Theileria spp., and Mycoplasma wenyonii in the blood of diseased animals (5).
M. wenyonii, first described in 1934, was formerly known as Eperythrozoon wenyonii (1, 13). The species was recently reclassified within the group of hemotropic mycoplasmal species based on the 16S rRNA gene sequence (11-13). M. wenyonii is a cell wall-free bacterium that parasitizes bovine red blood cells (11). In our study of the above-mentioned outbreak, we reported two distinct hemotropic mycoplasma species: M. wenyonii and a second, unknown, agent (5). The 16S rRNA gene of the second agent was shorter than that in M. wenyonii and was 95% identical to the 16S rRNA gene found in Mycoplasma haemofelis, the causative agent of feline infectious anemia (24, 25). A similar bovine hemoplasma species has since been detected in China and Japan using molecular assays, and the name “Candidatus Mycoplasma haemobos” has been proposed (20). Other bovine hemoplasmas that were found to be distinct from M. wenyonii using other methods are described elsewhere (3, 30-32). Characterization included morphological and immunogenic differences, as well as different localization of the agents within the host (28, 29, 31, 32, 38).
The clinical relevance of M. wenyonii is controversial (16, 18); in the United States, infection with M. wenyonii is considered to be of low pathogenicity. However, a study with splenectomized calves showed that a preexisting M. wenyonii infection followed by an A. marginale superinfection led to severe anemia, with packed-cell volumes (PCV) dropping from 30% to 9.5% when M. wenyonii was found in the blood and about 2 weeks before A. marginale appeared (13). The clinical relevance of “Candidatus Mycoplasma haemobos” remains unclear (14, 20).
The aims of the present study were to characterize the two bovine hemotropic mycoplasma species identified in 2002 using molecular techniques, to develop specific real-time TaqMan PCR assays for the detection and quantification of these species, and to determine the occurrence of the two bovine hemoplasmas in sick and healthy cattle in order to evaluate their clinical importance.
MATERIALS AND METHODS
Samples.
We studied a total of 216 EDTA-anticoagulated blood samples collected from diseased animals. In total, 159 samples came from cattle originating from a large dairy herd that had experienced an anaplasmosis disease outbreak in Switzerland in August 2002 (5). The remaining 57 samples originated from cows that were brought to the clinic for farm animals at the University of Zurich between January and May 2004 for the treatment of diseases other than anaplasmosis. In addition, whole blood samples were available from 61 healthy animals originating from different Swiss farms. Furthermore, we also tested EDTA-anticoagulated blood samples from 12 healthy calves from a herd owned by the University of Zurich as well as 35 calves from 20 different herds in northeastern Switzerland.
PCR analysis and sequencing.
DNA was purified from 200 μl of whole blood with a MagNA Pure LC DNA isolation kit I (Roche Diagnostics, Rotkreuz, Switzerland).
The near-complete sequence of the 16S rRNA gene of the new hemoplasma isolate was determined from three cows. Two of the cows were involved in the outbreak; one was infected only with the new hemoplasma isolate (isolate CH311), while the second was coinfected with the new isolate and with A. marginale (isolate CH307). The third cow originated from the group of animals that were transported to the University of Zurich clinic (isolate CH88). Briefly, the species-specific primers MHBforw (5′-GAA TTA ATG CTG ATG GTA TGC CTA A-3′, 25 bp) and MHBrev (5′-CCA ATC AGA ATG TTC ACT CTA GAT GC-3′, 26 bp) were used to amplify 2.5 μl of template DNA in a 25-μl reaction mixture containing 5× Phusion HF buffer (Finnzymes, Espoo, Finland), 500 nM each primer, 200 nM each deoxynucleoside triphosphate (dNTP) (Sigma-Aldrich, Buchs, Switzerland), and 1 U Phusion DNA polymerase (Finnzymes). The thermal program comprised 98°C for 3 min; 35 cycles of 98°C for 10 s, 52°C for 30 s, and 72°C for 1 min; and finally 72°C for 10 min. PCR products of 1,393 bp were gel purified and cloned into the vector pCR4-TOPO (Invitrogen, Basel, Switzerland). Sequencing was performed with vector-specific M13 forward and reverse primers and two internal primers, Mwen_short.forw (5′-CCA TGT GAA CGA AGA AGG TCT TT-3′, 23 bp) and Mwen_short.rev (5′-AGT TTG CTG TCA CTT ATT CAT GAG GTA-3′, 27 bp), using the BigDye Terminator cycle sequencing ready reaction kit v1.1 (Applied Biosystems, Rotkreuz, Switzerland). Sequences were then analyzed on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). Three clones from each animal were sequenced.
For phylogenetic and molecular evolutionary analysis, the sequences were aligned with known hemoplasma sequences from GenBank using ClustalW (26) and manually adjusted when necessary. Only nucleotides that were available for all included sequences were used in the phylogenetic analysis. A bootstrap phylogenetic tree was used to demonstrate the relationship of the new bovine species to other hemoplasma species. The tree was created by the neighbor-joining method (17) using a distance matrix corrected for nucleotide substitutions based on the Kimura two-parameter model. The data set was resampled 1,000 times to generate bootstrap values. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (21).
Development of real-time PCR assays specific for the two bovine hemotropic mycoplasma species.
To develop specific real-time PCR assays, two sets of primers and probes were designed. The first set, specific for M. wenyonii, was designed based on alignments of M. wenyonii 16S rRNA gene sequences from five cattle (5) (accession numbers GQ259756 to GQ259760) and on a published sequence (AY946266): MwenyoniiF (5′-CCA CGT GAA CGA TGA AGG TCT T-3′, 22 bp), MwenyoniiR (5′-GGC ACA TAG TTA GCT GTC ACT TAT TCA A-3′, 28 bp), and Mweny_P (5′-FAM-AGT ACC ATC AAG GCG CGC TCA TTT CCT AG-TAMRA-3′, 29 bp). Sequence comparisons revealed that the assay may potentially amplify Mycoplasma ovis (AF338268) and also a swine hemotropic mycoplasma (DQ346727) but not the feline and canine hemoplasma species or “Candidatus Mycoplasma haemobos.” The second set of primers was designed based on alignments of 16S rRNA gene sequences from the bovine mycoplasma isolates obtained from three animals (5) (GQ259761 to GQ259763), which were compared to the 16S rRNA sequence of M. haemofelis (AY069948 and U88563) and M. haemocanis (AF407208 and AF197337). The assay was designed to amplify “Candidatus Mycoplasma haemobos,” but not the feline and canine sequences or the common M. wenyonii, using the following primers and probe: Mwen_short.forw (5′-CCA TGT GAA CGA TGA AGG TCT TT-3′, 23 bp), Mwen_short.rev (5′-AGT TTG CTG TCA CTT ATT CAT GAG GTA-3′, 27 bp), and Mwen_short.p (5′-VIC-CTA TCA GTT RTT ATC CCT CAT AA-MGB-3′, 23 bp). For both systems, the 25-μl PCR mixture comprised 12.5 μl of 2× quantitative PCR (qPCR) Mastermix (Eurogentec), 900 nM each primer, 250 nM probe, and 5 μl of template DNA. Assays were performed using the ABI PRISM 7700 sequence detection system (Applied Biosystems).
DNA standards for absolute quantitation.
The sequenced pCR4-TOPO plasmids (see above) containing the almost-complete 16S rRNA genes of “Candidatus Mycoplasma haemobos” (isolates CH311 and CH307) and M. wenyonii (isolate CH82), including the target sequences encompassed by the real-time PCR assays, were linearized by restriction digestion using NotI and gel purified (QIAquick gel extraction kit; Qiagen). The plasmid DNA copy numbers were then calculated by spectrophotometry (NanoDrop ND-1000; Witec Ag, Littau, Switzerland) and agarose gel electrophoresis (Gene Tools; Syngene, Cambridge, United Kingdom). Ten-fold serial dilutions (108 to 100 copies/reaction) of the DNA standards were prepared and analyzed as previously described (22).
Analytic specificities, sensitivities, and amplification efficiencies of the real-time PCR assays.
The specificities of both assays were evaluated by testing template DNA from the hemotropic mycoplasma species “Candidatus Mycoplasma haemominutum” and M. haemofelis (feline), M. haemocanis and “Candidatus Mycoplasma heamatoparvum” (canine), and “Candidatus Mycoplasma haemolamae” (camelidae), as well as the nonhemotropic mycoplasma species M. arginini, M. agalactiae, M. penetrans, and M. pneumoniae. The sensitivities of the assays were assessed by analyzing 10-fold dilutions of standard DNA templates produced using the cloned 16S rRNA sequences. The sensitivity of the assay was assessed in an endpoint dilution experiment using standard DNA template as previously described (22). The amplification efficiency was determined by assessing the slope (s) of the curve (threshold cycle versus dilution) obtained by PCR amplification of serial 10-fold dilutions of the standard DNA and a positive DNA sample in triplicates. Amplification efficiency was calculated as 101/−s − 1 (7).
Statistical analysis.
Data were compiled and analyzed with Excel 2007 (Microsoft), Analyze-it clinical laboratory version 2.20 (Analyze-it Software, Ltd., Leeds, United Kingdom), and GraphPad Prism version 3.00 (GraphPad Software, Inc., San Diego, CA). The frequency of infection between groups was assessed using a two-tailed chi-square test. Blood loads (copy numbers) were tested for statistically relevant differences among cattle groups using a Kruskal-Wallis one-way analysis of variance (ANOVA) by ranks and a Dunn posttest for multiple comparisons.
Nucleotide sequence accession numbers.
The nucleotide sequences have been submitted to GenBank under accession numbers EF616467, EF616468, and GQ259756 to GQ259763.
RESULTS
Molecular characterization of the second bovine hemoplasma species.
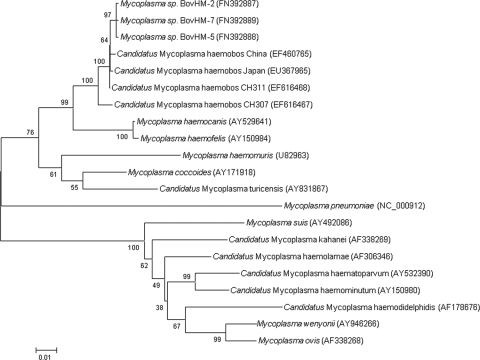
The second, unknown bovine hemoplasma isolate identified during the outbreak of fatal anemia (5) was characterized by a shorter 16S rRNA gene fragment (Fig. 1). Sequencing of the amplified PCR products (193 and 172 bp) revealed that that the longer amplicon (193 bp; accession numbers GQ259756 to GQ259760) was >99% identical to published M. wenyonii sequences (AF016546). Interestingly, the shorter amplicon (172 bp; accession numbers GQ259761 to GQ259763) was 95% identical to M. haemofelis (U88563 and U95297). The sequences of nearly the entire 16S rRNA genes of three isolates (CH88, CH307, and CH311) were determined. The sequence of the CH88 isolate was 100% identical to that of isolate CH311 (EF616468). Isolates CH307 (EF616467) and CH311 were 98% identical to each other. Comparisons of the gene sequences obtained with those in the GenBank database showed the highest identity (Table 1) to “Candidatus Mycoplasma haemobos” from China and Japan (EF460765 and EU367965) and to three bovine mycoplasma sequences from German cattle (FN392887 to FN392889). Identities to other hemotropic mycoplasma species are summarized in Table 1. A bootstrap phylogenetic tree confirmed the close evolutionary relationship between the Swiss isolates, “Candidatus Mycoplasma haemobos” from China and Japan, and the bovine German isolates, which formed a separate cluster (Fig. 2). Together with M. haemocanis and M. haemofelis, they branched away from the group of M. wenyonii.
FIG. 1.
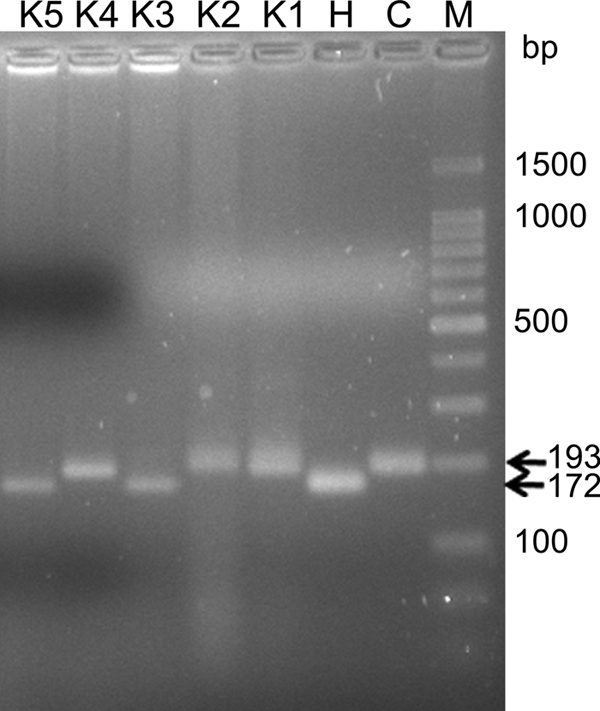
Bovine hemoplasma 16S rRNA gene products identified using conventional PCR. Agarose gel electrophoresis (3%) of PCR products that were amplified with primers specific for both feline and bovine hemoplasma 16S rRNAs (MychaeF and MychaeR [25]) is shown. Abbreviations: M, molecular size marker (100-bp ladder); C, “Candidatus Mycoplasma haemominutum”; H, M. haemofelis; K1 to K5, five different samples from cows from the herd with fatal anemia (5).
TABLE 1.
Sequence identity between “Candidatus Mycoplasma haemobos” Swiss isolates and other hemotropic mycoplasma species
| Organism (accession no.) | % Sequence identity with “Candidatus Mycoplasma haemobos” isolate (accession no.): |
|
|---|---|---|
| CH307 (EF616467) | CH311 (EF616468) | |
| “Candidatus Mycoplasma haemobos,” China (EF460765) | 98 | 99 |
| “Candidatus Mycoplasma haemobos,” Japan (EU367965) | 98 | 99 |
| Mycoplasmasp. strain BovHM-2, Germany (FN392887-FN392889) | 98 | 99 |
| M. haemocanis (AY529641) | 94 | 94 |
| M. haemofelis (AY150984) | 94 | 94 |
| M. coccoides (AY171918) | 90 | 90 |
| “Candidatus Mycoplasma turicensis” (AY831867) | 88 | 88 |
| M. haemomuris (U82963) | 87 | 87 |
| “Candidatus Mycoplasma haemominutum” (AY150980) | 85 | 82 |
| “Candidatus Mycoplasma haemolamae” (AF306346) | 85 | 85 |
| “Candidatus Mycoplasma kahanei” (AF338269) | 85 | 85 |
| “Candidatus Mycoplasma hematoparvum” (AY532390) | 84 | 84 |
| M. wenyonii (AY946266) | 84 | 84 |
| M. suis (AY492086) | 83 | 84 |
| “Candidatus Mycoplasma haemodidelphidis” (AF178676) | 83 | 84 |
| M. ovis (AF338268) | 83 | 84 |
FIG. 2.
Phylogenetic analysis of near-complete 16S rRNA gene sequences from “Candidatus Mycoplasma haemobos” and related hemoplasmas. A bootstrap phylogenetic tree demonstrating the relationship of the Swiss “Candidatus Mycoplasma haemobos” isolates (CH307 and CH311) to other hemoplasma species using the neighbor-joining method is shown. The numbers at the nodes were generated from 1,000 bootstrap resamplings. The bar represents the mean number of differences per 100 sites. GenBank accession numbers are shown in parentheses.
Real-time PCR assays.
We developed two real-time PCR assays for the specific detection of M. wenyonii and “Candidatus Mycoplasma haemobos.” We tested the specificities of the assays using isolates from different mycoplasma species, as noted in Materials and Methods. Neither of the assays amplified any of the other tested mycoplasmas. The assay for M. wenyonii did not amplify “Candidatus Mycoplasma haemobos” and vice versa (data not shown). Amplification of 10-fold serial dilutions of both standards (“Candidatus Mycoplasma haemobos” and M. wenyonii) showed a linear range over 8 orders of magnitude. The highest dilution still yielding a positive signal contained an average of one copy per reaction; in an endpoint dilution experiment, 9 out of 13 replicates (69%) for “Candidatus Mycoplasma haemobos” and 7 out of 12 replicates (58%) for M. wenyonii of this dilution were positive. The sensitivity of both assays was thus approximately one 16S rRNA gene copy per reaction, corresponding to 200 16S rRNA genome equivalents per ml blood. The amplification efficiency of the real-time PCR assay using the standard dilutions was 96.6% (slope of the curve, −3.41) for “Candidatus Mycoplasma haemobos” and 92.9% (slope of the curve, −3.50) for M. wenyonii.
Occurrence of M. wenyonii and “Candidatus Mycoplasma haemobos” in healthy and sick animals.
To assess the potential clinical importance of bovine hemotropic mycoplasmas, we tested different populations of cattle for M. wenyonii and “Candidatus Mycoplasma haemobos”. Both bovine hemoplasmas were frequently detected in diseased and healthy adult cattle, but just a few calves (3 out of 47) were found to be infected (Table 2). In the cattle herd with fatal anemia, the frequencies of M. wenyonii and “Candidatus Mycoplasma haemobos” infection were significantly higher than those in cattle with unrelated diseases (P [χ2] < 0.0001 and P [χ2] = 0.0044, respectively) and or in healthy cows (P [χ2] = 0.0007 and P [χ2] = 0.0467, respectively) (Table 2). The frequency of infection was significantly higher in the herd with fatal anemia when coinfection with both M. wenyonii and “Candidatus Mycoplasma haemobos” was taken into account. This was also true when the frequency of either infection was compared to that in cattle with unrelated diseases (P [χ2] = 0.0285 and P [χ2] < 0.0001, respectively) and healthy cows (P [χ2] = 0.0307 and P [χ2] = 0.0001, respectively) (Table 2). We identified no significant difference between cattle with unrelated diseases and healthy animals when we assessed the frequencies of both hemoplasma species considered either individually or together (P [χ2] > 0.05 in both cases). In the diseased cattle presented at our clinic, we failed to identify an association between the occurrence of the hemoplasmas and low PCV (data not shown).
TABLE 2.
Frequency distribution of the two bovine hemoplasma species in the examined cattle populations, assessed using real-time PCR
| Organism(s) | No. of occurrences (total no. of cows), %, in: |
|||
|---|---|---|---|---|
| Cows from herd with fatal anemia | Cows with unrelated diseases | Healthy cows | Healthy calves | |
| M. wenyonii | 133 (159), 84 | 33 (57), 58 | 38 (61), 62 | 2 (47), 4 |
| “Candidatus Mycoplasma haemobos” | 104 (159), 65 | 25 (57), 44 | 31 (61), 51 | 1 (47), 2 |
| M. wenyonii and “Candidatus Mycoplasma haemobos” | 91 (159), 57 | 23 (57), 40 | 25 (61), 41 | 0 (47), 0 |
| M. wenyonii and/or “Candidatus Mycoplasma haemobos” | 146 (159), 92 | 35 (57), 61 | 44 (61), 72 | 3 (47), 6 |
Bacterial loads of M. wenyonii and “Candidatus Mycoplasma haemobos” in healthy and sick animals.
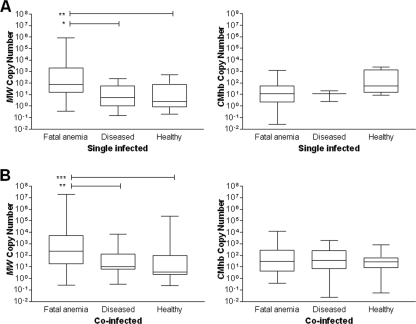
We quantified the mycoplasmal blood load in all samples that tested positive using real-time PCR, and we compared the loads for the following three cattle populations: herd with fatal anemia, cattle with unrelated diseases, and healthy cows. When comparing singly infected and coinfected animals from all groups, we observed no significant difference in the bacterial load for either M. wenyonii or “Candidatus Mycoplasma haemobos” (data not shown). To assess the contributions of the single groups, we separately analyzed singly infected and coinfected animals. In the case of M. wenyonii, the bacterial blood load was significantly higher in the cattle herd with fatal anemia than in cattle with unrelated diseases and healthy animals, when considering both singly and coinfected animals (Kruskal-Wallis P < 0.001 and Dunn's posttest P < 0.05 for single infection; Kruskal Wallis P < 0.001 and Dunn's test P < 0.01 for coinfection) (Fig. 3). No significant differences were found in the bacterial loads of “Candidatus Mycoplasma haemobos” among the three groups (Fig. 3).
FIG. 3.
Box plot of the distribution of bacteremia as assessed using real-time qPCR in the following three examined cattle populations: animals from a herd with a fatal anemia outbreak, cows with unrelated diseases, and healthy animals. (A) Singly infected animals; (B) coinfected animals. MW, M. wenyonii; CMhb, “Candidatus Mycoplasma haemobos.” Boxes extend from the 25th to the 75th percentile, a horizontal line represents the median, and the error bars extend down to the smallest and up to the largest value. Only hemoplasma-positive cattle were included in our analysis. Groups were compared using Kruskal-Wallis statistics and Dunn's posttests. A significant difference in the bacterial loads (copy number) was apparent among the three groups for M. wenyonii in both singly infected and coinfected animals. Significant differences in bacterial loads between groups are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
In this study we characterized a second hemoplasma species found in Swiss cattle, which was characterized by a shorter 16S rRNA gene than in M. wenyonii (5). Sequence analysis of the almost-complete 16S rRNA gene showed that it shared more than 98% identity with “Candidatus Mycoplasma haemobos” (20) (synonym, “Candidatus Mycoplasma haemobovis” [6, 33, 36]). It further showed more than 94% identity with mycoplasmas of feline and canine origin (M. haemofelis and M. haemocanis) and less than 80% identity with M. wenyonii. Furthermore, we developed two species-specific real-time qPCR assays to differentiate the two mycoplasmas. A similar real-time PCR assay based on melting curve analysis has been published by a Japanese group (14) since the submission of this paper.
In cats and dogs, different hemotropic mycoplasmas are characterized by different pathogenic potentials (2, 4, 10, 19, 34, 35), and we hypothesized that the same may also hold true for the two bovine hemoplasmas. Furthermore, bovine hemoplasmas were significantly more common in animals with severe anemia than in cattle with less pronounced anemia (5). Therefore, we analyzed three cattle populations, namely, the herd that was affected by the fatal anemia outbreak (5), animals with unrelated diseases, and healthy animals. M. wenyonii infection was found at a high frequency in the Swiss cattle population. This is in accordance with PCR-based observations from the United States and from Great Britain (11-13). In addition, “Candidatus Mycoplasma haemobos” was also frequently present in Swiss cattle. “Candidatus Mycoplasma haemobos” was reported in clinically ill cattle from Japan (20); however, no healthy animals were included in that study. Whereas the prevalence of M. wenyonii in Japanese cattle (14) was found to be comparable to the prevalence in Swiss cattle, both the prevalence of “Candidatus Mycoplasma haemobos” and the frequency of coinfected animals were found to be much higher in our study. This might reflect geographical differences and/or different sensitivities of the assays.
Additionally, M. wenyonii was found more often and the bacterial load was significantly higher in animals from the herd with fatal anemia than in any other animals tested. This may be due to the epidemiological and clinical contexts of the herd. These cattle were coinfected with a large number of pathogens and infested with potential arthropod vectors. We identified several factors that may have facilitated the rapid spread of the infectious agents from one animal to another in this herd: the animals had experienced an infestation of ticks and lice, and were roaming free within a limited area, and had access to automatically rotating brushes (5). Little is known about the means of transmission of M. wenyonii and other mycoplasma species, but there is some evidence that flies, lice, and fleas may serve as vectors for mechanical transmission (5, 15), while blood-sucking arthropods such as ticks may serve as biological vectors (8, 23, 37). The fact that just a very small portion of the analyzed calves was found to be infected might substantiate this hypothesis, as they are normally less exposed to vectors in the first months of life. Alternatively this could reflect an immunological phenomenon, e.g., protection due to maternally derived antibodies.
The exact role of each pathogen in the herd with fatal anemia is currently unclear; however, in animals infected with different pathogens, the immune system may be overwhelmed or even impaired (e.g., due to Theileria or Anaplasma infections [9, 27]). This may have favored other infections and/or could have increased the pathogenic potential of hemoplasmas or other infectious agents.
In cattle with unrelated diseases that were seen at our clinic, we were unable to identify a correlation between the occurrence of the hemoplasmas and a decrease in PCV. However, in these cattle, the stage of infection was unknown. For other hemoplasma species, it has been hypothesized that during chronic carrier stages, clinical signs may be absent, while acute infection may lead to anemia in infected animals (10). Thus, the cattle that were referred to our clinic for unrelated reasons might have been in the chronic phase, while the cattle in the herd with the acute fatal anemia outbreak may have been in the acute stage of infection.
In conclusion, we identified and molecularly characterized “Candidatus Mycoplasma haemobos” in the Swiss cattle population. Using real-time qPCR, we were able to detect both bovine hemoplasmal species in all tested groups of cattle, independent of their clinical status. The pathogenic potential of bovine hemoplasmas is still difficult to interpret; the presence of coinfections may contribute to their pathogenicity in a synergistic manner, in particular when leading to immunosuppression.
Acknowledgments
We thank Y. Bosshart, C. Brönnimann, E.-E. Gönczi, U. Egger, U. Frick, B. Glaus, E. Grässli, M. Huder, S. Keo, H. A. Knorr, B. Lange, B. Pineroli, T. Meili Prodan, M. Nussbaumer, E. Schuler, E. Rogg, J. Wälchli, B. Weibel, and B. Wenger for their excellent technical assistance. We are grateful to W. Regli, DIAVET Labor AG, Bäch, Switzerland, for providing blood samples from healthy animals. Laboratory work was performed using the logistics of the Center for Clinical Studies at the Vetsuisse Faculty of the University of Zurich.
This work was financially supported by the Swiss Organization for Ruminant Medicine (SVW), the Swiss Federal Veterinary Office (BVET) (Bern, Switzerland), and UBS AG. R.H.L. is the recipient of a professorship from the Swiss National Science Foundation (grants PP00B-102866 and PP00P3-119136).
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Adler, S., and V. Ellenbogen. 1934. A note on two new blood parasites of cattle, Eperythrozoon and Bartonella. J. Comp. Pathol. Ther. 47:219-221. [Google Scholar]
- 2.Berent, L. M., J. B. Messick, and S. K. Cooper. 1998. Detection of Haemobartonella felis in cats with experimentally induced acute and chronic infections, using a polymerase chain reaction assay. Am. J. Vet. Res. 59:1215-1220. [PubMed] [Google Scholar]
- 3.Donatien, A., and F. Lestoquard. 1934. Sur une Bartonella nouvelle du boeuf, Bartonella bovis n. sp. Bull. Soc. Pathol. Exot. 27:652-654. [Google Scholar]
- 4.Foley, J. E., S. Harrus, A. Poland, B. Chomel, and N. C. Pedersen. 1998. Molecular, clinical, and pathologic comparison of two distinct strains of Haemobartonella felis in domestic cats. Am. J. Vet. Res. 59:1581-1588. [PubMed] [Google Scholar]
- 5.Hofmann-Lehmann, R., M. L. Meli, U. M. Dreher, E. Gonczi, P. Deplazes, U. Braun, M. Engels, J. Schupbach, K. Jorger, R. Thoma, C. Griot, K. D. Stark, B. Willi, J. Schmidt, K. M. Kocan, and H. Lutz. 2004. Concurrent infections with vector-borne pathogens associated with fatal hemolytic anemia in a cattle herd in Switzerland. J. Clin. Microbiol. 42:3775-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornok, S., M. L. Meli, A. Erdos, I. Hajtos, H. Lutz, and R. Hofmann-Lehmann. 2009. Molecular characterization of two different strains of haemotropic mycoplasmas from a sheep flock with fatal haemolytic anaemia and concomitant Anaplasma ovis infection. Vet. Microbiol. 136:372-377. [DOI] [PubMed] [Google Scholar]
- 7.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20:291-299. [DOI] [PubMed] [Google Scholar]
- 8.Lappin, M. R., B. Griffin, J. Brunt, A. Riley, D. Burney, J. Hawley, M. M. Brewer, and W. A. Jensen. 2006. Prevalence of Bartonella species, haemoplasma species, Ehrlichia species, Anaplasma phagocytophilum, and Neorickettsia risticii DNA in the blood of cats and their fleas in the United States. J. Feline Med. Surg. 8:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen, H. J., G. Overnes, H. Waldeland, and G. M. Johansen. 1994. Immunosuppression in sheep experimentally infected with Ehrlichia phagocytophila. Res. Vet. Sci. 56:216-224. [DOI] [PubMed] [Google Scholar]
- 10.Messick, J. B. 2003. New perspectives about Hemotrophic mycoplasma (formerly, Haemobartonella and Eperythrozoon species) infections in dogs and cats. Vet. Clin. North Am. Small Anim. Pract. 33:1453-1465. [DOI] [PubMed] [Google Scholar]
- 11.Neimark, H., K. E. Johansson, Y. Rikihisa, and J. G. Tully. 2001. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of ‘Candidatus Mycoplasma haemofelis', ‘Candidatus Mycoplasma haemomuris’, ‘Candidatus Mycoplasma haemosuis’ and ′Candidatus Mycoplasma wenyonii’. Int. J. Syst. Evol. Microbiol. 51:891-899. [DOI] [PubMed] [Google Scholar]
- 12.Neimark, H., K. E. Johansson, Y. Rikihisa, and J. G. Tully. 2002. Revision of haemotrophic Mycoplasma species names. Int. J. Syst. Evol. Microbiol. 52:683. [DOI] [PubMed] [Google Scholar]
- 13.Neimark, H., and K. M. Kocan. 1997. The cell wall-less rickettsia Eperythrozoon wenyonii is a Mycoplasma. FEMS Microbiol. Lett. 156:287-291. [DOI] [PubMed] [Google Scholar]
- 14.Nishizawa, I., M. Sato, M. Fujihara, S. Sato, and R. Harasawa. 2010. Differential detection of hemotropic Mycoplasma species in cattle by melting curve analysis of PCR products. J. Vet. Med. Sci. 72:77-79. [DOI] [PubMed] [Google Scholar]
- 15.Prullage, J. B., R. E. Williams, and S. M. Gaafar. 1993. On the transmissibility of Eperythrozoon suis by Stomoxys calcitrans and Aedes aegypti. Vet. Parasitol. 50:125-135. [DOI] [PubMed] [Google Scholar]
- 16.Purnell, R. E., D. W. Brocklesby, and E. R. Young. 1976. Eperythrozoon wenyoni, a possible cause of anaemia in British cattle. Vet. Rec. 98:411. (Letter.) [DOI] [PubMed] [Google Scholar]
- 17.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 18.Smith, J. A., M. A. Thrall, J. L. Smith, M. D. Salman, S. V. Ching, and J. K. Collins. 1990. Eperythrozoon wenyonii infection in dairy cattle. J. Am. Vet. Med. Assoc. 196:1244-1250. [PubMed] [Google Scholar]
- 19.Sykes, J. E., L. M. Ball, N. L. Bailiff, and M. M. Fry. 2005. ‘Candidatus Mycoplasma haematoparvum’, a novel small haemotropic mycoplasma from a dog. Int. J. Syst. Evol. Microbiol. 55:27-30. [DOI] [PubMed] [Google Scholar]
- 20.Tagawa, M., K. Matsumoto, and H. Inokuma. 2008. Molecular detection of Mycoplasma wenyonii and ‘Candidatus Mycoplasma haemobos’ in cattle in Hokkaido, Japan. Vet. Microbiol. 132:177-180. [DOI] [PubMed] [Google Scholar]
- 21.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 22.Tandon, R., V. Cattori, M. A. Gomes-Keller, M. L. Meli, M. C. Golder, H. Lutz, and R. Hofmann-Lehmann. 2005. Quantitation of feline leukaemia virus viral and proviral loads by TaqMan real-time polymerase chain reaction. J. Virol. Methods 130:124-132. [DOI] [PubMed] [Google Scholar]
- 23.Taroura, S., Y. Shimada, Y. Sakata, T. Miyama, H. Hiraoka, M. Watanabe, K. Itamoto, M. Okuda, and H. Inokuma. 2005. Detection of DNA of ‘Candidatus Mycoplasma haemominutum’ and Spiroplasma sp. in unfed ticks collected from vegetation in Japan. J. Vet. Med. Sci. 67:1277-1279. [DOI] [PubMed] [Google Scholar]
- 24.Tasker, S., S. H. Binns, M. J. Day, T. J. Gruffydd-Jones, D. A. Harbour, C. R. Helps, W. A. Jensen, C. S. Olver, and M. R. Lappin. 2003. Use of a PCR assay to assess the prevalence and risk factors for Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ in cats in the United Kingdom. Vet. Rec. 152:193-198. [DOI] [PubMed] [Google Scholar]
- 25.Tasker, S., C. R. Helps, M. J. Day, T. J. Gruffydd-Jones, and D. A. Harbour. 2003. Use of real-time PCR to detect and quantify Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ DNA. J. Clin. Microbiol. 41:439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuomi, J. 1967. Experimental studies on bovine tick-borne fever. 1. Clinical and haematological data, some properties of the causative agent, and homologous immunity. Acta Pathol. Microbiol. Scand. 70:429-445. [PubMed] [Google Scholar]
- 28.Tuomi, J., and R. Tanskanen. 1980. Antigenic non-relationship of two bovine Eperithrozoa demonstrated by the immunofluorescent method. Acta Vet. Scand. 21:699-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuomi, J., and C.-H. von Bonsdorff. 1967. Ultrastructure of a microorganism associated with bovine platelets. Experientia 23:111-114. [DOI] [PubMed] [Google Scholar]
- 30.Uilenberg, G. 2009. ‘Candidatus Mycoplasma haemobos’. Vet. Microbiol. 138:200-201. [DOI] [PubMed] [Google Scholar]
- 31.Uilenberg, G. 1967. Eperythrozoon tuomii, n.sp. (Rickettsiales), the 3rd species of Eperythrozoon of cattle in Madagascar. Rev. Elev. Med. Vet. Pays Trop. 20:563-569. [DOI] [PubMed] [Google Scholar]
- 32.Uilenberg, G. 1965. Notes on the Eperythrozoon of cattle in Madagascar. Rev. Elev Med. Vet. Pays Trop. 18:73-81. [PubMed] [Google Scholar]
- 33.Wengi, N., B. Willi, F. S. Boretti, V. Cattori, B. Riond, M. L. Meli, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2008. Real-time PCR-based prevalence study, infection follow-up and molecular characterization of canine hemotropic mycoplasmas. Vet. Microbiol. 126:132-141. [DOI] [PubMed] [Google Scholar]
- 34.Westfall, D. S., W. A. Jensen, W. J. Reagan, S. V. Radecki, and M. R. Lappin. 2001. Inoculation of two genotypes of Hemobartonella felis (California and Ohio variants) to induce infection in cats and the response to treatment with azithromycin. Am. J. Vet. Res. 62:687-691. [DOI] [PubMed] [Google Scholar]
- 35.Willi, B., F. S. Boretti, C. Baumgartner, S. Tasker, B. Wenger, V. Cattori, M. L. Meli, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2006. Prevalence, risk factor analysis, and follow-up of infections caused by three feline hemoplasma species in cats in Switzerland. J. Clin. Microbiol. 44:961-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willi, B., F. S. Boretti, S. Tasker, M. L. Meli, N. Wengi, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2007. From Haemobartonella to hemoplasma: molecular methods provide new insights. Vet. Microbiol. 125:197-209. [DOI] [PubMed] [Google Scholar]
- 37.Woods, J. E., M. M. Brewer, J. R. Hawley, N. Wisnewski, and M. R. Lappin. 2005. Evaluation of experimental transmission of ‘Candidatus Mycoplasma haemominutum’ and Mycoplasma haemofelis by Ctenocephalides felis to cats. Am. J. Vet. Res. 66:1008-1012. [DOI] [PubMed] [Google Scholar]
- 38.Zwart, D., P. Leeflang, and C. J. A. H. V. van Vorstenbaosch. 1969. Studies on the Eperithrozoon associated with bovine thrombocytes. Zentralbl. Bakteriol. Orig. 210:82-105. [PubMed] [Google Scholar]