Abstract
Herpes simplex virus type 2 (HSV-2) is the most common cause of genital ulcer disease and is a cofactor for HIV-1 acquisition and transmission. We analyzed specimens from three separate phase III trials of acyclovir (ACV) for prevention of HIV-1 acquisition and transmission to determine if failure of ACV to interrupt HIV acquisition and transmission was associated with genotypic ACV resistance. Acyclovir (400 mg twice daily) or placebo was provided to HSV-2-infected persons at risk of HIV-1 infection in the Mwanza and HPTN 039 trials and to persons dually infected with HSV-2 and HIV-1 who had an HIV-negative partner in the Partners in Prevention study. We extracted HSV DNA from genital ulcer swabs or cervicovaginal lavage fluids from 68 samples obtained from 64 participants randomized to ACV and sequenced the HSV-2 UL23 gene encoding thymidine kinase. The UL23 sequences were compared with published and unpublished data. Variants were observed in 38/1,128 (3.4%) nucleotide positions in the UL23 open reading frame, with 58% of these encoding amino acid changes. No deletions, insertions, or mutations known to be associated with resistance were detected. Thirty-one of the variants (81.5%) are newly reported, 15 of which code for amino acid changes. Overall, UL23 is highly polymorphic compared to other loci in HSV-2, but no drug resistance mutations were detected that could explain the failure to reduce HIV incidence or to prevent HIV-1 transmission in these studies.
Herpes simplex virus type 2 (HSV-2) is one of the most common sexually transmitted infections. The prevalence of infection is higher in many parts of the developing world, especially in some settings in sub-Saharan Africa, where up to 75 to 80% of women may be infected by the age of 35 years (26).
The standard treatment for symptomatic primary and recurrent genital herpes and for suppression of genital herpes has primarily been acyclovir (ACV), a guanosine nucleoside analogue that inhibits replication of HSV. The inactive prodrug is phosphorylated to ACV triphosphate by HSV-encoded thymidine kinase. The active ACV triphosphate then inhibits viral DNA polymerase. Thymidine kinase is encoded by the HSV UL23 gene. The frequency of ACV resistance is increased in ACV-treated, immunosuppressed individuals and may reach 5% (25). However, ACV resistance has also occasionally been documented in immunocompetent persons in both the absence and presence of ACV therapy (8, 28a). Up to 95% of clinically significant ACV resistance is caused by mutations in the UL23 gene that limit thymidine kinase enzymatic activity and thus prevent initial phosphorylation of the drug (1a). These are typically insertions or deletions of a single nucleotide in a poly(G) or poly(C) tract, leading to a frameshifted open reading frame with a carboxy-terminal nonsense polypeptide and premature termination (1a). Point mutations that prevent or reduce recognition of ACV as a substrate occur at lower frequencies (1a). Compared to the much greater understanding of human cytomegalovirus (CMV) and ganciclovir resistance, and HIV-1 and resistance to HIV antiretroviral therapy, the total database of resistance-associated HSV UL23 point mutations is small. Therefore, hot spots for ACV resistance-associated point mutations have not yet been identified in UL23 of HSV-2.
Until recently, ACV therapy was used predominantly in developed countries. However, with the recognition that an increasing proportion of genital ulcers in developing countries are now caused by HSV (3), the World Health Organization has recommended that treatment for HSV now be included in first-line therapy for genital ulcer disease (32). In addition HSV-2 shares a synergic relationship with HIV, since it enhances HIV acquisition and transmission (1, 30). Suppression of HSV-2 was therefore evaluated to determine if this could be a potential strategy to reduce HIV acquisition or transmission, or both.
Three phase III randomized controlled trials (RCTs) of HSV-2-suppressive therapy for the control of HIV have recently been completed in North and South America and Africa. Two of these trials, the Mwanza HSV trial and HPTN 039, examined the hypothesis that ACV (400 mg twice daily) given to HSV-2-seropositive, HIV-seronegative individuals would prevent HIV acquisition. The Mwanza HSV trial also examined the effects of ACV on the HIV and HSV genital viral load in dually HSV-2-infected, HIV-seropositive women. The third trial, the Partners in Prevention HSV/HIV Transmission Study, enrolled HIV-serodiscordant couples and examined the effect of the same regimen of ACV given to HIV/HSV-2 dually infected persons on HIV transmission to their non-HIV-infected partners. The trials found no effect of suppressive treatment with ACV on HIV acquisition or transmission (4, 4a, 31). Given these results, we conducted detailed examination of the UL23 sequence of a representative subset of HSV-2 specimens from subjects in the ACV arm. This is the first study to examine ACV resistance in developing countries in the setting of HSV-suppressive therapy.
MATERIALS AND METHODS
Each of the three RCTs received approval from the appropriate institutional review boards (IRB) (4, 4a, 31). IRB approval for use of specimens for the studies described in this paper was either explicitly obtained or inherent in the originating trials. Specifically, the Mwanza trial protocol was approved by the National Ethics Committee of Tanzania and the London School of Hygiene and Tropical Medicine, and the HPTN 039 and Partners in Prevention studies were approved at each participating trial site and by the University of Washington Human Subjects Review Committee.
Study enrollment. (i) Mwanza HSV trial (International Standard Randomized Controlled Trial number [ISRCTN] 35385041).
Women aged 16 to 35 years working as food handlers or in bars, guesthouses, and similar facilities in 19 communities in northwest Tanzania were invited to attend a mobile study clinic for HSV-2 and HIV-1 antibody screening, as previously described (29a, 31). The women returned 8 to 12 weeks later for their HSV-2 results and were eligible for enrollment if they were aged 16 to 35 years at screening, were HSV-2 seropositive, and planned to stay in the area. Women were not enrolled if they were pregnant. Knowledge of HIV status was not required for enrollment, and women were enrolled regardless of HIV status. The participants were randomized to ACV (400 mg twice daily) or matching placebo tablets. They attended the study clinic every 3 months. CD4 T-cell counts were collected at some visits and, if available, were reported from the visit closest to collection of the HSV sample analyzed in this study. Cervicovaginal lavage (CVL) specimens were collected at enrollment and at the 6-, 12-, and 24-month visits, and participants were followed for up to 30 months.
(ii) HPTN 039 (ClinicalTrials.gov number NCT00076232).
A total of 3,127 HIV-negative, HSV-2 antibody-positive participants were enrolled in HPTN 039. Men who had sex with men (MSM) were enrolled at sites in the United States and Peru, and women were enrolled at sites in Harare, Zimbabwe; Lusaka, Zambia; and Johannesburg, South Africa, as detailed previously (4a).
The participants were randomized to receive ACV (400 mg twice daily) or matching placebo tablets and were seen monthly for 12 to 18 months. Clinicians performed a genital examination at all quarterly study visits and at any monthly or interim study visit if a genital ulcer was reported. The study staff obtained swabs from lesions clinically consistent with a herpes recurrence.
(iii) Partners in Prevention (ClinicalTrials.gov number NCT00194519).
HIV-1-seropositive persons who were also seropositive for HSV-2 were enrolled in an RCT of ACV as HSV-2-suppressive therapy, along with their HIV-1-seronegative heterosexual partners (4). HIV-1-positive persons were randomized to ACV (400 mg twice daily) or matching placebo tablets. The Partners in Prevention HSV/HIV Transmission Study was conducted at 14 sites in 7 African countries (Botswana, Kenya, Rwanda, South Africa, Tanzania, Uganda, and Zambia), and the participants were followed for up to 24 months. At the time of enrollment, the HIV-1- infected partners were ≥18 years of age, were HIV-1 and HSV-2 seropositive, had a CD4 count of ≥250 cells/mm3 and no history of AIDS-defining conditions, and were not receiving antiretroviral therapy (ART). The HIV-1-infected partners were seen monthly for provision of the study drug, evaluation of clinical status, and behavioral risk assessment. Clinicians performed a genital examination at the quarterly study visits and at any monthly or interim visit if symptoms of genital ulcers were reported. The study staff obtained swabs from lesions clinically consistent with a herpes recurrence.
Clinic procedures and sample selection. (i) Mwanza HSV trial.
After centrifugation at 3,500 rpm for 10 min, CVL supernatant aliquots were frozen at −20°C for 2 to 3 weeks and later at −80°C. HSV DNA was quantified in the CVL supernatants by real-time PCR (31). Enrollment HSV DNA-positive CVL samples were typed (3a), and all were HSV-2. HSV DNA quantifications were done with an ABI 7300 PCR instrument (Applied Biosystems, Courtaboeuf, France) and a commercial HSV-1/HSV-2 typing/quantification standard (HSV-1/HSV-2 Clear QC Panel; Argene, Verniolle, France).
CVL supernatant samples were selected for UL23 sequencing from women who were randomized to ACV, who had not been withdrawn from tablets, and whose genital HSV load was >104 per milliliter. Samples were selected at enrollment to assess whether resistance to ACV was present at baseline and from the 12- and 24-month visits. At enrollment, 3 HIV-negative and 19 HIV-seropositive women met the selection criteria. Baseline CVL samples from each HIV-negative woman and seven random HIV-seropositive women were analyzed. At the 12- and 24-month visits, during ACV therapy, 5 HIV-negative and 15 HIV-seropositive women met the selection criteria, and CVL samples from each were tested. Overall, 10 baseline and 20 follow-up CVL samples from 8 HIV-negative and 22 HIV-seropositive subjects were selected for testing.
(ii) HPTN 039 and Partners in Prevention.
Swabs obtained from genital lesions clinically consistent with a herpes recurrence were obtained at enrollment and periodically thereafter as described previously (4, 4a). These swabs were placed into PCR media, frozen, and shipped to the University of Washington Virology Laboratory. We selected archived swab-derived DNA from specimens with previously detectable HSV-2 DNA using quantitative real-time PCR (21) from time points at which subjects were receiving ACV.
Laboratory methods.
For Mwanza CVL specimens, replicate 1-ml aliquots were transported to the University of Washington, and DNA was prepared manually from 200 μl of specimen using 96-well high-throughput blood kits and the instructions for cell-free fluids (Qiagen, Valencia, CA). This kit extracts principally DNA. The presence of HSV-2 DNA was evaluated using our in-house PCR targeting the glycoprotein B gene exactly as described previously (14). A portion of the DNA from each CVL specimen yielding a positive result for HSV-2 DNA was submitted for sequencing. HPTN 039 and Partners in Prevention genital swabs were collected, and DNA was extracted and analyzed for HSV-2 DNA as described previously (4, 4a). A portion of the extracted DNA from selected positive swabs was submitted for sequencing.
The full-length sequence of the UL23 gene was determined. The primers primarily used for amplification were 5′GTGGCGTTGAACTCCCGCACCTC3′ and 5′CTGTTCTTTTATTGCCGTCATCG3′. PCR mixtures (50 μl) contained 5 μl High Fidelity Taq buffer, 1 μl of 10 mM deoxynucleoside triphosphates (dNTPs), 2 μl of 50 mM MgSO4 providing a final Mg2+ concentration of 2.2 mM, 0.2 μl High Fidelity Taq Platinum thermostable polymerase compound containing Pyrococcus GB-D polymerase, 20 picomoles of each primer (all from Invitrogen, Carlsbad, CA), 5 to 10 μl of template DNA, 3 μl dimethyl sulfoxide (DMSO), and water as needed. The tubes were heated to 95°C for 30 s and then cycled 35 times for 30 s at 95°C, 30 s at 56°C, and 2 min at 68°C, followed by terminal extension for 7 min at 68°C. PCR products were checked for the predicted molecular weight by gel electrophoresis and gel purified (Qiaquick; Qiagen, Valencia, CA). Separate sequencing reactions used the following amplification primers and additional primers: 5′CTGACGAACATCTACAACACGC3′, 5′GATATGAGGAGCCAAAACGGCGTC3′, 5′CTTGACCTGGCCATGCTGTC3′, and 5′CTGCAGGTACCGCACCGTGTTG3′. Some templates were amplified by nested PCR using the original amplification primers. This first round used either the amplification primers listed above or the laterally flanking primers 5′CGAGGTCCACTTCGCATATT3′ and 5′CGCTTATGGACACACCACAC3′. PCR products from the first round were gel or spin column purified (Qiaquick), and a portion was submitted for second-round amplification as described above. For both one-round and nested PCRs, a water control template was always submitted and coprocessed through each step. All amplicons from any run in which water controls yielded a PCR product were discarded. Sequencing by dideoxy chain termination with BigDye3.0 kits (ABI, Foster City, CA) was performed at the Fred Hutchinson Cancer Research Center Shared Resource laboratory, and all regions received at least double coverage.
Contigs were assembled, ambiguities were resolved by manual review of chromatograms, and alignments were done (Clustal W algorithm) using Lasergene V8 software (DNAStar, Madison, WI). To ascertain previous HSV-2 UL23 sequences, GenBank accession numbers NC_001798, AB178228, AB178229, AB178230, AB009256, AB009257, AB009261, AB009262, AB009263.2, AY038368, AY038369, DQ372963, EF522120, M29940, M29941, M29943, X01712, S63520, and S46174 and partial sequence information available from 40 strains that have not been submitted to GenBank (12, 16, 27, 27a) were manually reviewed. Unpublished complete coding sequences of UL23 genes from HSV-2 strains 26 through 54 (6) were kindly provided by Doris Chibo, Victorian Infectious Diseases Reference Laboratory, North Melbourne, Victoria, Australia. Nucleotide and amino acid variations are named AXB/CXY or AXB/NC, where A is the nucleotide in reference strain HG52 (10); X is the numerical position in the sequence related to A in the ATG start codon; B is the nucleotide in the study strain; C, X, and Y are similar but refer to amino acids; and NC indicates noncoding. We used strain HG52 as our reference strain for variation naming and numbering. The ratio of nonsynonymous to synonymous nucleotide changes (average dN/average dS) was calculated using SNAP (17; http://www.hiv.lanl.gov). A maximum-likelihood phylogenetic tree using a general time reversible plus gamma distribution plus invariant sites (GTR+G+I) model and a tree-bisection-reconnection swapping algorithm with laboratory strain HG52 as the outgroup was estimated in PAUP* version 4.0b10 (Sinauer Associates, Inc., Sunderland, MA).
Nucleotide sequence accession numbers.
UL23 gene sequences were uploaded to GenBank under accession numbers GU057918 to GU057982.
RESULTS
Full-length HSV-2 UL23 sequences were obtained from 68 specimens from 64 subjects (Table 1). Specimens with sequence data were obtained from diverse locations in Africa, as well as from three locations in Peru, with a single specimen from North America.
TABLE 1.
Demographic characteristics of subjects from whom HSV-2 UL23 sequences were obtained
| Study | Location | HIV statusa | No. of subjects | Gender (%) | Age (mean yr) | ACV therapy (%)b | Duration of ACV (mean no. of moc) | Baseline CD4 (meand) |
|---|---|---|---|---|---|---|---|---|
| HPTN039 | Peru | Neg | 17e | Male (100) | 28 | 100 | 9.1 | |
| HPTN039 | San Francisco | Neg | 1 | Male (100) | 35 | 100 | 8.2 | |
| HPTN039 | South Africa | Neg | 3 | Female (100) | 25 | 100 | 6.9 | |
| HPTN039 | Zambia | Neg | 6e | Female (100) | 32 | 100 | 12.2 | |
| HPTN039 | Zimbabwe | Neg | 8e | Female (100) | 30 | 100 | 11.7 | |
| PIPf | Kenya | Pos | 3 | Female (67) | 35 | 67 | 20.4 | 550 |
| Male (33) | ||||||||
| PIP | Uganda | Pos | 2 | Female (100) | 30 | 100 | 6.0 | 484 |
| Mwanza | Tanzania | Neg | 3 | Female (100) | 23 | 33 | 12 | |
| Mwanza | Tanzania | Pos | 21 | Female (100) | 27 | 67 | 17.1 | 352 |
At the time of specimen collection. Pos, positive; Neg, negative.
Percentage of subjects assigned to receive ACV at the time of specimen collection.
Among subjects assigned to receive ACV at the time of specimen collection. The totals in boldface exclude one Partners in Prevention subject, two Mwanza HIV-negative subjects, and seven Mwanza HIV-positive subjects who had not been started on acyclovir when the specimen was collected. These subjects were not included in calculations of means.
Among subjects who were HIV infected at the time of specimen collection. CD4 counts were available for all Partners in Prevention subjects and 15 of the 21 HIV-positive subjects from the Mwanza study.
One subject each from Zambia and Zimbabwe contributed two specimens each, and one subject from Peru contributed three specimens.
PIP, Partners in Prevention.
Overall, 19 of 64 participants (29.7%) were men and 45 (70.3%) were women. At the time of specimen collection, 26 (40.6%) were HIV infected. CD4 counts were available for 20 of the 26 (77%) HIV-infected subjects. The overall mean CD4 level was 395 cells/μl (range, 102 cells/μl to 1,037 cells/μl). Fifty-four participants (84.4%) were receiving ACV at the time of specimen collection. Because some subjects had more than one specimen (below), a total of 58 of 68 (85.3%) specimens were collected during ACV administration. Among the 58 specimens collected from the 54 subjects who were receiving ACV at the time of collection, the mean duration of ACV therapy prior to specimen collection was 12.1 months. Among the subset of 26 specimens obtained from HIV-infected subjects, 14 source subjects (54%) were receiving ACV at the time of specimen collection, with a mean duration of 16.4 months. The 10 specimens collected from subjects prior to initiation of ACV therapy were from nine persons (seven HIV positive) from Mwanza and one HIV-positive subject from a Partners in Prevention site in Kenya.
There were 18 additional samples originally selected for analysis from which UL23 sequence data were not obtainable. They included 17 samples in which we could not amplify UL23 for sequencing: 1 from HPTN 039, 11 from Partners in Prevention, and 5 from Mwanza. These specimens had between 6 and 14,144 HSV-2 DNA copies/specimen. The 68 specimens that were successfully sequenced ranged between 27 × 1010 and 2.4 × 1010 copies/specimen. One additional CVL fluid specimen from Mwanza contained no detectable HSV-2 DNA when analyzed in the University of Washington laboratory (21), so sequencing was not attempted.
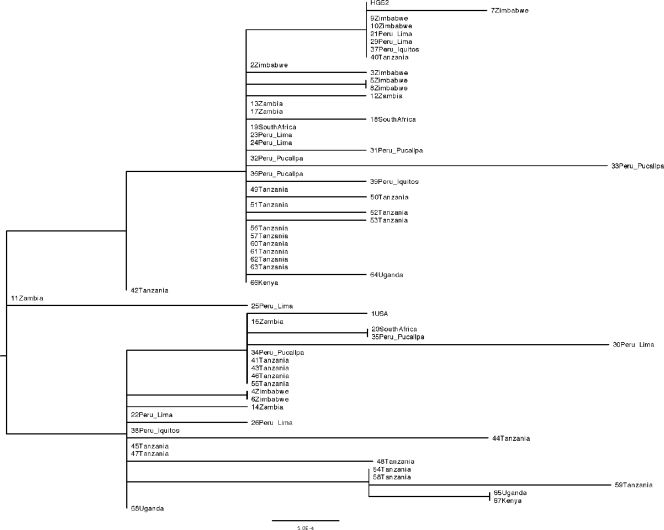
We observed that the UL23 protein-coding region was 1,128 bp in length for each of the 68 sequences we obtained. This is in agreement with HSV reference strain HG52 (10). Overall, 6 of the 68 sequences, obtained from 6 of the 64 participants, were identical to that of HG52 (Fig. 1). In the remaining 62 sequences, we detected changes in 38 of 1,128 (3.4%) nucleotide positions compared with HG52. Overall, 22 (58%) of the 38 variant nucleotide loci encoded amino acid changes, while the remaining 16 (42%) were noncoding variants. We did not detect any deletions or insertions.
FIG. 1.
Neighbor-joining dendrogram for UL23 sequences from this study and reference strain HG52. The bar represents 0.05% variation between sequences. The country and, if available, the region of origin are indicated. Identical sequences from the same subject are included only once.
Seven (18%) of the variants observed have been previously reported either in GenBank (19 full-length HSV-2 UL23 sequences), in partial sequence information published for a further 36 specimens (12, 27), or in full-length sequences of 29 of these strains made available by D. Chibo (see Materials and Methods). In contrast, 31 (82%) of the variants detected were not previously reported. The previously described variants, at nucleotides 79, 85, 116, 232, 420, 476, and 643, each encode an amino acid change: A27T, S29A, G39E, N78D, L140F, T159I, and A215D, respectively (Table 2 ). Among the 31 newly reported changes, 15 coded for amino acid changes and 16 were noncoding. At three variable loci, the allele frequenciesy of the rare alleles were greater than 10%. These loci are A232G, G420T, and G116A, in each of which the HG52 laboratory strain has the rare allele.
TABLE 2.
Nucleotide variations in HSV-2 UL23 detected in this study
| Nucleotidea | Amino acida | No. of strains (of 68) in which variant was detected | Frequency of rare allele (%) |
|---|---|---|---|
| Previously detected variants | |||
| G116A | G39E | 58 | 14.7b |
| A232G | N78D | 31 | 45.6 |
| G420T | L140F | 11 | 16.2 |
| G79A | A27T | 3 | 4.4 |
| G643A | A215T | 2 | 2.9 |
| C476T | T159I | 1 | 1.5 |
| T85G | S29A | 1 | 1.5 |
| Newly detected variants | |||
| T90C | NCc | 5 | 7.4 |
| A660G | NC | 5 | 7.4 |
| C783T | NC | 3 | 4.4 |
| G207A | NC | 2 | 2.9 |
| A307G | N103D | 2 | 2.9 |
| G449A | G150D | 2 | 2.9 |
| C582T | NC | 2 | 2.9 |
| C52T | R18C | 1 | 1.5 |
| C87T | NC | 1 | 1.5 |
| G183T | NC | 1 | 1.5 |
| G199A | A67T | 1 | 1.5 |
| A256C | M86L | 1 | 1.5 |
| G286A | E96K | 1 | 1.5 |
| C471A | NC | 1 | 1.5 |
| C493G | H165D | 1 | 1.5 |
| T565C | NC | 1 | 1.5 |
| T621C | NC | 1 | 1.5 |
| C663A | NC | 1 | 1.5 |
| C682A | L228I | 1 | 1.5 |
| C729T | NC | 1 | 1.5 |
| C787T | NC | 1 | 1.5 |
| C811A | P271T | 1 | 1.5 |
| C842G | S281C | 1 | 1.5 |
| C895T | NC | 1 | 1.5 |
| C942T | NC | 1 | 1.5 |
| G1013A | R338Q | 1 | 1.5 |
| G1039A | G347R | 1 | 1.5 |
| A1063G | T355A | 1 | 1.5 |
| C1068T | NC | 1 | 1.5 |
| C1101T | NC | 1 | 1.5 |
| A1125C | NC | 1 | 1.5 |
The nucleotide and amino acid in strain HG52 (10) is listed before the relevant position in the HG52 sequence, followed by the sequence detected in this study.
The rare allele, G at position 116, was present in the prototype HG52 strain.
NC, noncoding variation.
The 22 coding sequence variations detected in this survey were compared to the available medical literature and GenBank sequences in order to determine if they had been previously associated with possible ACV resistance. Several point mutations have been implicated in the development of resistance to ACV. They include R34C, G56E, T131P, R177W, S182D, T202A, R223H, D229H, T288M, and C336Y; a contiguous triad of R271V/P272S/D273R; and a dispersed triad of P85S, N100H, and V192M (6a, 8, 12, 16, 23a, 25a, 27, 29). We did not detect any exact overlap between the coding variants in our specimen set and these previously reported resistance-associated sequences. We then searched for variants close in the linear amino sequence to known resistance-associated mutations. One of the variants we detected, L228I, immediately adjacent to the resistance-associated mutation D229H (27), was detected in a baseline specimen from a 17-year-old HIV-negative woman from Masumbwe, southwest of Mwanza, Tanzania, before she was randomized to ACV or placebo. The L228I mutation is quite conservative, in contrast to D229H, which carries a change in the charge of the amino acid residue and thus is probably less likely to be associated with ACV resistance. A second mutation, R338Q, was present 2 amino acids away from the resistance-associated mutation C336Y (12). This variant was detected in a sample from an HIV-negative man from Iquitos, Peru, who had been on ACV for almost 17 months. The R338Q mutation from a positively charged to a polar uncharged amino acid residue is quite nonconservative.
Among the 31 separate UL23 sequence variants that are newly reported, 24 are present in only 1 of the 68 specimens (Table 2). We manually resequenced six random specimens, each of which contained one or two unique sequence variants not present in any of the other specimens or in the literature. The variants at C87T, G183T, G199A, C842G, C663A, and C682A (present in the same specimen), G1039A, and C1101T were each confirmed as present.
We had a limited opportunity to examine UL23 among paired specimens from individuals. From one participant from Zambia, 2 specimens were collected 6 months apart, and from one participant from Lima, Peru, 3 specimens were collected over 10 months. The UL23 sequences were identical for each specimen from the same subject. However, for a participant from Zimbabwe, differences between the two strains were noted at 3 loci (nucleotides 207, 232, and 449) for specimens collected 10 weeks apart. Detailed anatomic site information is not available for these specimens. Repeat sequencing confirmed these differences. Similar to previous reports (28), these data are consistent with infection with more than one strain of HSV-2.
A phylogenetic tree was constructed to determine if the UL23 sequences clustered in a pattern that might reflect the geographic origins of the specimens (Fig. 1). The sequence of laboratory strain HG52 (10) was included as a reference. Overall, there was pronounced admixture of strains from South America and various locations within sub-Saharan Africa. The one North American sequence was unique but clustered with selected strains from South Africa and Peru. Several identical sequences were recovered from various locations in Africa and Peru. Among the four strains that each had several unique nucleotide changes compared to the others (strains 25, 30, 33, and 59 [Fig. 1]), three were from Peru and one was from Tanzania. The average dN/average dS ratio for the set of 65 unique sequences from 64 persons, including the HG52 reference strain, was 0.71, implying the presence of evolutionary constraints on amino acid changes in thymidine kinase.
DISCUSSION
This is the first evaluation of the UL23 thymidine kinase gene of HSV-2, associated with ACV resistance, that focuses on specimens from the developing world, mostly Africa. Most of the subjects were receiving daily suppressive ACV therapy, for a mean duration of 12.1 months, at the time that HSV-2 DNA was recovered from a genital site. Slightly under half (40.6%) of the subjects had a known risk factor for ACV resistance, HIV infection, at the time of specimen collection (25). We found no evidence of known genotypic mutations associated with decreased susceptibility to ACV among 68 specimens collected from 64 individuals during three phase III RCTs. Although we did not detect genotypic evidence of ACV resistance in any sample, we documented marked genetic variation that was higher than that observed in any other HSV locus.
The mainstays of anti-HSV therapy are ACV; its prodrug, valacyclovir; and the related drug famciclovir. ACV resistance is readily diagnosed by classical virologic methods, using graded concentrations of ACV and one of several cell culture-based readouts (23). However, such testing for resistance is feasible only if a live viral isolate is available. For our study, only DNA specimens were obtained, due to storage and transport issues at geographically remote sites. This limited our ability to easily interpret the significance of the amino acid coding variants that we observed. One strength of analyzing DNA collected directly from subjects, however, is that potential changes in DNA sequences associated with passage in vitro, as has been documented for some HSV loci (20), are avoided.
Even with the use of direct swabs, it is still possible for sequence errors to result from PCR. The reported error rate for Taq polymerase is on the order of 1 error per 10−4 nucleotides per duplication cycle (or less) (2). With 35 cycles of amplification and an amplicon length of 1,128 bp, we estimated about 3.5 errors per amplicon. The proofreading function in specific thermostable polymerase products used in this project is reported to decrease the error rate by 6- to 8-fold, leading to an estimated 0.5 errors per final sequence. We believe that most of our newly reported variants are accurate for the following reasons. First, the newly reported sequence variants that we observed independently in more than one specimen (T90C, A660G, C783T, G207A, A307G, G449A, and C582T) (Table 2) are likely to be real polymorphisms, because identical PCR errors are very unlikely to have arisen independently in separate reactions. Second, some of the variants observed only once, such as T621C, C811A, and C895T, were all found in the same specimen from Pucallpa in mid-Amazonia in Peru (strain 33 in Fig. 1). It is very unlikely that PCR would introduce three separate changes into a single amplicon of this length. This is likely a unique, geographically isolated strain. Third, some of the UL23 variants we observed only once (G79A, T85G, C476C, and G643A) were also detected in a previous survey (6) of Australian HSV-2 cultures that used a less stringent polymerase. While PCR artifacts occur nonrandomly, it is unlikely that the same exact errors would occur independently.
It is also possible that we missed some mutations associated with acyclovir resistance because of our focus on UL23. Changes in the HSV DNA polymerase gene can alter its inhibition by ACV triphosphate. These mutations make a minor contribution to clinical ACV resistance (1a).
We did not detect any obvious resistance-associated frameshift mutations, but we did detect a large number of novel coding mutations. Coding point mutations associated with ACV resistance have also been described (6a, 8, 12, 23a, 23b, 25a, 29), but unlike in the example of human CMV and ganciclovir resistance (18), obvious clustering of such resistance-associated point mutations in hot spots has not yet been recognized. We did detect two point mutations (L228I and R338Q) that map near known ACV resistance-associated sequence variants (D229H and C336Y, respectively) (12, 27). In addition, the H165D amino acid substitution, which carries a charge change from positive to negative, observed in a specimen collected in Zambia from an HIV-seronegative subject in the HPTN 039 study after 267 days of ACV therapy, is near the putative nucleoside binding domain of HSV-2 thymidine kinase at amino acids 169 to 177 (29). Unfortunately, technical limitations precluded the storage or direct recovery of viable virus strains suitable for direct testing of ACV susceptibility. There are technologies available to determine the possible functional significance of the coding variations we have detected. They include the expression of recombinant protein and ACV phosphorylation assays in vitro (13) or the creation of chimeric viruses containing the UL23 sequences from this study inserted into a thymidine kinase-deficient strain, followed by phenotypic resistance testing.
Studies from developed countries have documented in vitro ACV resistance in approximately 0.28 to 0.33% of HSV isolates from immunocompetent individuals (5, 7, 23, 24). Interestingly, ACV use is not generally associated with drug resistance in immunocompetent persons at either the individual or population level (1a). The frequency of resistance is typically higher in immunocompromised individuals, including those with HIV infection, where 4 to 7% of isolates in developed countries may be resistant to ACV (25), although most source patients are also without clinical evidence of resistant disease. These strains are generally felt to arise due to selective pressure from ACV in the absence of a strong antiviral contribution from host immunity.
We did not detect sequence clusters that correlated with the geographic origins of our specimens. This contrasts with data from Norberg et al., who sequenced regions of the HSV-2 genes US7 and US8 from 20 isolates from Scandinavia and 27 isolates from Tanzania. Two straggling clades were detected, one of which contained only Tanzanian strains while the other included sequences from both regions (22). In our data set, sequences from Peru were admixed with African sequences in all major branches of the UL23 neighbor-joining tree. Kaneko et al. sequenced 36 HSV-2 genes from 36 subjects in Japan (15). They analyzed UL3, US1, and US4, their most divergent loci, as trees and noted that they had differing topologies. These data, and coanalysis of US4 data with the US7/US8 data set by Norberg et al., are consistent with crossovers and recombination in HSV-2 molecular evolution (22).
The UL23 locus studied in this report seems particularly useful for strain tracking, with variants detected at 3.4% of nucleotides among 68 sequences. The most divergent strains, 31 (Pucallpa, Peru) and 57 (Mwanza, Tanzania), differed at 0.7% of the nucleotides. Several variable loci have rare alleles that are reasonably prevalent, so that single-nucleotide polymorphism (SNP) typing schemes could be useful. This variation is higher than the maximal 0.4% variance noted by Norberg et al. among 47 strains sequenced in the US4, US7, and US8 regions. Kaneko et al. noted that many HSV-2 genes were completely invariant in Japan (e.g., UL4, UL5, UL12, and 9 other genes) (15). The low dN/dS ratio observed for UL23 is consistent with active selection to preserve certain amino acid sequences despite the apparent tolerance for amino acid changes in many locations.
The HIV-infected persons we studied had relatively preserved CD4 T-cell counts, in contrast to the advanced immune deficiency typically reported in HIV/HSV-2-coinfected persons who develop clinically ACV-resistant HSV-2 (9). These findings are encouraging, given the recommendation by WHO to add ACV to the treatment regimen for genital ulcer disease (32). Although the clinical trials from which our specimens came did not detect a decrease in HIV transmission, there were beneficial effects of ACV, such as decreased disease progression among HIV-infected persons being treated with ACV in the Partners in Prevention trial and a reduced frequency of genital ulcerations (19). Recent biological findings that may explain the poor biological activity of ACV at the doses used in the recent trials include the observation of frequent shedding of HSV-2 in the genital tract despite administration of standard doses of antiherpesviral drugs (A. Wald, C. Johnston, M. Saracino, L. Olin, K. Mark, S. Selke, M. Huang, and L. Corey, presented at the 18th Meeting of the International Society for Sexually Transmitted Diseases Research, London, England, 28 June to 1 July 2009) and the persistence of HIV-susceptible, CCR5- or CXCR4-expressing, HSV-2-specific CD4 T cells in the genital skin at sites of previous HSV-2 lesions despite several months of antiherpesvirus therapy (33). In addition, ACV suppression of genital shedding of HSV-2 was not as effective at South American and African sites as at North American sites within the HPTN 039 study, for unknown reasons (11). Improved antiherpesvirus regimens might show activity against HIV transmission, and clinical trials should continue to monitor for the possible development of resistance to anti-HSV drugs in geographically distinct populations.
Acknowledgments
We thank the study participants and the entire investigative team for each of the trials providing specimens. Geoffrey Gottlieb assisted with phylogenetic-tree analysis. Meei-Li Huang provided DNA extraction and HSV-2 DNA PCR measurement. Ellen Wilcox assisted with IRB coordination. Stacy Selke and Jim Hughes assisted with database management and specimen selection.
This work was supported in part by USPHS grants AI30731, AI52054, AI27757, and AI46749;, the Bill and Melinda Gates Foundation; the Wellcome Trust; the United Kingdom Medical Research Council; and the United Kingdom Department for International Development. Acyclovir for the HPTN 039 study only was provided by GlaxoSmithKline. A.W., C.C., and D.W.-J. received grant support from GlaxoSmithKline.
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Abu-Raddad, L. J., A. S. Magaret, C. Celum, A. Wald, I. M. Longini, Jr., S. G. Self, and L. Corey. 2008. Genital herpes has played a more important role than any other sexually transmitted infection in driving HIV prevalence in Africa. PLoS One 3:e2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Bacon, T. H., M. J. Levin, J. J. Leary, R. T. Sarisky, and D. Sutton. 2003. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin. Microbiol. Rev. 16:114-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bracho, M. A., A. Moya, and E. Barrio. 1998. Contribution of Taq polymerase-induced errors to the estimation of RNA virus diversity. J. Gen. Virol. 79:2921-2928. [DOI] [PubMed] [Google Scholar]
- 3.Brankin, A. E., A. A. Tobian, O. Laeyendecker, T. R. Suntoke, A. Kizza, B. Mpoza, G. Kigozi, F. Nalugoda, B. Iga, M. Z. Chen, R. H. Gray, M. J. Wawer, T. C. Quinn, and S. J. Reynolds. 2009. Aetiology of genital ulcer disease in female partners of male participants in a circumcision trial in Uganda. Int. J. STD AIDS 20:650-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Burrows, J., A. Nitsche, B. Bayly, E. Walker, G. Higgins, and T. Kok. 2002. Detection and subtyping of Herpes simplex virus in clinical samples by LightCycler PCR, enzyme immunoassay and cell culture. BMC Microbiol. 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celum, C., A. Wald, J. R. Lingappa, A. S. Magaret, R. S. Wang, N. Mugo, A. Mujugira, J. M. Baeten, J. I. Mullins, J. P. Hughes, E. A. Bukusi, C. R. Cohen, E. Katabira, A. Ronald, J. Kiarie, C. Farquhar, G. J. Stewart, J. Makhema, M. Essex, E. Were, K. H. Fife, G. de Bruyn, G. E. Gray, J. A. McIntyre, R. Manongi, S. Kapiga, D. Coetzee, S. Allen, M. Inambao, K. Kayitenkore, E. Karita, W. Kanweka, S. Delany, H. Rees, B. Vwalika, W. Stevens, M. S. Campbell, K. K. Thomas, R. W. Coombs, R. Morrow, W. L. Whittington, M. J. McElrath, L. Barnes, R. Ridzon, and L. Corey. 2010. Acyclovir and transmission of HIV-1 from persons infected with HIV-1 and HSV-2. N. Engl. J. Med. 362:427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Celum, C., A. Wald, J. Hughes, J. Sanchez, S. Reid, S. Delany-Moretlwe, F. Cowan, M. Casapia, A. Ortiz, J. Fuchs, S. Buchbinder, B. Koblin, S. Zwerski, S. Rose, J. Wang, and L. Corey. 2008. Effect of aciclovir on HIV-1 acquisition in herpes simplex virus 2 seropositive women and men who have sex with men: a randomised, double-blind, placebo-controlled trial. Lancet 371:2109-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., C. Scieux, V. Garrait, G. Socié, V. Rocha, J. M. Molina, D. Thouvenot, F. Morfin, L. Hocqueloux, L. Garderet, H. Espérou, F. Sélimi, A. Devergie, G. Leleu, M. Aymard, F. Morinet, E. Gluckman, and P. Ribaud. 2000. Resistant herpes simplex virus type 1 infection: an emerging concern after allogeneic stem cell transplantation. Clin. Infect. Dis. 31:927-935. [DOI] [PubMed] [Google Scholar]
- 6.Chibo, D., J. Druce, J. Sasadeusz, and C. Birch. 2004. Molecular analysis of clinical isolates of acyclovir resistant herpes simplex virus. Antiviral Res. 61:83-91. [DOI] [PubMed] [Google Scholar]
- 6a.Chibo, D., A. Mijch, R. Doherty, and C. Birch. 2002. Novel mutations in the thymidine kinase and DNA polymerase genes of acyclovir and foscarnet resistant herpes simplex viruses infecting an immunocompromised patient. J. Clin. Virol. 25:165-170. [DOI] [PubMed] [Google Scholar]
- 7.Christophers, J., J. Clayton, J. Craske, R. Ward, P. Collins, M. Trowbridge, and G. Darby. 1998. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob. Agents Chemother. 42:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czartoski, T., C. Liu, D. M. Koelle, S. Schmechel, A. Kalus, and A. Wald. 2006. Fulminant, acyclovir-resistant, herpes simplex virus type 2 hepatitis in an immunocompetent woman. J. Clin. Microbiol. 44:1584-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danve-Szatanek, C., M. Aymard, D. Thouvenot, F. Morfin, G. Agius, I. Bertin, S. Billaudel, B. Chanzy, M. Coste-Burel, L. Finkielsztejn, H. Fleury, T. Hadou, C. Henquell, H. Lafeuille, M. E. Lafon, A. Le Faou, M. C. Legrand, L. Maille, C. Mengelle, P. Morand, F. Morinet, E. Nicand, S. Omar, B. Picard, B. Pozzetto, J. Puel, D. Raoult, C. Scieux, M. Segondy, J. M. Seigneurin, R. Teyssou, and C. Zandotti. 2004. Surveillance network for herpes simplex virus resistance to antiviral drugs: 3-year follow-up. J. Clin. Microbiol. 42:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, J., C. Celum, J. Wang, J. Hughes, J. Sanchez, F. Cowan, S. Reid, S. Delany-Moretlwe, L. Corey, and A. Wald. 2010. Clinical and virologic efficacy of herpes simplex virus type 2 suppression by acyclovir in a multicontinent clinical trial. J. Infect. Dis. 201:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudreau, A., E. Hill, H. H. Balfour, Jr., A. Erice, and G. Boivin. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297-303. [DOI] [PubMed] [Google Scholar]
- 13.Horsburgh, B. C., S. H. Chen, A. Hu, G. B. Mulamba, W. H. Burns, and D. M. Coen. 1998. Recurrent acyclovir-resistant herpes simplex in an immunocompromised patient: can strain differences compensate for loss of thymidine kinase in pathogenesis? J. Infect. Dis. 178:618-625. [DOI] [PubMed] [Google Scholar]
- 14.Jerome, K. R., M. L. Huang, A. Wald, S. Selke, and L. Corey. 2002. Quantitative stability of DNA after extended storage of clinical specimens as determined by real-time PCR. J. Clin. Microbiol. 40:2609-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko, H., T. Kawana, K. Ishioka, E. Fukushima, and T. Suzutani. 2008. Discrimination of herpes simplex virus type 2 strains by nucleotide sequence variations. J. Clin. Microbiol. 46:780-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kit, S., M. Sheppard, H. Ichimura, S. Nusinoff-Lehrman, M. N. Ellis, J. A. Fyfe, and H. Otsuka. 1987. Nucleotide sequence changes in thymidine kinase gene of herpes simplex virus type 2 clones from an isolate of a patient treated with acyclovir. Antimicrob. Agents Chemother. 31:1483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korber, B. 2000. Computational analysis of HIV molecular sequences, p. 55-72. In A. G. Rodrigo, and G. H. Learn (ed.), HIV signature and sequence variation analysis. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 18.Limaye, A. P., G. Raghu, D. M. Koelle, J. Ferrenberg, M.-L. Huang, and M. Boeckh. 2002. High incidence of ganciclovir-resistant cytomegalovirus infection among lung transplant recipients receiving preemptive therapy. J. Infect. Dis. 185:20-27. [DOI] [PubMed] [Google Scholar]
- 19.Lingappa, J. R., J. M. Baeten, A. Wald, J. P. Hughes, K. K. Thomas, A. Mujugira, N. Mugo, E. A. Bukusi, C. R. Cohen, E. Katabira, A. Ronald, J. Kiarie, C. Farquhar, G. J. Stewart, J. Makhema, M. Essex, E. Were, K. H. Fife, G. de Bruyn, G. E. Gray, J. A. McIntyre, R. Manongi, S. Kapiga, D. Coetzee, S. Allen, M. Inambao, K. Kayitenkore, E. Karita, W. Kanweka, S. Delany, H. Rees, B. Vwalika, A. S. Magaret, R. S. Wang, L. Kidoguchi, L. Barnes, R. Ridzon, L. Corey, and C. Celum. 2010. Daily aciclovir for HIV-1 disease progression in people dually infected with HIV-1 and herpes simplex virus type 2: a randomised placebo-controlled trial. Lancet 375:824-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maertzdorf, J., L. Remeijer, A. Van Der Lelij, J. Buitenwerf, H. G. M. Niesters, A. D. M. E. Osterhaus, and G. M. G. M. Verjans. 1999. Amplification of reiterated sequences of herpes simplex virus type 1 (HSV-1) genome to discriminate between clinical HSV-1 isolates. J. Clin. Microbiol. 37:3518-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magaret, A. S., A. Wald, M. L. Huang, S. Selke, and L. Corey. 2007. Optimizing PCR positivity criterion for detection of herpes simplex virus DNA on skin and mucosa. J. Clin. Microbiol. 45:1618-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norberg, P. 2010. Divergence and genotyping of human alpha-herpesviruses: an overview. Infect. Genet. Evol. 10:14-25. [DOI] [PubMed] [Google Scholar]
- 23.Nugier, F., J. N. Colin, M. Aymard, and M. Langlois. 1992. Occurrence and characterization of acyclovir-resistant herpes simplex virus isolates: report on a two-year sensitivity screening survey. J. Med. Virol. 36:1-12. [DOI] [PubMed] [Google Scholar]
- 23a.Palu, G., G. Gerna, F. Bevilacqua, and A. Marcello. 1992. A point mutation in the thymidine kinase gene is responsible for acyclovir-resistance in herpes simplex virus type 2 sequential isolates. Virus Res. 25:133-144. [DOI] [PubMed] [Google Scholar]
- 23b.Pereira, C. F., K. Rutten, R. Stranska, M. C. Huigen, P. C. Aerts, R. J. de Groot, H. F. Egberink, R. Schuurman, and H. S. Nottet. 2005. Spectrum of antiviral activity of o-(acetoxyphenyl)hept-2-ynyl sulphide (APHS). Int. J. Antimicrob. Agents 25:419-426. [DOI] [PubMed] [Google Scholar]
- 24.Reyes, M., N. S. Shaik, J. M. Graber, R. Nisenbaum, N. T. Wetherall, K. Fukuda, W. C. Reeves, and the Task Force on Herpes Simplex Virus Resistance. 2003. Acyclovir-resistant genital herpes among persons attending sexually transmitted disease and human immunodeficiency virus clinics. Arch. Intern. Med. 163:76-80. [DOI] [PubMed] [Google Scholar]
- 25.Safrin, S., T. Elbeik, L. Phan, D. Robinson, J. Rush, A. Elbaggari, and J. Mills. 1994. Correlation between response to acyclovir and foscarnet therapy and in vitro susceptibility result for isolates of herpes simplex virus from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 38:1246-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Shimada, Y., M. Suzuki, F. Shirasaki, E. Saito, K. Sogo, M. Hasegawa, K. Takehara, J. Phromjai, T. Chuhjo, and K. Shiraki. 2007. Genital herpes due to acyclovir-sensitive herpes simplex virus caused secondary and recurrent herpetic whitlows due to thymidine kinase-deficient/temperature-sensitive virus. J. Med. Virol. 79:1731-1740. [DOI] [PubMed] [Google Scholar]
- 26.Smith, J. S., and N. J. Robinson. 2002. Age-specific prevalence of infection with herpes simplex virus types 2 and 1: a global review. J. Infect. Dis. 186(Suppl. 1):S3-S28. [DOI] [PubMed] [Google Scholar]
- 27.Stránská, R., R. Schuurman, E. Nienhuis, I. W. Goedegebuure, M. Polman, J. F. Weel, P. M. Wertheim-Van Dillen, R. J. Berkhout, and A. M. van Loon. 2005. Survey of acyclovir-resistant herpes simplex virus in the Netherlands: prevalence and characterization. J. Clin. Virol. 32:7-18. [DOI] [PubMed] [Google Scholar]
- 27a.Stranska, R., A. M. van Loon, M. Polman, M. F. Beersma, R. G. Bredius, A. C. Lankester, E. Meijer, and R. Schuurman. 2004. Genotypic and phenotypic characterization of acyclovir-resistant herpes simplex viruses isolated from haematopoietic stem cell transplant recipients. Antivir. Ther. 9:565-575. [PubMed] [Google Scholar]
- 28.Sucato, G., A. Wald, E. Wakabayashi, J. Vieira, and L. Corey. 1998. Evidence of latency and reactivation of both herpes simplex virus (HSV)-1 and HSV-2 in the genital region. J. Infect. Dis. 177:1069-1072. [DOI] [PubMed] [Google Scholar]
- 28a.Swetter, S. M., E. L. Hill, E. L. Kern, D. M. Koelle, C. M. Posavad, and S. Safrin. 1998. Chronic vulvar ulceration in an immunocompetent woman due to acyclovir-resistant, thymidine-kinase-deficient herpes simplex virus. J. Infect. Dis. 177:543-550. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, S., Y. Toh, and R. Mori. 1993. Molecular analysis of a neurovirulent herpes simplex virus type 2 strain with reduced thymidine kinase activity. Arch. Virol. 131:61-73. [DOI] [PubMed] [Google Scholar]
- 29a.Tanton, C., H. A. Weiss, M. Rusizoka, J. Legoff, J. Changalucha, K. Baisley, K. Mugeye, D. Everett, L. Belec, T. C. Clayton, D. A. Ross, R. J. Hayes, and D. Watson-Jones. 2010. Long-term impact of acyclovir suppressive therapy on genital and plasma HIV RNA in Tanzanian women: a randomized controlled trial. J. Infect. Dis. 201:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wald, A., and K. Link. 2002. Risk of human immunodeficiency virus infection in herpes simplex virus type 2-seropositive persons: a meta-analysis. J. Infect. Dis. 185:45-52. [DOI] [PubMed] [Google Scholar]
- 31.Watson-Jones, D., H. A. Weiss, M. Rusizoka, J. Changalucha, K. Baisley, K. Mugeye, C. Tanton, D. Ross, D. Everett, T. Clayton, R. Balira, L. Knight, I. Hambleton, J. Le Goff, L. Belec, and R. Hayes. 2008. Effect of herpes simplex suppression on incidence of HIV among women in Tanzania. N. Engl. J. Med. 358:1560-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. WHO guidelines for the management of sexually transmitted infections, in press. World Health Organization, Geneva, Switzerland.
- 33.Zhu, J., F. Hladik, A. Woodward, A. Klock, T. Peng, C. Johnston, M. Remington, A. Magaret, D. M. Koelle, A. Wald, and L. Corey. 2009. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat. Med. 15:886-892. [DOI] [PMC free article] [PubMed] [Google Scholar]