Abstract
The recently introduced Xpert MTB/RIF assay (Xpert) has point-of-care potential, but its capacity for biohazard containment remained to be studied. We compared the bioaerosols generated by the Xpert assay to acid-fast bacillus (AFB) microscope slide smear preparation. The Xpert assay sample treatment reagent (SR) was also studied for its sterilizing capacity, stability, and effect on assay sensitivity after prolonged treatment. During the preparation of AFB smears, sputum samples spiked with Mycobacterium bovis BCG at 5 × 108 CFU/ml produced 16 and 325 CFU/m3 air measured with an Andersen impactor or BioSampler, respectively. In contrast, neither the sample preparation steps for the Xpert assay nor its automated processing produced any culturable bioaerosols. In testing of SR sterilizing capacity, clinical sputum samples from strongly smear-positive tuberculosis patients treated with SR at a 2:1 ratio eliminated Mycobacterium tuberculosis growth in all but 1/39 or 3/45 samples cultured on solid or liquid medium, respectively. These few unsterilized samples had a mean 13.1-day delay in the time to positive culture. SR treatment at a 3:1 ratio eliminated growth in all samples. SR retained a greater than 6-log-unit killing capacity despite storage at temperatures spanning 4 to 45°C for at least 3 months. The effect of prolonged SR sample treatment was also studied. Spiked sputum samples could be incubated in SR for up to 3 days without affecting Xpert sensitivity for M. tuberculosis detection and up to 8 h without affecting specificity for rifampin resistance detection. These results suggest that benchtop use of the Xpert MTB/RIF assay limits infection risk to the user.
The recently introduced Xpert MTB/RIF assay (Cepheid, Sunnyvale, CA) simultaneously detects the presence of Mycobacterium tuberculosis and its susceptibility to the important first-line drug rifampin (RIF) in less than 2 h (14). The assay is very easy to perform and almost entirely automated, requiring only two manual steps. The simplicity of the assay and its “on-demand” features make it amenable to point-of-care testing by personnel with minimal training.
Point-of-care tests should be safe to use in their intended environments. Tests that expose testing personnel to relatively low numbers of bacilli, such as a direct acid-fast bacillus (AFB) smear examination, pose a much smaller biohazard risk (7, 13, 22) than do tests that include a biologic amplification step (cultivation) (17). Though there is no epidemiologic data linking smear preparation with tuberculosis risk (International Union against Tuberculosis and Lung Disease, Paris, France), many laboratory guidelines suggest that sputum handling and AFB smear preparation should be performed inside a biological safety cabinet (BSC) (11, 16). However, smear preparation is often performed on open benchtops in resource-poor settings (4, 21). Industrialized nations with stringent biosafety standards usually permit sputum samples from suspected tuberculosis patients to be processed for routine Gram staining and (nontuberculosis) bacterial culture in clinical microbiology laboratories with biological safety level 2 (BSL-2) biocontainment, although a BSC is normally used. A good tuberculosis point-of-care test should be no more risky than an AFB smear examination, and an ideal test would eliminate all infectious risk.
Several aspects particular to the Xpert MTB/RIF assay suggest that it might present a very low risk to testing personnel. The Xpert MTB/RIF assay starts with the addition of a highly tuberculocidal sample treatment reagent (SR) to each sputum sample (or concentrated sputum pellet). Used as recommended, SR treatment complies with international decontamination standards (3, 24), reducing the viability of M. tuberculosis in sputum by at least 6 log10 units after 15 min of incubation (14). Longer incubation periods appear to completely sterilize samples spiked with very high numbers of M. tuberculosis. After the 15-min incubation, the sputum-SR mixture is transferred to the plastic assay cartridge, the cartridge lid is closed, and the cartridge is placed into a GeneXpert instrument. The remainder of the assay is performed within the closed cartridge. The use of a closed-cartridge system can potentially further reduce or eliminate biohazard risk by performing sample processing in an aerosol-resistant enclosure.
We studied the amounts of potentially infectious aerosols that could be generated by the manual and automated steps of the Xpert MTB/RIF assay. We studied whether any of the assay steps posed a greater risk to testing personnel than the preparation of an AFB smear. We also determined whether it was possible to process a cartridge loaded with sputum plus SR without generating any infection risk. To confirm the critical tuberculocidal activity of SR, we performed a series of SR killing studies using various lots of SR stored under different conditions and performed a quantitative assessment of killing on strongly smear-positive clinical sputum samples. Finally, we investigated the effect of prolonged SR treatment on analytic assay sensitivity, to determine whether increased SR incubation time (which enhances killing) had any adverse affects on the assay performance.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and clinical samples.
The Mycobacterium bovis BCG Pasteur and Mycobacterium smegmatis Mc2155 strains used in all experiments were a kind gift of William Jacobs, Jr., Howard Hughes Medical Institute, Bronx, NY. M. tuberculosis strain H37Rv was obtained from the American Tissue Culture Collection (ATCC), Manassas, VA. Mycobacteria were cultured on 7H10 solid medium (BD Diagnostic Systems, Sparks, MD) supplemented with 0.5% glycerol and 10% albumin-dextrose-catalase with sodium chloride (Middlebrook ADC Enrichment; BD Diagnostic Systems). Each 4-ml mycobacterial growth indicator tube (MGIT) (BD Diagnostic Systems) was supplemented with 0.5 ml MGIT oleic acid-albumin-dextrose-catalase (OADC) and 0.1 ml PANTA antibiotic (BD), supplied with the kit. The Middlebrook 7H9 liquid medium (BD Diagnostic Systems) used in the BioSampler studies was supplemented with 10% ADC (BD Diagnostic Systems) and 0.05% Tween 80 (Sigma-Aldrich, St. Louis, MO). M. bovis BCG (ATCC 35743; TMC 1028) was acquired from the ATCC by the independent testing service (Microbiotest, Sterling, VA).
BCG organisms were inoculated into Middlebrook 7H9 liquid medium and incubated at 37 ± 2°C with gentle agitation for 15 to 25 days or until a pellicle was formed. This material was used as the inoculum for the quantitative tuberculocidal test as follows. Solid-medium cultures were wrapped in aluminum foil, incubated at 37°C for up to 6 weeks, and checked periodically for growth. MGITs were cultured and analyzed in a Bactec MGIT 960 automated culture system (BD) according to the manufacturer's recommendations, and the number of days between MGIT inoculation and a positive culture (time-to-positive days [TpD]) was recorded for each sample. Negative cultures were discarded after 6 weeks. M. tuberculosis was confirmed in all positive MGIT cultures by the observation of a positive AFB smear and a positive immunochromatographic lateral-flow assay for MPB64 (Capilia TB Neo) performed according to the manufacturer's instructions (Tauns Laboratories, Inc., Shizuoka, Japan). Solid cultures on 7H10 plates were identified as contaminated when the colonies present did not have the typical morphological appearance of M. tuberculosis, and representative colonies were confirmed to be Capilia TB Neo test negative. Liquid cultures that were indicated as positive by the MGIT 960 system were identified as contaminated if both the Capilia TB Neo test and the AFB smear from the culture medium were negative.
The origins of the sputum samples varied in accordance with study objectives. Analytical studies that required the addition of known numbers of BCG CFU into tuberculosis-negative sputum were performed with sputa obtained from patients at New Jersey Medical School—UMDNJ University Hospital, Newark, NJ, who were not suspected of having tuberculosis, as described previously (14), and/or with sputum pools from confirmed negative patients obtained from Montefiore Medical Center, Bronx, NY. Sputum samples for the clinical SR killing studies were obtained from patients being evaluated for tuberculosis at Hinduja Hospital, India, between January and February 2009. The Hinduja patients were required to have symptoms suggestive of tuberculosis and grade 2+ to 3+ AFB smears on prescreening (where 2+ represents 1 to 9 AFB per field and 3+ represents 10 or more AFB per field). Patients were excluded if they had received any antituberculosis therapy within the past 60 days. Sputum samples were collected prospectively from each patient, frozen, and then sent to a laboratory in the United States for SR killing studies. Sputum samples were obtained only after approval by the Hinduja and UMDNJ institutional review boards.
Detection of viable aerosols.
As there is no standardized reference method for detecting bioaerosol contamination from new devices or during procedures, we aimed to detect both total and culturable aerosol generation under simulated conditions. Aerosolization studies during smearing and pipetting were performed in a 6-foot (2-m) class II biological safety cabinet (BSC-2) (NU-425-600; NuAire, Plymouth, MN). The GeneXpert aerosolization studies were performed in a 4-foot (1.2- m) BSC-2 (NU-425-400; NuAire) with the airflow turned off and the sash pulled down into an incomplete airtight seal. The limit-of-detection (LOD) (2) studies were performed inside a custom-built aerosol generation and sampling chamber based on the design of a chamber successfully used for similar aerosol studies (15). This was essentially a closed wind tunnel kept under negative pressure, which permitted safe aerosolization and sampling of infectious particles. The chamber was operated inside the biological safety cabinet to control the large numbers of infectious BCG that were aerosolized during the experiment. For all aerosol studies, viable-aerosol production was measured using both a single-stage N-6 viable Andersen cascade impactor (Thermo Scientific, Waltham, MA) and a BioSampler (SKC Inc., Eighty Four, PA) loaded with 20 ml Middlebrook 7H9 liquid medium. The Andersen impactor was operated at calibrated vacuum flow rates of 28.3 liters per minute, and the BioSampler was operated at 12.8 liters per minute. Before sampling, the airflow rates for all samplers were calibrated with a DryCal DC-lite Calibrator (SKC Inc., Eighty Four, PA). The aerosol sampling time for each study is noted below. The BioSampler's collection liquid was plated in triplicate on 7H10 agar plates. All the agar plates were incubated at 37°C for up to 5 weeks for CFU counts. The total airborne bacterial concentrations were expressed per cubic meter of air.
The total airborne concentration (CFU/m3) for the Andersen impactor, CANDERSEN, was calculated as follows: CANDERSEN = 1,000 × Ntotal/Qsts, where Ntotal is the total number of CFU on the agar plate, Qs is the sampling flow rate per minute, and ts is the sampling time (min).
The total airborne concentration (CFU/m3) for the BioSampler, CBIOSAMPLER, was calculated as follows: CBIOSAMPLER = 1,000 × (Ntotal/Qsts) × (VLIQ/VPLIQ), where VLIQ is the volume of liquid left after collection and VPLIQ is the volume of liquid used for plating.
The total numbers of particles present in the BSCs before, during, and after the sample preparation steps were also studied as a second measure of potentially infectious aerosol generation. Particles were detected using an aerodynamic particle sizer spectrometer (APS model 3321; TSI Inc., St. Paul, MN).
Limits of detection of the aerosol samplers.
7H9 medium was spiked with log dilutions of BCG ranging from 10 to 104 CFU/ml. Approximately 0.8 ml of each mixture was then aerosolized using a Sparging Liquid Aerosol Generator (CH Technologies, Inc., Westwood, NJ) over 10 min within the custom-built aerosol generation and sampling chamber. Total aerosol particle counts within the chamber were measured using an APS Spectrometer. Aerosol collection was begun after the total aerosol particle counts were stable and then continued for 1 min using the Andersen impactor and for 15 min using the BioSampler.
Aerosolization during AFB smear and Xpert MTB/RIF assay setup.
Pooled human sputa from nontuberculous patients were spiked with BCG at a final concentration of 5 × 108 CFU/ml of sputum. SR was added to the BCG-sputum mixture at an SR/sputum ratio of 2:1. The “incorrect use of SR” was simulated by loading the SR-sputum mixture immediately into Xpert MTB/RIF cartridges without performing the recommended 15-min SR-sputum incubation. Instead, after SR introduction, the SR-sputum container was briefly shaken, and then the sputum was immediately pipetted in and out of three Xpert MTB/RIF cartridges for 15 min, simulating the loading of at least 30 cartridges. All pipetting steps used the disposable plastic pipettes included in the Xpert MTB/RIF test kit. The “correct use of SR” was simulated by briefly shaking the SR-sputum mixture and incubating the mixture for 15 min at room temperature before pipetting the mixture in and out of three Xpert MTB/RIF cartridges for 15 min as described above. AFB smear preparation was performed by smearing a loopful (10 μl) of the BCG-spiked sputum (without SR) onto three microscope slides for 10 min, simulating the preparation of 20 sputum AFB slides. Smears were made using disposable plastic loops. Aerosol collection using the Andersen impactor and BioSampler was begun at the start of each procedure and continued for 2 min and 10 min, respectively.
Air sampling during automated sample processing.
The production of viable aerosols generated by the GeneXpert instrument during sample-processing stages in Xpert MTB/RIF cartridges was studied by loading Xpert MTB/RIF cartridges with several different sample types and measuring viable-aerosol production during an assay run. Cartridges were loaded with one of three sample types: (i) water spiked with 2 ml of 5 × 108 BCG CFU/ml without SR treatment, (ii) 2 ml of sputum spiked with BCG (5 × 108 CFU/ml) treated for 15 min with SR at a 2:1 ratio, or (iii) 2 ml of sputum spiked with M. smegmatis (7 × 108 CFU/ml) treated for 15 min with SR at a 2:1 ratio. For each sample type studied, the GeneXpert instrument was loaded with three cartridges and the air was sampled for 2 and 15 min using two Andersen samplers and a BioSampler, respectively, during the Xpert MTB/RIF assay run. The first Andersen sampler was operated simultaneously with the start of the BioSampler, and the second Andersen sampler was operated at the 13th minute of the BioSampler's run time. Each assay was run three times, individually, for a total of nine cartridges tested under each condition.
SR killing studies.
Fifty M. tuberculosis-positive sputum samples collected from the Hinduja study were thawed and vortexed for 1 min. Five of the samples did not have the required minimum 3-ml volume and were discarded. The remaining 45 samples were thoroughly mixed with a pipette and then split into three portions of 1 ml each. The three portions were randomly assigned as follows. The first 1-ml portion was decontaminated by a standard NALC-NaOH (N-acetyl-l-cysteine-NaOH) method, using a MycoPrep kit (BD Diagnostic Systems) according to the manufacturer's instructions and resuspended in 700 μl of phosphate-buffered saline (PBS), pH 7.4. A 200-μl volume of each sample was inoculated onto 7H10 plates, and 500 μl was inoculated into MGITs supplemented with PANTA and OADC according to the manufacturer's instructions. Sputum samples from six tuberculosis-negative patients were also split, and 1 ml was cultured as described above. A second and a third 1-ml portion of each sputum sample was treated with SR at an SR/sputum ratio of 2:1 or 3:1, respectively. The SR-sputum mixtures were briefly shaken and incubated at room temperature for 15 min. A 1.5-ml volume of each sample was then transferred to a 50-ml centrifuge tube, which was filled to the top with PBS and centrifuged for 20 min at 4,000 × g. The supernatant was then discarded, and the remaining pellet was resuspended in 700 μl of PBS. Two hundred microliters of each sample was inoculated onto 7H9 medium, and 500 μl was inoculated into an MGIT. Positive growth in the MGIT was confirmed by both Capilia TB-Neo assay (Tauns, Shizuoka, Japan) and by AFB smear examination; 7H10 colonies were confirmed for M. tuberculosis bacilli by Capilia TB-Neo assay only.
SR stability at varied temperatures.
SR was stored under different conditions for at least 3 months. The killing efficiency of SR under each storage condition was evaluated in a manner similar to that described above. Briefly, negative-control sputum was spiked with 109 CFU/ml BCG cells, and SR was added at the recommended 2:1 ratio (SR/sputum) to the sample. The preparation was shaken for 5 s, incubated for 15 min at ambient temperature, and again shaken manually for 5 s. PBS was added to make up the volume to 45 ml and mixed well, and the tube was centrifuged for 15 min at 4,000 × g. The pellet was resuspended in 1 ml of fresh PBS, and 100 μl was plated on 7H10 plates in triplicate. Four individual experiments were run to evaluate the average killing efficiency of these stored SR. The average log kill was expressed as follows: log10(average initial CFU/ml) − log10(average test CFU/ml).
Quantitative tuberculocidal tests performed by an independent test service.
Testing was performed by Microbiotest (Sterling, VA). BCG organisms were macerated with a tissue grinder at 0 to 4°C, and the homogenized suspension was diluted with 7H9 medium to a concentration between 107 and 108 CFU/ml, as determined by spectrophotometric methods. Two milliliters of BCG suspension was added to 4.7 ml of sputum and vortexed to mix them, and then the mixture was added to 13.3 ml of SR in sterile glass tubes. Duplicate preparations were made. After incubation for 15 min, aliquots of the mixture were removed and mixed with aliquots of sterile saline to make 10-fold serial dilutions. Duplicates of each dilution were filtered through a 47-mm-diameter cellulose nitrate membrane filter with a pore size of 0.45 μm, and the filters were washed with sterile saline. The filters were aseptically transferred to the surface of a Middlebrook 7H11 plate and incubated at 37 ± 2°C for 15 to 25 days. Colonies were counted using a dissecting microscope with lateral lighting, and the average number of CFU/ml was calculated. Initial counts of BCG organisms were determined by mixing 2 ml of M. bovis BCG suspension with 18 ml of sterile saline and serially diluting 1-ml aliquots with sterile saline blanks. These control samples were processed similarly to the test samples, and the plates were incubated along with the test plates. BCG colonies were confirmed by AFB staining and colony morphology. The log reduction in viability was calculated as explained above: log10(average initial CFU/ml) − log10(average test CFU/ml).
Stability of samples after prolonged SR treatment.
M. tuberculosis-negative sputum samples were spiked with M. tuberculosis strain H37Rv at a final concentration of 60 CFU/ml. Negative controls were run for each sputum pool and at every time point. Samples were incubated at room temperature with SR at a 2:1 ratio for 15 min, 5 h, 8 h, 24 h, 3 days, and 7 days, and the Xpert MTB/RIF assay was performed in a GeneXpert system according to the manufacturer's instructions. Assay amplicons were assessed for mutations by DNA sequencing as described previously (C. C. Boehme, P. Nabeta, D. Hillemann, M. Nicol, S. Shennai, F. Krapp, J. Allen, R. Tahirli, R. Blakemore, R. Rustomjee, A. Milovic, M. Jones, S. M. O'Brien, D. H. Persing, S. Ruesch-Gerdes, E. Gotuzzo, C. Rodrigues, D. Alland, and M. D. Perkins, submitted for publication).
Statistical analysis.
Statistical analysis by t test was performed using Sigmaplot version 8.0 (Systat Software Inc., San Jose, CA), and Fisher's exact t test was performed using SAS version 9.1 (SAS, Cary, NC).
RESULTS
Validation of the air samplers.
We determined the limits of detection of our viable-aerosol air-sampling methods by nebulizing known concentrations of BCG CFU ranging from 104 CFU/ml down to 10 CFU/ml in 7H9 medium over 10 min. Cultivable aerosols were measured by sampling within the chamber using an Andersen impactor loaded with solid Middlebrook 7H10 medium plates and a BioSampler loaded with liquid Middlebrook 7H9 medium. As shown in Table 1, the system was able to detect less than 10 aerosolized CFU (Andersen impactor) or less than 100 CFU (BioSampler), demonstrating high sensitivity.
TABLE 1.
Ability to detect viable aerosols
| Concn of BCG (CFU/ml)a | CFU/m3 airb |
|
|---|---|---|
| Andersen impactor | BioSampler | |
| 104 | 712 ± 97.6 | 10,416 ± 2604 |
| 103 | 169 ± 13.8 | 2,976 ± 1704 |
| 102 | 91 ± 5.2 | 578 ± 501 |
| 10 | 28 ± 12.8 | 0 |
A total of 0.8 ml of mixture was aerosolized in each experiment.
Mean and standard deviation of three experiments.
Sample preparation.
We compared the number of viable aerosols that were generated by the process of loading 2 ml of SR-treated sputum into the Xpert MTB/RIF cartridge versus the number of aerosols that were generated when a loopful (10 μl) of untreated sputum was smeared on microscope slides until almost dry to make direct AFB smears. BCG added to unprocessed human sputum at a final concentration of 5 × 108 CFU/ml was used for these studies. We simulated the “incorrect use of SR” in the first study to assess what would occur if the tester did not incubate the sputum in SR for the recommended 15 min before loading the cartridge. In the next study, we simulated the manufacturer's recommended 15-min SR incubation before cartridge loading. We simulated direct AFB slide smearing in the third study. Each study protocol was performed three times on separate occasions, and viable-aerosol production was measured using both the Andersen impactor and the BioSampler. As shown in Table 2, incorrect SR usage produced a mean of 6 BCG CFU/m3 air as measured by the Andersen impactor or a mean of 67 BCG CFU/m3 air as measured by the BioSampler. Importantly, correct SR usage did not produce any measurable BCG CFU in any of the three experiments. In contrast, AFB smear preparation produced a mean of 16 BCG CFU/m3 air as measured by the Andersen impactor and a mean of 325 BCG CFU/m3 air as measured by the BioSampler.
TABLE 2.
Viable-aerosol generation during manual sample preparation
| Sputum preparation method | CFU/m3 airb |
|
|---|---|---|
| Andersen impactor | BioSampler | |
| SR added to sputum, then cartridge loaded without incubationa | 6 ± 5.5 | 67 ± 115.4c |
| SR added to sputum, then cartridge loaded after 15-min incubationa | 0 | 0 |
| Sputum smeared directly on microscope slides; no SR | 16 ± 2.3 | 325 ± 112.5 |
After SR addition, samples were pipetted in and out of three Xpert MTB/RIF cartridges (equivalent to loading >30 cartridges).
Mean and standard deviation of three experiments.
Two of the three replicates did not show any growth in the 7H9 medium.
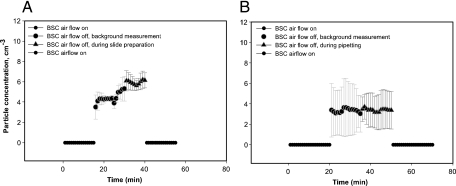
In parallel studies, we also measured the number of particles of any type generated by each procedure. As shown in Fig. 1 A, AFB slide preparation resulted in a small but measurable increase in the particle concentration from approximately 4.1 particles/cm3 air to approximately 5.8 particles/cm3 air. In contrast, the average particle number in the air of the hood was not changed during pipetting of the sputum-SR mixture (Fig. 1B), although the interexperiment variability was somewhat larger at each time point.
FIG. 1.
Particles generated during manual sample preparation. Total airborne particles were measured for AFB smear preparation (A) or pipetting and cartridge loading (B). Measurements were first taken either with the BSC airflow on or with the BSC airflow off and the sash closed as much as possible before and during each sample preparation procedure. Each time point represents the mean of three experiments. The error bars indicate ±1 standard deviation.
Automated sample processing.
We measured the viable aerosols generated by the GeneXpert instrument while processing Xpert MTB/RIF cartridges loaded with samples. Three studies were performed. In the first study, a final concentration of 5 × 108 BCG CFU/ml was made in water. Two milliliters of water was then placed directly into each Xpert MTB cartridge without SR treatment. SR was not used in this study so that a very high number of viable cells (many log units higher than could possibly be present in SR-treated sputum) would be processed by the GeneXpert instrument. In the second study, raw sputum was spiked with BCG at a final concentration of 5 × 108 CFU/ml sputum. One milliliter of sputum was then mixed with 2 ml of SR according to the manufacturer's recommendation, and then 2 ml of the mixture was placed into each Xpert MTB/RIF assay cartridge. In the final study, raw sputum was spiked with M. smegmatis instead of BCG at a final concentration of 7 × 108 CFU/ml sputum and then treated with a 2:1 ratio of SR. The sputum was then treated and loaded into the cartridge as in the second study. The GeneXpert instrument was loaded with three cartridges, and viable-aerosol production was measured using both the Andersen impactor and the BioSampler during the assay run. Each assay was run three times for a total of nine cartridges tested under each condition. We did not detect viable aerosols in any assay run in any of the three studies. These results suggest that infectious aerosols were not generated even when Xpert MTB/RIF cartridges were loaded with high numbers of live bacteria not treated with SR.
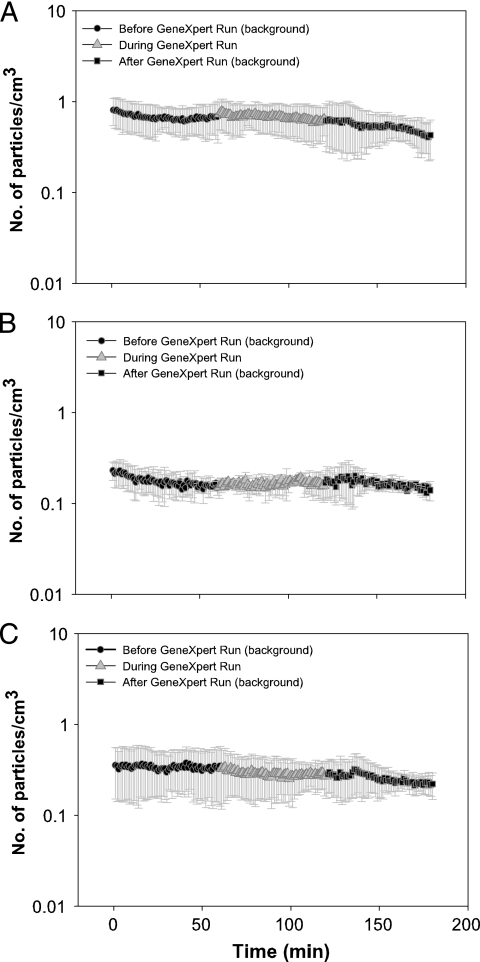
In parallel studies, we also measured the total numbers of all particle types generated by the GeneXpert as it processed cartridges loaded with each type of sample. As shown in Fig. 2 A to C, the number of particles generated when the GeneXpert processed cartridges containing samples did not increase compared to the number of particles generated when the GeneXpert was run without any cartridges loaded into the system. Together, these results suggest that the Xpert MTB/RIF assay is not associated with a measurable infection risk when performed properly.
FIG. 2.
Particles generated during automated sample processing. Total airborne particles were measured during the sample-processing portion of the Xpert MTB/RIF assay, which is performed automatically by the GeneXpert instrument. During each sample-processing run, the GeneXpert was loaded with three cartridges containing water spiked with 5 × 108 CFU/ml BCG (A), sputum spiked with 5 × 108 CFU/ml BCG and then treated with SR (B), or sputum spiked with 7 × 108 CFU/ml M. smegmatis and treated with SR (C). Measurements were first taken with the BSC airflow off and the sash closed as much as possible before and after or during each sample-processing run. Each time point represents the mean of three experiments. The error bars indicate ±1 standard deviation.
Inactivation study with clinical samples.
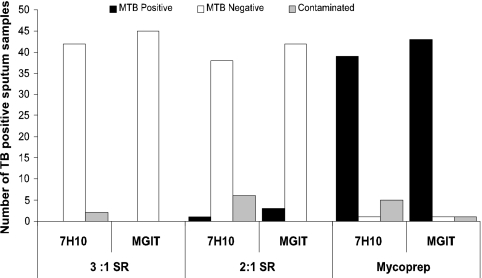
We tested the tuberculocidal activity of SR in clinical sputum samples from 45 highly smear-positive patients with confirmed pulmonary tuberculosis. Two different ratios were tested, the 2:1 SR sputum ratio recommended by the manufacturer and the 3:1 ratio that we have observed is commonly added by testers when visually estimating sputum volumes. None of the samples treated with the 3:1 ratio showed any positives for M. tuberculosis on either 7H10 or MGIT (Fig. 3). Two contaminants observed on 7H10 were confirmed negative by the Capilia TB Neo assay. Of the 45 samples treated with the 2:1 SR ratio, three were culture positive in MGIT and one was culture positive on 7H10. However, the TpD for the three MGIT-positive samples was 17.8 days, showing that only a very small number of bacteria remained viable (27). The single culture-positive 7H10 plate grew only nine M. tuberculosis colonies. Of the 45 sputum aliquots treated by the standard NALC-NaOH method and not treated with SR, 43/45 were culture positive for M. tuberculosis on either solid or liquid culture, with an average TpD culture in the MGIT system of only 6.2 ± 0.6 days. The control sputum aliquots from the three samples that were culture positive in MGIT after SR treatment had an average TpD culture of only 4.7 days when processed by the standard NaOH method, indicating very high numbers of CFU in the original sputa.
FIG. 3.
SR killing activity in strongly smear-positive clinical sputum samples from tuberculosis patients. Forty-five clinical tuberculosis sputum samples were decontaminated with either SR/sputum ratios of 2:1 or 3:1 or Mycoprep (BD, Sparks, MD) as indicated. The washed samples were then cultured on 7H10 agar plates or in MGITs. The number of sputum samples culture positive for M. tuberculosis (MTB), culture negative for M. tuberculosis, or contaminated by other bacteria is shown for each culture type.
SR stability studies.
Changes in the SR that might occur through prolonged storage at high or low temperatures could potentially impact its tuberculocidal activity. SR aliquots were stored at 4°C, 25°C, and 45°C under controlled conditions for at least 3 months and then tested for their sustained killing efficiencies. Killing studies demonstrated that all of the SR aliquots produced average killing of 6 to 7 log10 (Table 3). There was no statistical difference in killing activity among the different storage conditions.
TABLE 3.
Effect of SR storage temperature on M. bovis BCG killing
| SR storage temp (°C) | Storage time (mo) | Testing method | Avg log10reduction |
|---|---|---|---|
| 25 | 3 | Centrifugation/wash | 6.8 ± 0.9 |
| 45 | 3 | Centrifugation/wash | 6.7 ± 1.0 |
| 4 | >3 | Centrifugation/wash | 7.0 ± 1.1 |
| 45 | 9 | Filter/washa | >6 |
| 35 | 9 | Filter/wash | >6 |
| 25 | 18 | Filter/wash | >6 |
| 4 | 9 | Filter/wash | >6 |
Filter and wash tests were performed by an independent testing agency (Microbiotest, Sterling, VA).
Quantitative tuberculocidal test results.
An independent testing agency was contracted to repeat the SR killing studies following U.S. Environmental Protection Agency (EPA) guidelines (http://www.epa.gov/oppbead1/methods/atmpmethods/MB-16-00.pdf). The SR used in this study had been stored for 9 months at fixed temperatures of 4°C, 35°C, and 45°C and for 18 months at 25°C. The results confirmed that SR achieved at least a 6-log-unit kill when mixed 2:1 with sputum containing BCG under every tested storage condition (Table 3).
Effect of prolonged SR treatment.
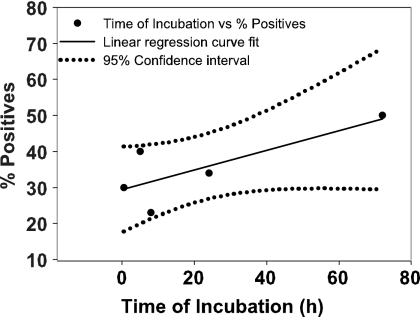
The manufacturer recommends incubating sputum samples with SR at a 2:1 SR-to-sample ratio for a minimum of 15 min but not more than 12 h (Xpert MTB/RIF product insert) before loading them into the Xpert MTB/RIF cartridge. Because the tuberculocidal effect of SR increases with the length of exposure (for at least 3 h) (14), prolonged SR treatment could be considered a way to ensure sterility of the samples used for testing and for sample transport. However, the effect of prolonged SR incubation on assay sensitivity had not been carefully studied. We examined the effect of prolonged SR treatment in this study. Tests were performed on sputum samples spiked with only 60 CFU/ml of M. tuberculosis to maximize the chance of detecting a minimal drop in assay sensitivity. M. tuberculosis was expected to be detected in <50% of the tests after 15 min of SR treatment at this sub-LOD target concentration (14). We detected M. tuberculosis in 13/59 (22%), 55/145 (38%), 9/38 (23%), and 41/118 (34%) test samples after 15 min (0 h), 5 h, 8 h, and 24 h of SR incubation (Fig. 4). SR incubation could be prolonged up to 72 h (3 days) without further decrease in M. tuberculosis detection by GeneXpert. Fisher's exact test showed no statistical difference in performance at different time points (P > 0.05). None of the samples stored for 7 days were positive for M. tuberculosis. We noted that 4/41 M. tuberculosis-positive samples identified after 24 h of SR incubation were detected as RIF resistant by the Xpert MTB/RIF assay. DNA sequencing of the three out of four amplicons extracted from the false RIF-resistant assay mixtures revealed that each contained a single nucleotide mutation at position 517 (CAG-TAG), 522 (TCG-TCA), or 523 (GGG-GAG). Interestingly, each mutation consisted of a C-to-T or G-to-A base change. This change is typical of mutations caused by NaOH (28, 29). Unfortunately, the fourth sample, which showed probe D and E delays in GeneXpert, could not be sequenced due to accidental loss of the sample. No false RIF-resistant assay results or mutations were identified when large numbers of samples were incubated with SR for periods from 15 min to 8 h. To evaluate the relevance of this finding for SR treatment periods closer to those recommended, 100 additional samples were spiked with 1,000 CFU/ml of BCG, treated by 5 h of SR incubation, and then tested with the Xpert MTB/RIF assay. None of the 100 positive assays generated a false RIF-resistant test result (data not shown).
FIG. 4.
SR stability study in sputa spiked with 60 CFU/ml of M. tuberculosis H37Rv. Sputum spiked with 60 CFU/ml M. tuberculosis was treated with a 2:1 SR-to-sample ratio and incubated for up to 7 days. Samples were tested by Xpert MTB/RIF assay at 15 min (n = 59), 5 h (n = 145), 8 h (n = 38), 24 h (n = 118), 3 days (n = 20), and 7 days (n = 11) (n represents the number of replicates run at each time point). The percentage of positives detected at each time point was plotted. Linear regression curve fitting (r2 = 0.6) was performed, and 95% upper and lower confidence intervals were determined using SigmaPlot 8.0. Fisher's t test at a P value of >0.05 showed no significant difference at different time points.
DISCUSSION
Tuberculosis is known to spread primarily through aerosols of live bacteria. Surface contact or dried fomites (e.g., linen, furniture, books, and floors) are not considered significant infection risks (1, 6, 8). To limit the risk to laboratory workers, tuberculosis tests must minimize the number of live bacteria that are manipulated by the tester and effectively control aerosolization of the bacteria that remain. Sample preparation methods such as centrifugation, vortexing, and vigorous shaking generate a significant amount of aerosols (12, 16, 20, 26) and are thus likely to present a biohazard in tuberculosis detection assays. The current study was performed to evaluate the safety of the Xpert MTB/RIF assay. We studied whether SR treatment could eliminate the biohazard risk by killing more than 6 log units of the M. tuberculosis present in sputum and whether the closed-cartridge sample-processing system used in the Xpert MTB/RIF assay could provide additional aerosol risk reduction. Our study used sensitive detection methods to demonstrate that no viable aerosols or additional particles were generated by either the manual or automated portion of the Xpert MTB/RIF assay. It is notable that even incorrect use of SR (where the 15-min incubation step was omitted) produced fewer cultivable aerosols than did smear preparation using untreated sputum inoculated with the same burden of AFB. Our results suggest that the Xpert MTB/RIF assay poses a smaller biohazard risk than the performance of a direct AFB smear. Any type of sputum manipulation, including AFB smear preparation, can generate potentially infectious aerosols from a sputum sample containing a sufficient number of bacteria.
Prior studies of SR had demonstrated outstanding tuberculocidal activity on sputum samples spiked with M. tuberculosis grown in culture (14). However, it was important to test SR activity in sputa from actual tuberculosis patients. All of the patient samples tested in the current study were highly AFB smear positive. Furthermore, the MGIT cultures of NALC-NaOH-treated samples were positive in an average of 6.2 days, indicating that the clinical samples had very high titers of live M. tuberculosis. SR treatment performed very well under these circumstances by effectively sterilizing almost all of the sputum samples. The few samples that remained culture positive after SR treatment initially contained very high bacterial titers (approximately 107 CFU/ml as estimated by MGIT time to positive) and were reduced by SR treatment to counts of viable M. tuberculosis CFU that were similar to those found in low-CFU smear-negative samples. The single sample that was positive on solid medium contained only nine viable colonies after SR treatment. The few liquid cultures that were positive after SR treatment required an average of 17.8 days to become positive in the MGIT system. This much delayed time-to-positive result indicates that only small quantities of AFB remained viable after SR treatment (27).
It was also important to evaluate the stability of SR stored under conditions that might exist outside the laboratory. We tested SR that had been subjected to 3 months of cold (4°C) or heat (45°C) stress, verifying that the tuberculocidal activity was not affected. These results were validated for even longer storage periods by an independent testing laboratory: there was no decrease in tuberculocidal activity even when the SR was stored at 25°C for 18 months or at 35°C or 45°C for 9 months (Table 3).
Our studies with prolonged incubation of sputum samples with SR suggest that sputum samples can be incubated in SR for at least 72 h (3 days) without a detectable decrease in Xpert assay sensitivity. Thus, some users may wish to prolong SR treatment to ensure complete sample sterility or to safely transport specimens to a central laboratory for testing even though prolonged SR incubation beyond 12 h is not recommended by the manufacturer. However, our results suggest that prolonged SR incubation should be used with caution, as it appears that at least paucibacillary samples cannot be incubated with SR for ≥24 h without increasing the risk of a false RIF resistance determination due to chemically induced mutations. DNA can be deaminated by endogenous and environmental agents (5, 9, 18, 19, 23, 25), including NaOH (28). This may result in postreplicative mutations, producing G-to-T and A-to-C changes, if not recovered by DNA repair systems (5, 9, 10). The falsely RIF-resistant amplicons produced by 24-h SR treatment contained exactly these mutations (in locations where they are not seen in clinically RIF-resistant M. tuberculosis strains), suggesting that false RIF resistance was caused by the NaOH component of the SR buffer. Therefore, samples stored for prolonged periods with SR should be retested if RIF resistance is observed.
In conclusion, the combination of an effective tuberculocidal SR and the closed configuration of the Xpert MTB/RIF cartridge effectively reduces the risk of infectious-aerosol formation to below that of AFB smear preparation. These features help to make the Xpert MTB/RIF assay suitable for near-patient detection of tuberculosis and drug resistance in settings where biocontainment facilities are not available.
Acknowledgments
This work was supported by National Institutes of Health grants R41-AI52523 and R42-AI52523 and by a grant from the Foundation for Innovative New Diagnostics.
D.A. is one of a group of coinvestigators who invented molecular beacon technology and who receive income from licensees, including a license to Cepheid for M. tuberculosis detection.
Footnotes
Published ahead of print on 18 August 2010.
REFERENCES
- 1.Ad Hoc Committee for the Guidelines for Preventing the Transmission of Tuberculosis in Canadian Health Care Facilities and Other Institutional Settings. 1996. Guidelines for preventing the transmission of tuberculosis in Canadian health care facilities and other institutional settings. Can. Commun. Dis. Rep. 22(Suppl. 1):i-iv, 1-55. [PubMed] [Google Scholar]
- 2.An, H. R., G. Mainelis, and M. Yao. 2004. Evaluation of a high-volume portable bioaerosol sampler in laboratory and field environments. Indoor Air 14:385-393. [DOI] [PubMed] [Google Scholar]
- 3.Association for the Advancement of Medical Instrumentation. 2003. Sterilization of health care products—requirements for products labeled “sterile.” Standard ANSI/AAMI ST67. Association for the Advancement of Medical Instrumentation, Arlington, VA.
- 4.Bonnet, M., A. Ramsay, W. Githui, L. Gagnidze, F. Varaine, and P. J. Guerin. 2008. Bleach sedimentation: an opportunity to optimize smear microscopy for tuberculosis diagnosis in settings of high prevalence of HIV. Clin. Infect. Dis. 46:1710-1716. [DOI] [PubMed] [Google Scholar]
- 5.Caulfield, J. L., J. S. Wishnok, and S. R. Tannenbaum. 1998. Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J. Biol. Chem. 273:12689-12695. [DOI] [PubMed] [Google Scholar]
- 6.CDC. 2005. Guidelines for preventing the transmission of Mycobacterium tuberculosis in health-care settings, 2005. MMWR Morbid. Mortal. Wkly. Rep. 54:1-141. [PubMed] [Google Scholar]
- 7.Chedore, P., C. Th'ng, D. H. Nolan, G. M. Churchwell, D. E. Sieffert, Y. M. Hale, and F. Jamieson. 2002. Method for inactivating and fixing unstained smear preparations of Mycobacterium tuberculosis for improved laboratory safety. J. Clin. Microbiol. 40:4077-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker, C. F. 2007. Tuberculosis and infection control measures. Dis. Mon. 53:55-58. [DOI] [PubMed] [Google Scholar]
- 9.Dong, L., R. Mi, R. A. Glass, J. N. Barry, and W. Cao. 2008. Repair of deaminated base damage by Schizosaccharomyces pombe thymine DNA glycosylase. DNA Repair 7:1962-1972. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, B. K., and J. H. Miller. 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287:560-561. [DOI] [PubMed] [Google Scholar]
- 11.Forbes, B. A., N. Banaiee, K. G. Beavis, B. A. Brown-Elliot, P. D. Latta, L. B. Elliott, G. S. Hall, B. Hanna, M. D. Perkins, S. H. Siddiqui, R. J. Wallace, and N. G. Warren (ed.). 2008. Laboratory detection and identification of mycobacteria. Approved guideline. M48-A, vol. 28. CLSI, Wayne, PA.
- 12.Ganoza, C. A., J. N. Ricaldi, J. Chauca, G. Rojas, C. Munayco, J. Agapito, J. C. Palomino, and H. Guerra. 2008. Novel hypertonic saline-sodium hydroxide (HS-SH) method for decontamination and concentration of sputum samples for Mycobacterium tuberculosis microscopy and culture. J. Med. Microbiol. 57:1094-1098. [DOI] [PubMed] [Google Scholar]
- 13.Grzybowski, S., G. D. Barnett, and K. Styblo. 1975. Contacts of cases of active pulmonary tuberculosis. Bull. Int. Union Tuberc. 50:90-106. [PubMed] [Google Scholar]
- 14.Helb, D., M. Jones, E. Story, C. Boehme, E. Wallace, K. Ho, J. Kop, M. R. Owens, R. Rodgers, P. Banada, H. Safi, R. Blakemore, N. T. N. Lan, E. C. Jones-Lopez, M. Levi, M. Burday, I. Ayakaka, R. D. Mugerwa, B. McMillan, E. Winn-Deen, L. Christel, P. Dailey, M. D. Perkins, D. H. Persing, and D. Alland. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, P. A., W. F. Todd, G. N. Davis, and P. V. Scarpio. 1992. Evaluation of eight bioaerosol samplers challenged with aerosols of free bacteria. Am. Ind. Hyg. Assoc. J. 53:660-667. [DOI] [PubMed] [Google Scholar]
- 16.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology. A guide for the level III laboratory. CDC, Atlanta, GA.
- 17.Kim, S. J., S. H. Lee, I. S. Kim, H. J. Kim, S. K. Kim, and H. L. Rieder. 2007. Risk of occupational tuberculosis in National Tuberculosis Programme laboratories in Korea. Int. J. Tuber. Lung Dis. 11:138-142. [PubMed] [Google Scholar]
- 18.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 19.Merchant, K., H. Chen, T. C. Gonzalez, L. K. Keefer, and B. R. Shaw. 1996. Deamination of single-stranded DNA cytosine residues in aerobic nitric oxide solution at micromolar total NO exposures. Chem. Res. Toxicol. 9:891-896. [DOI] [PubMed] [Google Scholar]
- 20.Morcillo, N., B. Imperiale, and J. C. Palomino. 2008. New simple decontamination method improves microscopic detection and culture of mycobacteria in clinical practice. Infect. Drug Resist. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rattan, A., K. Kishore, S. Singh, M. Jaber, I. Xess, and R. Kumar. 1994. Evaluation of a safe sputum processing method for detecting tuberculosis. J. Clin. Pathol. 47:411-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieder, H. L., A. V. Deun, K. M. Kam, S. J. Kim, T. M. Chonde, A. Trébucq, and R. Urbanczik. 2007. Priorities for tuberculosis bacteriology services in low-income countries, 2nd ed. International Union against Tuberculosis and Lung Disease, Paris, France.
- 23.Routledge, M. N., D. A. Wink, L. K. Keefer, and A. Dipple. 1994. DNA sequence changes induced by two nitric oxide donor drugs in the supF assay. Chem. Res. Toxicol. 7:628-632. [DOI] [PubMed] [Google Scholar]
- 24.Rutala, W. A., and D. J. Weber. 2001. New disinfection and sterilization methods. Emerg. Infect. Dis. 7:348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shapiro, R. 1981. Damage to DNA caused by hydrolysis, p. 3-18. In E. Seeberg and K. Kleppe (ed.), Chromosome damage and repair. Plenum Press, New York, NY.
- 26.Tomita, M., H. Takeno, S. Yoshida, K. Suzuki, and M. Sakatani. 2008. Comparison of BBL Mycoprep and 2% NaOH decontamination procedures for MGIT. Kekkaku 83:471-473. [PubMed] [Google Scholar]
- 27.Wallis, R. S., S. Patil, S. H. Cheon, K. Edmonds, M. Phillips, M. D. Perkins, M. Joloba, A. Namale, J. L. Johnson, L. Teixeira, R. Dietze, S. Siddiqi, R. D. Mugerwa, K. Eisenach, and J. J. Ellner. 1999. Drug tolerance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, P., and J. Ren. 2004. Separation of purine and pyrimidine bases by capillary electrophoresis using [beta]-cyclodextrin as an additive. J. Pharm. Biomed. Anal. 34:277-283. [DOI] [PubMed] [Google Scholar]
- 29.Wang, R. Y., K. C. Kuo, C. W. Gehrke, L. H. Huang, and M. Ehrlich. 1982. Heat- and alkali-induced deamination of 5-methylcytosine and cytosine residues in DNA. Biochim. Biophys. Acta 697:371-377. [DOI] [PubMed] [Google Scholar]