Abstract
This report describes for the first time Corynebacterium macginleyi as a cause of conjunctivitis in Canada, where menaquinone analysis was done as part of the strain characterization. This species is typically isolated from ocular surfaces of patients from Europe and Japan. The isolate was resistant to erythromycin and clindamycin.
CASE REPORT
A 54-year-old microbiologist was seen by an ophthalmologist in Winnipeg, Manitoba, Canada, for evaluation of bilateral conjunctivitis. She had no history of eye injury or prior eye complaints, nor was a laboratory-acquired infection suspected. None of her family members had similar symptoms. She had no recent travel outside North America. Swabs from each eye were sent within several hours to a hospital laboratory for direct microscopy and bacterial culture. Until culture results were known, the patient was told to apply an over-the-counter “tear refresher” to the eyes to provide some relief. Approximately 5 days later, when microbiological results became apparent, the patient was prescribed 1 drop of 0.5% moxifloxacin hydrochloride in each eye 3 times per day for 7 days. The infection by that point had begun to resolve, but the patient did comply with the entire treatment course and since has had no recurrence. No follow-up culture was done to see if the organism had been eradicated from the eyes.
At the hospital laboratory, after ∼48 h of incubation under aerobic and facultatively anaerobic conditions at 35°C on 5% sheep blood agar, the culture from each sample grew fine (∼1-mm-diameter), round, and convex colonies that were slightly alpha-hemolytic. Growth in broth was enhanced by the addition of ∼1% (vol/vol) sterile Tween 80, and so a lipophilic corynebacterium was suspected. Staining revealed Gram-positive coccobacilli in short chains and clusters. A few larger Gram-positive bacillary forms (thicker at one end) were also found interspersed among the coccobacilli. No other bacterial types were recovered. Isolates from each eye were deemed to be identical to each other, and both strains were catalase positive, oxidase negative, and facultatively anaerobic. The API Coryne system (bioMérieux, Montreal, Canada) was used for identification by the testing laboratory, generating API code 5100305 after a 24-h incubation, and the bacterium could be identified as Corynebacterium macginleyi with a 99.5% probability by that system.
One strain was referred to the Canadian National Microbiology Laboratory (NML) for confirmation of identification (strain NML 080212). Because such isolates have not been previously described as being recovered in Canada, the strain was extensively characterized. Conventional carbohydrate broth sugars enhanced with ∼1% serum (vol/vol) and other biochemical testing procedures were used as reviewed previously (2). This strain slowly fermented glucose, sucrose, and ribose, but in contrast to the results of Riegel et al. (14) also slowly fermented glycogen and maltose as well as mannose, glycerol, and fructose but not galactose, lactose, mannitol, raffinose, trehalose, or xylose. When the strain was tested using the API Coryne strip, only glucose, ribose, and sucrose were reactive, similar to findings by Riegel et al. By conventional methods, the strain reduced nitrate to nitrite but did not degrade urea, casein, tyrosine, starch, or gelatin. With the API Coryne strip, the strain produced alkaline phosphatase and pyrrolidonyl arylamidase but not pyrazinamidase, β-galactosidase, β-glucuronidase, α-glucosidase, or N-acetyl-β-glucosaminidase. By the API ZYM strip (bioMérieux), the isolate was reactive only with alkaline phosphatase, acid phosphatase, and naphthol-AS-BI-phosphohydrolase.
Cellular fatty acid composition analysis was done using the Sherlock system (MIDI, Newark, DE; with operating system version 4.5), by which predominately straight-chain and monounsaturated cellular fatty acids (CFAs), but not tuberculostearic acid, were detected, similar to the results described for other members of the genus (1). Menaquinones for C. macginleyi were not elucidated as part of the species novum description (14), and so this assay was undertaken for NML 080212, and it was analyzed by liquid chromatography-mass spectrometry (LC-MS), as described previously (4). Minor amounts of MK-8 (H2) were detected, which is consistent with results for the genus Corynebacterium (6). Small volumes of lactic, succinic, and acetic but not propionic acid were detected as metabolic products, as found previously for this species (2).
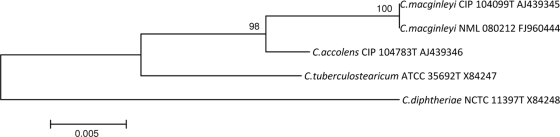
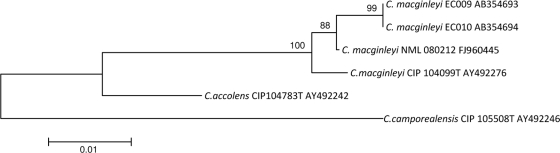
The 16S rRNA gene was amplified as described previously (3), aligned using Clustal W, and compared with other similar sequences using neighbor-joining software found in MEGA4. The resulting 1,483-bp sequence was found to be 100% homologous to C. macginleyi CIP 104099T (GenBank accession no. AJ439345) (Fig. 1). However, as the identities of C. accolens and C. macginleyi cannot be adequately determined by 16S rRNA gene sequencing alone (12), partial rpoB gene sequencing was also done. rpoB sequences were first aligned using Clustal W, and then the entire (∼3,100-bp) rpoB sequences for C. accolens AY492242, C. macginleyi AY492276, and C. camporealensis AY492246 were trimmed to ∼400 bp, prior to comparison, using the neighbor-joining software found in MEGA 4, with the partial sequence (403 bp) generated for NML 080212. This strain had 98.5% identity with C. macginleyi CIP 104099T (accession no. AY492276) but only 92.7% identity with C. accolens CIP104783T (accession no. AY492242), thus providing definitive identification between these otherwise closely related lipophilic species (Fig. 2).
FIG. 1.
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences, showing the relationship of NML 080212 to its closest Corynebacterium species. Percentages at the nodes are bootstrap values based on 1,000 replications. The sequence from the type strain of the type species, C. diphtheriae, was used as the outgroup. Bar, 0.005 substitutions per nucleotide position.
FIG. 2.
Neighbor-joining phylogenetic tree based on partial rpoB gene sequences, showing the relationship of NML 080212 to its closest Corynebacterium species. Percentages at the nodes are bootstrap values based on 1,000 replications. Two sequences derived from the Japanese outbreak involving C. macginleyi (8) were also included as close relatives, with the sequence from distantly related C. camporealensis used as an outgroup. Bar, 0.01 substitutions per nucleotide position.
Antimicrobial susceptibility and MICs were determined by the broth microdilution method using Mueller-Hinton medium containing 2.5% (vol/vol) lysed horse blood according to CLSI methods and breakpoint standards (5) with commercial Sensititre plates and antibiotics as described previously (4). The strain was resistant to erythromycin and clindamycin. ermX, the gene associated with this phenotype, was detected using methods outlined by Rosato et al. (15). Otherwise, the strain was susceptible to ampicillin, cefepime, cefotaxime, ceftriaxone, cefuroxime, chloramphenicol, ciprofloxacin, daptomycin, ertapenem, gatifloxacin, gentamicin, levofloxacin, linezolid, meropenem, moxifloxacin, penicillin, quinupristin-dalfopristin, rifampin, telithromycin, tetracycline, tigecycline, trimethoprim-sulfamethoxazole, and vancomycin.
Riegel et al. were the first to describe Corynebacterium macginleyi among the lipophilic corynebacterium genomospecies (14). Since then, several clinically relevant isolates have been detected, almost exclusively from ocular surfaces of symptomatic patients in Europe (e.g., Switzerland, Germany, and Italy) (9-11). This species was only very rarely recovered from a study of healthy eyes (20). Extraocular involvements, such as urinary tract infection, central venous catheter infection, endocarditis, and septicemia, have also been reported (7, 13, 18, 19). In 2007, suture-related keratitis caused by this agent was described by Suzuki et al. (17), and in 2008, Eguchi et al. described C. macginleyi isolates from among ocular infections studied in Japan (8). In this instance, the patient was clearly symptomatic, with a bilateral ocular infection, and the same bacterium was recovered from both eyes, with no other aerobic or facultatively anaerobic organism being found. Although no follow-up culture of the eyes was done after the antibiotic course was completed, these observations suggested that the C. macginleyi isolates recovered were the causative agents of infection. This is the first such case of conjunctivitis reported in Canada and the second in North America, as a case involving C. macginleyi recovered from a corneal ulcer scraping has recently been described (16), suggesting a worldwide distribution of this organism. The strain described here was consistent with the description of C. macginleyi, and its identity was determined by a polyphasic (biochemical, genetic, and chemotaxonomic [including the first description of menaquinones for this species]) rather than a single identification approach.
Joussen et al. reported C. macginleyi in 13 of 107 (12%) cases of culture-positive conjunctivitis, indicating its importance in ocular surface infections, especially in immunocompromised hosts (11). Eguchi et al. showed C. macginleyi to be the only bacterium recovered from their cultured conjunctivitis cases (8).
The initial reports by Funke et al. and Joussen et al. showed almost uniform susceptibility of C. macginleyi to commonly used topical antibiotics, with some resistance to erythromycin and clindamycin being observed (9, 11). Additional resistance phenotypes, however, are emerging, including to fluoroquinolone, co-trimoxazole, clindamycin, tobramycin, and erythromycin (8, 18). Our isolate was resistant to erythromycin and clindamycin, supported by ermX gene expression.
It was unexpected that reports describing the recovery of C. macginleyi from ocular infections from North American sources did not exist prior to 2010 (this study and reference 16). This could be attributed to the frequent practice of treating acute conjunctivitis without obtaining cultures. Our findings, along with the cited literature, make culture and susceptibility testing highly advisable. Clinicians also empirically prescribing topical antibiotics without performing culture may encounter resistant strains (8).
Nucleotide sequence accession number.
The sequence for NML 080212 has been deposited under GenBank accession numbers FJ960444 (16S rRNA gene) and FJ960445 (partial rpoB gene).
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Bernard, K. A., M. Bellefeuille, and E. P. Ewan. 1991. Cellular fatty acid composition as an adjunct to the identification of asporogenous, aerobic gram-positive rods. J. Clin. Microbiol. 29:83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard, K. A., C. Munro, D. Wiebe, and E. Ongsansoy. 2002. Characteristics of rare or recently described Corynebacterium species recovered from human clinical material in Canada. J. Clin. Microbiol. 40:4375-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard, K. A., L. Shuttleworth, C. Munro, J. C. Forbes-Faulkner, D. Pitt, J. H. Norton, and A. D. Thomas. 2002. Propionibacterium australiense sp. nov. derived from granulomatous bovine lesions. Anaerobe 8:41-47. [Google Scholar]
- 4.Bernard, K. A., D. Wiebe, T. Burdz, A. Reimer, B. Ng, C. Singh, S. Schindle, and A. L. Pacheco. 2010. Assignment of Brevibacterium stationis (ZoBell and Upham 1994) Breed 1953 to the genus Corynebacterium as Corynebacterium stationis comb. nov. and emended description of the genus Corynebacterium to include isolates which can alkalinize citrate. Int. J. Syst. Evol. Microbiol. 60:874-879. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. M45-A. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Approved guideline. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed]
- 6.Collins, M. D., and C. S. Cummins. 1986. Genus Corynebacterium, p. 1266-1276. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, 1st ed., vol. 2. The Williams & Wilkins Co., Baltimore, MD. [Google Scholar]
- 7.Dobler, G., and I. Braveny. 2003. Highly resistant Corynebacterium macginleyi as cause of intravenous catheter-related infection. Eur. J. Clin. Microbiol. Infect. Dis. 23:72-73. [DOI] [PubMed] [Google Scholar]
- 8.Eguchi, H., T. Kuwahara, T. Miyamoto, H. Nakayama-Imaohji, M. Ichimura, T. Hayashi, and H. Shiota. 2008. High-level fluoroquinolone resistance in ophthalmic clinical isolates belonging to the species Corynebacterium macginleyi. J. Clin. Microbiol. 46:527-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funke, G., M. Pagano-Niederer, and W. Bernauer. 1998. Corynebacterium macginleyi has to date been isolated exclusively from conjunctival swabs. J. Clin. Microbiol. 36:3670-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giammanco, G. M., V. DiMarco, I. Priolo, A. Intrivici, F. Grimont, and P. A. D. Grimont. 2002. Corynebacterium macginleyi isolation from conjunctival swab in Italy. Diagn. Microbiol. Infect. Dis. 44:205-207. [DOI] [PubMed] [Google Scholar]
- 11.Joussen, A. M., G. Funke, F. Joussen, and G. Herbertz. 2000. Corynebacterium macginleyi: a conjunctiva specific pathogen. Br. J. Ophthalmol. 80:1420-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khamis, A., D. Raoult, and B. La Scola. 2004. rpoB gene sequencing for identification of Corynebacterium species. J. Clin. Microbiol. 42:3925-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pubill Sucarrat, M., X. Martinez-Costa, G. Sauca Subias, and J. A. Capdevila Morell. 2003. Corynebacterium macginleyi as an exceptional cause of endocarditis: a case report. An. Med. Interna 20:654-655. [In Spanish.] [PubMed] [Google Scholar]
- 14.Riegel, P., R. Ruimy, D. de Briel, G. Prevost, F. Jehl, R. Christen, and H. Monteil. 1995. Genomic diversity and phylogenetic relationships among lipid-requiring diphtheroids from humans and characterization of Corynebacterium macginleyi sp. nov. Int. J. Syst. Bacteriol. 45:128-133. [DOI] [PubMed] [Google Scholar]
- 15.Rosato, A. E., B. S. Lee, and K. A. Nash. 2001. Inducible macrolide resistance in Corynebacterium jeikeium. Antimicrob. Agents Chemother. 45:1982-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruoff, K. L., C. M. Toutain-Kidd, M. Srinivasan, P. Lalitha, N. R. Acharya, M. E. Zegans, and J. D. Schwartzman. 2010. Corynebacterium macginleyi isolated from a corneal ulcer. Infect. Dis. Rep. 2:7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki, T., H. Iihara, T. Uno, Y. Hara, K. Ohkusu, H. Hata, M. Shudo, and Y. Ohashi. 2007. Suture-related keratitis caused by Corynebacterium macginleyi. J. Clin. Microbiol. 45:3833-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villamil-Cajoto, I., L. Rodriguez-Otero, M. J. Villacian-Vicedo, M. A. Garcia-Zabarte, A. Aguilera-Guirao, C. Garcia-Riestra, and B. J. Regueiro. 2008. Septicemia caused by Corynebacterium macginleyi: a rare form of extraocular infection. Int. J. Infect. Dis. 12:333-335. [DOI] [PubMed] [Google Scholar]
- 19.Villanueva, J. L., A. Dominguez, M. J. Rios, and C. Iglesias. 2002. Corynebacterium macginleyi isolated from urine in a patient with a permanent bladder catheter. Scand. J. Infect. Dis. 34:699-700. [DOI] [PubMed] [Google Scholar]
- 20.von Graevenitz, A., U. Schumacher, and W. Bernauer. 2001. The corynebacterial flora of the normal human conjunctiva is lipophilic. Curr. Microbiol. 42:372-374. [DOI] [PubMed] [Google Scholar]