Abstract
High rates and transmission of multidrug-resistant (MDR) tuberculosis (TB) have been associated with the Mycobacterium tuberculosis complex (MTBC) Beijing lineage, pointing to the importance of pathogen genetic factors for the modulation of infection outcome and epidemiology. We present here an in-depth analysis of the population structure of MTBC strains from the Republic of Georgia, a high-incidence setting at the Black Sea Coast. Phylogenetic lineages were identified based on 24-locus MIRU-VNTR (for mycobacterial interspersed repetitive unit-variable number tandem repeat) and spoligotyping analysis. Clusters of strains with identical genotyping profiles were determined as an indicator for the rate of recent transmission. Among the 183 M. tuberculosis isolates investigated, the most prominent lineage found was Beijing (26%), followed by the LAM (18%), Ural (12%), and Haarlem (5%) strains. A closely related previously undefined phylogenetic group (62 strains) showed a genotyping pattern similar to laboratory strain H37RV and was denominated as “Georgia-H37RV-like.” Although isoniazid resistance was found among strains of different lineages, MDR TB was nearly completely restricted to Beijing strains (P < 0.0001). Approximately 50% of the isolates were grouped in clusters, indicating a high rate of recent transmission. Our data indicate that, in addition to the confirmation of the importance of Beijing genotype strains for the TB epidemiology in former Soviet Union countries, a high-population diversity with strains of the LAM, Ural, Haarlem, and a previously undefined lineage represents nearly two-thirds of the strains found in Georgia. Higher rates among previously treated and MDR TB patients point to a higher potential of lineage Beijing to escape therapy and develop MDR TB.
Drug-resistant Mycobacterium tuberculosis complex (MTBC) strains have emerged worldwide as a serious threat for tuberculosis (TB) control. Rates of multidrug-resistant (MDR) strains (i.e., resistance at least to isoniazid [INH] and rifampin [RIF]) have reached levels of up to 14% among patients never treated and up to 40% among previously treated patients in several MDR TB “hot spots” such as such as Karakalpakstan (Uzbekistan) and Kazakhstan in Eastern Europe (10, 37). Every year an estimated 489,000 cases of MDR TB arise globally (36). MDR TB is associated with much poorer treatment outcomes than for drug-susceptible TB, with a much higher risk of developing further resistances (2, 7). Prolonged periods of infectivity result in enhanced transmission of drug-resistant strains, further accelerating the rates of drug resistance (3). Even more worrisome is the emergence of a nearly untreatable form of TB, namely, extensively drug-resistant TB (XDR TB), which is defined as MDR plus additional resistance to any fluoroquinolone and at least one of three injectable drugs (i.e., amikacin, kanamycin, or capreomycin). A recent survey confirmed the worldwide presence of XDR strains, with rates of up 15% of MDR TB cases (30).
Considering the difficulties and problems associated with the treatment of resistant TB, high levels of MDR and XDR TB have the clear potential to jeopardize TB control on a local or national level. In addition to various measures for strengthening TB control such as rapid case detection, proper treatment, and rapid detection of drug resistance, the long-term effect of the emergence of drug-resistant strains on the worldwide TB epidemic also depends on the relative fitness of the MDR and XDR strains compared to susceptible strains (3).
In clinical MTBC strains, drug resistance results from chromosomal mutations in particular genes that confer resistance, which might also have an effect on bacterial fitness (5, 38). Although initial experiments indicated a lower fitness of e.g., INH-resistant strains, recent results confirm that the fitness of resistant strains depends on the kind of mutations, as well on the strain's genetic background (13). Furthermore, the initial adverse effects on bacterial fitness might be reversed by compensatory mutations occurring during long-term infection and ongoing transmission. In fact, MDR variants have been described to have even an enhanced fitness compared to susceptible progenitor strains (13). If these figures are used in models for prediction of the MDR TB epidemic, it turns out that even in the case of a well-functioning TB control system, small subpopulations of comparatively fit MDR clones might outcompete susceptible and less-fit resistant strains and become the dominant clones in future with dramatic consequences for TB treatment and control (5).
The most striking association between a mycobacterial genetic background and drug resistance documented thus far has been described for strains of the so-called Beijing lineage. These strains have been found to be involved in outbreaks and the transmission of MDR TB in several areas of the world (15). In Eastern Europe, a rising number of studies report a clear association between Beijing genotype infection and drug resistance (8, 11, 15, 28). Furthermore, large clusters of dominant clones have been determined that might indicate the development of “highly transmissible” MDR Beijing strains circulating in the community (28). Similar observations have been recently reported from South Africa, where a rapidly spreading highly resistant clone represents nearly half of all cases in the George subdistrict (35).
However, the overall picture of the correlation between bacterial genotype disease characteristics is incomplete. The majority of studies focused on particular strain types such as the Beijing genotype only, basically, because they are easy to recognize by applying genotyping techniques such as IS6110 DNA fingerprint and spoligotyping (34). Based on these markers, a variety of strains were not classifiable into phylogenetic lineages or clonal complexes since the genotyping information was not informative, e.g., due to homoplasy (4, 6). Therefore, the presence of particular genotypes might simply be overlooked and, consequently, the association with clinical characteristics could not be investigated or false associations have been obtained. Since this question is of scientific and public health relevance, further studies addressing the population structure of the MTBC applying more appropriate genetic markers are urgently needed. More recently, a new genotyping techniques based on mycobacterial interspersed repetitive unit-variable number tandem repeat (MIRU-VNTR) typing was developed that allows the simultaneous high-resolution discrimination of clinical isolates for epidemiological studies and a valid phylogenetic strain classification (26, 31).
In the present study, we used MIRU-VNTR typing and spoligotyping to investigate the population structure of strains obtained from patients living in the Republic of Georgia, where high rates of MDR TB have been recently reported (14, 20). We specifically analyzed the association between Beijing genotype and drug resistance. Furthermore, the data have been used to classify determine the whole variety of strains circulating in Georgia and to describe new clonal complexes and/or phylogenetic lineages. Phylogenetic strain classifications have been correlated with clinical characteristics.
MATERIALS AND METHODS
Laboratory methods.
Consecutive M. tuberculosis isolates recovered from patients referred to the National Center of Tuberculosis and Lung Diseases in Tbilisi, Republic of Georgia, between January and March 2006 were investigated. Collected clinical specimens were processed by standard methods and cultured on Lowenstein-Jensen medium for isolation of mycobacteria. Sample demographic and treatment information was also documented.
Susceptibility testing for antimycobacterial drugs was performed by the method of absolute concentration (23). The mycobacterial suspension was determined from the primary culture, and the turbidity was adjusted to 1 McFarland standard with sterile saline. A series of 10-fold dilutions were prepared, and 0.2 ml was inoculated onto media containing the following first-line TB drugs: streptomycin (SM; 4 μg/ml), RIF (40 μg/ml), and ethambutol (EBM; 2 μg/ml). The INH (0.2 μg/ml)-containing media were inoculated with 0.2 ml of a 100-fold dilution of the suspension. All inoculated sets were incubated at 37°C in an atmosphere of 5 to 10% CO2 for 28 days. MDR TB isolates were defined when they were resistant to INH and RIF (14).
DNA techniques.
A loopful of colony from the Lowenstein-Jensen medium was suspended in 200 μl of RNase-free water and boiled for 10 min as previously described (14). The suspension was centrifuged at 13,000 rpm for 10 min, and the supernatant was stored at −20°C until used.
All isolates were analyzed by the spoligotyping technique as described previously by Kamerbeek et al. (18a). For MIRU-VNTR genotyping, 24 loci were amplified by PCR as described previously (31). Briefly, analyses were performed by using multiplex PCRs, the Rox-labeled MapMarker 1000 size standard (BioVentures, Inc., Murfreesboro, VT), and the ABI 3130 XL sequencer with 16 capillaries (Applied Biosystems, Foster City, CA). Sizing of the PCR fragments and assignment of the various VNTR alleles were done by using customized GeneScan and Genotyper software packages (Applied Biosystems).
The molecular typing data were analyzed with the Bionumerics software (version 5.0; Applied Maths, Sint-Martens-Latem, Belgium) as instructed by the manufacturer. Similarities of spoligotyping and MIRU-VNTR patterns were calculated by using the categorical coefficient. A dendrogram was generated by using the unweighted pair group method with arithmetic averages (UPGMA). Minimum spanning tree analysis based on MIRU typing data was done by using the categorical coefficient. The priority rule was to link types first that had highest number of single-locus variants. Identification of MTBC genotypes was carried out by using the MIRU-VNTRplus database (1).
For the cluster analysis, a cluster was defined as a minimum of two strains harboring identical genotype pattern from different patients belonging to the study population.
Statistical analysis.
Categorical data were compared by the chi-square test or the Fisher exact test, when expected cell sizes (n) were smaller than 5. The Wilcoxon rank sum test was performed to determine whether the distribution of age as a continuous variable differed between the two groups. All tests were performed as two-sided tests. P values below 0.05 were considered statistically significant. Two models were then constructed in a logistic regression analysis with epidemiologically linked membership of a cluster and MDR as the respective outcome variables. Odds ratios (OR) and 95% confidence intervals (CI) were calculated by using logistic regression analysis, including demographic (age, sex, and ethnic group), epidemiologic (previous treatment, history of incarceration in a prison, and internal displacement), and microbiological variables (multiresistance, infection by a Beijing strain) in order to determine independent risk factors (e.g., adjusted for confounding). Statistical analyses were carried out by using SPSS software (version 15; SPSS, Inc., Chicago, IL).
RESULTS
The study sample comprising consecutive MTBC strains isolated from 196 patients was initially used to evaluate a new rapid test for the detection of drug resistance related mutations (14). We then utilized the available DNA preparations to carry out 24-locus MIRU-VNTR and spoligotyping analysis. For 13 strains, amplification was not successful, presumably due to degradation of the DNA. For the remaining 183 isolates, valid genotyping data were obtained and used for further analyses.
Among the final sample of 183 patients, the mean age ± the standard deviation was 39.1 ± 14.7 (range, 69 years), and most patients were male 151/183 (82.5%). Nearly all patients had a microscopically confirmed pulmonary TB (175 were sputum positive, 178 patients had pulmonary TB). Only 8.7% (16/183) patients were not of Georgian ethnicity.
Overall, 77 patients (42.1%) had a strain showing resistance against at least one drug tested. Thirty-nine strains (31.3%) were resistant to INH, sixteen (8.7%) were resistant to RIF, and thirteen (7.1%) were MDR (Table 1). Patients with MDR TB did not differ with respect to sex (not significant), nor with respect to age (multiresistant [39.9 ± 19.4] versus nonmultiresistant [39.1 ± 14.3; not significant), nor with respect to the proportion individuals with other ethnic groups (12/167 Georgians versus 1/16 non-Georgians) from non-MDR TB patients. As expected, previous treatment was significantly associated with MDR TB: 9/57 patients with a history of previous treatment were MDR, but only 4/126 patients without previous treatment (P = 0.004 [Fisher exact test]). On the contrary, a prison stay (2/26 versus 11/157; P > 0.05) was not associated with MDR TB.
TABLE 1.
Different categories of drug resistance stratified for MTBC genotypes
| Strain | Total no. | No. of strains (%)a |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| S | H | R | E | MDR | Cluster | PT | Male | ||
| Beijing | 46 | 17 (37) | 15 (33) | 11 (24) | 6 (13) | 10 (22) | 33 (72) | 20 (43) | 40 (87) |
| Haarlem | 10 | 3 (30) | 2 (20) | 0 | 0 | 0 | 2 (20) | 1 (10) | 6 (60) |
| Georgia-H37Rv-like | 62 | 30 (48) | 7 (11) | 2 (3) | 0 | 0 | 26 (42) | 15 (24) | 47 (76) |
| LAM | 34 | 6 (18) | 5 (15) | 0 | 0 | 0 | 21 (62) | 9 (26) | 28 (82) |
| Ural | 23 | 7 (30) | 7 (30) | 1 (4) | 0 | 1 (4) | 4 (17) | 8 (35) | 22 (96) |
| X-type | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| M. bovis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| No classification | 6 | 1 | 2 (33) | 2 (33) | 0 | 2 (33) | 0 | 2 (33) | 6 |
| All | 183 | 64 (35) | 39 (21) | 16 (9) | 6 (3) | 13 (7) | 86 (47) | 57 (31) | 151 (82) |
Abbreviations: H, isoniazid; R, rifampin; E, ethambutol; S, streptomycin; MDR, multidrug resistant; PT, previously treated.
According to the World Health Organization criteria, only 115 patients (62.8% of all patients) had been treated successfully (cured or completed). Although MDR patients had a lower treatment success rate (8 of the total of 13 patients were neither cured nor was treatment completed), in univariate analysis this association failed to reach statistical significance (8/68 without treatment success versus 5/115 with success; P < 0.076 [Fisher exact test]) due to the low total number of MDR strains.
Population structure and cluster analysis.
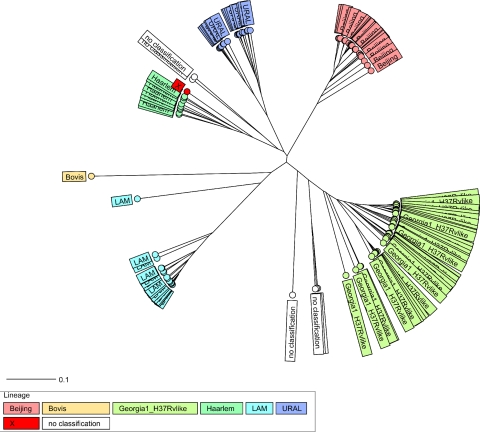
In a first step, 24-locus MIRU-VNTR profiles and spoligotyping patterns were used to classify the 183 strains from Georgia into main phylogenetic lineages by using the reference strain collection and identification tools available online at www.miru-vntrplus.org (1). Briefly, a stepwise identification procedure was carried out that based first on “simple match” analysis and second on phylogenetic tree identification (Fig. 1). In addition, SpoldB4 shared types were identified (4) and for each 24-locus MIRU pattern a unique MLVA 15-9 code was assigned by using the MIRU-VNTRplus nomenclature.
FIG. 1.
Radial UPGMA tree based on the copy numbers of 24 MIRU-VNTR loci (see Materials and Methods). The tree was calculated by using the MIRU-VNTRplus server. Abbreviations: LAM, M. tuberculosis Latin American Mediterranean.
Based on these analyses, 46 strains were classified as the M. tuberculosis Beijing genotype, 34 were classified as LAM the genotype (Latin American Mediterranean), 23 were classified as the Ural genotype, 10 were classified as the Haarlem genotype, 1 was classified as X-type, and 1 was classified as M. bovis (Fig. 1, Table 1). Interestingly, 62 strains were found to be closely related to laboratory strain H37Rv and appear to form a previously undefined branch that was denominated Georgia-H37RV-like. Only six strains could not be assigned to a known phylogenetic lineage or the new Georgia-H37Rv-like lineage (Table 1).
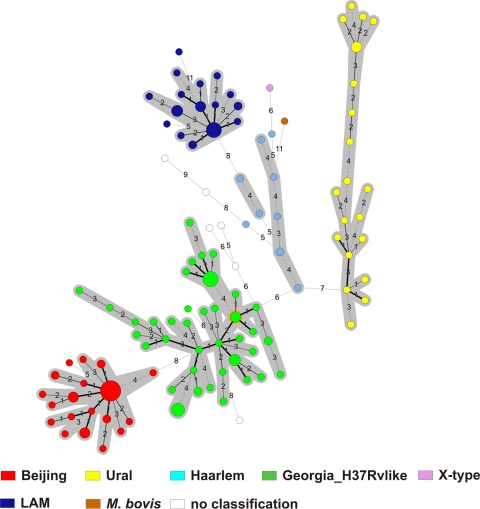
To confirm this strain classification, we calculated a minimum-spanning tree (MST) based on the 24-locus MIRU-VNTR data, which uses a maximum-parsimony algorithm to investigate phylogenetic relationships and to identify clonal complexes within a population. The MST (Fig. 2) strikingly confirmed the classification according to UPGMA tree-based analysis (Fig. 1) and by comparison with the reference database. All lineages suspected from dendrogram-based analysis were also detected as clonal complexes in the MST (Fig. 2), including the newly described Georgia-H37Rv-like genotype (Fig. 2). When the genetic diversity among isolates within one clonal complex is considered, it becomes obvious that Beijing and LAM strains are more closely related than strains of the other genotypes with the lowest intralineage average distances (0.10 and 0.13, respectively).
FIG. 2.
Minimum spanning tree based on the diversity of MIRU-VNTR data. The different complexes identified are shaded (maximum neighbor distance: four changes; minimum size: two MIRU-VNTR types) by the set of 24 loci among the 183 MTBC strains analyzed. The size of each circle is proportional to the number of MIRU-VNTR types belonging to a particular complex. LAM, M. tuberculosis Latin American Mediterranean.
This is also reflected by the cluster analysis. Overall, 86 of the 183 strains (47%) shared a genotyping pattern with at least one other isolate and were grouped in 22 clusters (shared types) ranging in size from 2 to 19 strains (see Fig. S1 in the supplemental material). The remaining strains were discriminated into 97 orphan types, all of which were assigned to MLVA MtbC15-9 types and SpolDB4 nomenclature if possible (see Fig. S1 in the supplemental material). The highest cluster rates were observed among Beijing and LAM strains (Table 1).
Among 23 patients of the 86 patients in clusters (26.7%) epidemiological links could be verified. Parameters such as being an inhabitant of the same region or treated in the same hospital without any additional known personal contact were not considered to establish an epidemiological link. The largest cluster (n = 19; cluster 19: MLVA MtbC15-9 type 94-32) is formed by a Beijing strain, followed by the second largest cluster formed by a strains of the Georgia-H37Rv-like genotype (n = 9, cluster 5: MLVA MtbC15-9 type 769-15), which might indicate ongoing transmission of these strains. Overall, patients with a Beijing strain were more often cluster members (33/86 in clusters versus 13/97 not in clusters; P < 0.0001), and they were often obtained from patients with verified epidemiological links within clusters: 15/23 patients with epidemiological link in clusters versus 18/63 patients without epilinks in clusters (P = 0.002). The risk of Beijing TB patients to be in a cluster with verified recent transmission within the very short study period was nearly 5-fold higher among Beijing patients (OR = 4.98; 95% CI = 2.4 to 17.1) and >4-fold higher among prison inmates (OR = 4.3; 95% CI = 1.2 to 27.3), both values being independent predictors for recent transmission. Multidrug resistance itself was no predictor for clustering (P > 0.05).
These data focused our interest to a more detailed analysis of characteristics of patients with Beijing strain infection. They did not differ with respect to sex, nor with respect to age 36.7 ± 15.4 versus 39.9 ± 14.3, nor with respect to patients with any drug resistance (19/77 [24.7%] with versus 27/106 [25.5%] without any resistance) from patients with non-Beijing strain infection. However, Beijing strains were more often spread in the native population (Georgians) than in the few foreign patients of other ethnicities (45/167 [27%] natives versus 1/16 [6%] non-Georgians); however, this does not reach statistical significance due to the small numbers (P = 0.076).
Among patients with previous treatment there were more patients with Beijing strains (20 Beijing among 57 previously treated individuals versus 26 Beijing among 126 not previously treated individuals; P = 0.037). Interestingly, there were more Beijing strain infections among patients with a prison history than among patients without any history of incarceration in a prison (15/26 versus 31/157; P < 0.0001), possibly indicating a better transmission of Beijing strains under crowded conditions. This could be confirmed by the fact the most patients in the largest cluster (19) have been in the same prison TB colony.
Factors associated with MDR TB.
If the distribution of different categories of drug resistance stratified for strains phylogenetic lineage is considered (Table 1), it becomes obvious that although strains of all lineages reach higher levels of INH or SM resistance (Table 1), the appearance of MDR is nearly completely restricted to Beijing strains. Accordingly, the MDR risk was 14-fold higher among Beijing patients (OR = 14.0, 95% CI = 3.1 to 63.0). Among several factors analyzed, only Beijing genotype infection and a history of previous treatment remain independently associated with MDR in multilogistic regression procedures.
DISCUSSION
In this study we present the first in-depth analysis of the population structure of MTBC strains from a high-incidence region in the Caucasus that is based on high-resolution 24-locus MIRU-VNTR typing. Our data confirm a highly diverse population structure that comprises, in addition to Beijing, four main phylogenetic lineages, one of which was not described before. Although these data indicate that TB epidemics in Eastern Europe might be more complex and not only driven by strains of the Beijing lineage, we also documented a clear association between Beijing strain infection and MDR TB. Furthermore, we present evidence of a clear association between the Beijing genotype and recent transmission, as well as a prison stay.
Only few in-depth analyses of the population structure of MTBC strains from high-incidence regions in Eastern Europe are available. Many investigations are limited by sample size, available epidemiological information, and/or by the genotyping technique used. The studies based on IS6110 DNA fingerprinting or spoligotyping described here are hampered by the inherent restrictions of both methods that allow the identification of some genotypes but fail to perform a valid phylogenetic classification of a large proportion of isolates in a certain area due to high rates of homoplasy, especially in the case of spoligotyping (6). This has also led to focused analysis of particular phylogenetic lineages such as Beijing or Haarlem, since these are easy to identify by spoligotyping.
Here, 24-locus MIRU-VNTR typing offers a clear advantage by providing high-resolution genotyping and valid phylogenetic classification, as well as screening for new phylogenetic lineages/clonal complexes (1). Furthermore, it allows for taking advantage from state-of-the-art analysis tools and the reference strain database available online on the MIRU-VNTRplus server that we have established previously (1). Scientific communication and tracking of particular strain types is facilitated by the implemented nomenclature server that assigns unique MLVA 15-9 codes to particular genotyping profiles. In addition to tree-based analyses, MIRU-VNTR typing data can also be used for generating parsimonious phylogenetic networks, e.g., minimum spanning trees (16).
Using these tools, we were able to classify nearly all strains investigated (97%) to MTBC lineages described before and one lineage that was discovered in our study. In concordance with a recently published study from Abkhazia, Beijing strains are not as dominant as in other parts of the former Soviet Union such as Uzbekistan, Turkmenistan, or several parts of Russia (9, 12, 22, 27). This might be due to the presence of strains of other genotype families, such as LAM or Ural, that were also reported from other Eastern European countries (17, 19, 24). In addition, we confirmed the presence of a previously unknown phylogenetic lineage named Georgia-H37Rv-like that was clearly defined by tree based, as well as by maximum-parsimony MST-based analysis. However, comparison with other studies is hampered by the fact that they are mainly based on IS6110 DNA fingerprint and spoligotyping analysis hindering a valid analysis of the population structure. Only a few studies, such as work by Moukrosov et al. (25) or Kovalev et al. (19) used a combination of MIRU-VNTR and spoligotyping, although with a reduced set of 12 MIRU-VNTR loci only. As such, the actual picture of MTBC population diversity in Eastern European high-incidence settings is largely incomplete and needs a systematic investigation with phylogenetic useful genotyping methods. Accordingly, we anticipate a larger study based on 24-locus MIRU-VNTR typing that shall comprise representative strain sets for several geographical settings in Eastern Europe.
A first phylogenetic network of Beijing strains based on 11 MIRU-VNTR loci was recently published by Mokrousov (21). When we compare the 11-locus pattern of our the Beijing strains of the present study to the reference patterns, it turned out that the main Beijing strains type in Georgia belonged to the M2 clone that is highly prevalent in the Russian populations. Together with LAM, the M2 strains from Georgia showed the lowest intralineage distances and were rather homogeneous. This argues for a more recent introduction of both lineages in Georgia (presumably from Russia) that might be followed by recent dissemination.
Interestingly, Beijing strains were found to be associated with recent transmission, especially in a prison setting. The largest cluster formed by a Beijing clone (MtbC15-9 type 94-32) could be related to an outbreak in a prison colony with, however, a spillover in the general population. This finding illustrates the high risk of accelerated TB transmission in the prison setting. Similar findings have been reported previously, e.g., from Samara (12) and Kyrgyztan (25), where Beijing strain infection was also found to be strongly associated with a history of imprisonment. Direct comparison of MIRU-VNTR profiles revealed a striking similarity of Beijing strains (type M2) spreading in Kyrgyzian and Georgian prison populations, also pointing to the particular propensity of Beijing strains to spread in a prison environment or in crowded living conditions in general. Considering prison systems and long-term hospitalization during TB treatment practiced in several Eastern European countries (32), this is likely to provide a selective advantage that might have contributed to the high rates of Beijing strains observed throughout former Soviet Union countries.
A further point of consideration is the strong association of Beijing strain infection with a history of previous treatment and MDR TB determined in the present study. This finding is in accordance with reports from our group, e.g., from Abkhazia, Turkmenistan, or Uzbekistan, or from other investigators, e.g., from Samara or Archangel Oblast (8, 12, 28, 32, 33). Taken together, these studies undoubtedly confirm Beijing strains as a major factor driving the MDR TB epidemic in Eastern Europe. The reasons for this phenomenon, however, are not yet well defined. Considering several reports that demonstrated recent transmission and outbreaks of MDR Beijing strains (8, 28), one key point appears to be the capability to spread after having MDR developed. Hence, fitness costs of MDR (and XDR) development in Beijing strains might be less than in non-Beijing strains and/or compensatory evolution might be more effective. A remarkable finding is the fact that the strong clonal expansion of some dominant MDR strains, e.g., documented in Abkhazia (28) or South Africa (18), has the potential to significantly influence the MDR epidemic in a certain region. These data also indicate that actual conditions in high-incidence settings, e.g., treatment policies, favor the selection of highly transmissible MDR strain variants that somehow have evolved unique pathogenic properties favoring efficient transmission.
The fact that in the present study strains of all major lineages showed higher levels of INH resistance, but only Beijing strains appear to progress efficiently to MDR TB, points to a higher capacity of Beijing strains to escape TB treatment in the case of underlying monoresistance (or in combination with SM). However, the proof of this hypothesis is difficult since longitudinal population-based studies addressing treatment success are necessary. In addition to valid documentation of patient data, e.g., treatment regimens, it is also mandatory to perform molecular typing of follow up culture since exogenous reinfection with MDR or even XDR strains is a frequent cause of treatment failure in high-incidence settings (9, 11). In two recent studies, higher failure rates among patients infected with Beijing strains were documented in Central Asia (9, 11) and Indonesia (29). The question of whether Beijing genotype infection per se can be seen as an independent risk factor for treatment failure and might require special treatment procedures is challenging and urgently requires further investigation.
In conclusion, our study confirms that the Beijing epidemic has reached Georgia and is associated with a high rate of recent transmission, especially in prison populations, and high MDR rates. On the other hand, the highly diverse population structure determined confirms the importance of strains of other genotypes, such as LAM and Ural, and the presence of as-yet-undefined phylogenetic lineages. Accordingly, the population structure of MTBC strains from high-incidence settings in Eastern Europe needs a more thorough investigation based on valid phylogenetic markers. To obtain a more detailed picture of the landscape of MTBC diversity in Eastern Europe, a concerted action of different research groups based on centralized databases and nomenclature, such as are available on the MIRU-VNTRplus server, is desirable. Based on this, more detailed investigations on genome diversity among strains of the different lineages and pathobiological consequences can be initiated.
Supplementary Material
Footnotes
Published ahead of print on 11 August 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Allix-Béguec, C., D. Harmsen, T. Weniger, P. Supply, and S. Niemann. 2008. Evaluation and user-strategy of MIRU-VNTRplus, a multifunctional database for on-line analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 46:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aziz, M. A., A. Wright, A. Laszlo, A. De Muynck, F. Portaels, A. Van Deun, C. Wells, P. Nunn, L. Blanc, and M. Raviglione. 2006. Epidemiology of antituberculosis drug resistance (the Global Project on Anti-Tuberculosis Drug Resistance Surveillance): an updated analysis. Lancet 368:2142-2154. [DOI] [PubMed] [Google Scholar]
- 3.Blower, S. M., and T. Chou. 2004. Modeling the emergence of the “hot zones”: tuberculosis and the amplification dynamics of drug resistance. Nat. Med. 10:1111-1116. [DOI] [PubMed] [Google Scholar]
- 4.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC. Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, T., and M. Murray. 2004. Modeling epidemics of multidrug-resistant Mycobacterium tuberculosis of heterogeneous fitness. Nat. Med. 10:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comas, I., S. Homolka, S. Niemann, and S. Gagneux. 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, H. S., S. Kalon, S. Allamuratova, V. Sizaire, Z. N. Tigay, S. Rusch-Gerdes, H. A. Karimovich, Y. Kebede, and C. Mills. 2007. Multidrug-resistant tuberculosis treatment outcomes in Karakalpakstan, Uzbekistan: treatment complexity and XDR-TB among treatment failures. PLoS One 2:e1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, H. S., T. Kubica, D. Doshetov, Y. Kebede, S. Rusch-Gerdess, and S. Niemann. 2005. The Beijing genotype and drug-resistant tuberculosis in the Aral Sea region of Central Asia. Respir. Res. 6:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, H. S., S. Niemann, G. Ismailov, D. Doshetov, J. D. Orozco, L. Blok, S. Rusch-Gerdes, and Y. Kebede. 2007. Risk of acquired drug resistance during short-course directly observed treatment of tuberculosis in an area with high levels of drug resistance. Clin. Infect. Dis. 44:1421-1427. [DOI] [PubMed] [Google Scholar]
- 10.Cox, H. S., J. D. Orozco, R. Male, S. Ruesch-Gerdes, D. Falzon, I. Small, D. Doshetov, Y. Kebede, and M. Aziz. 2004. Multidrug-resistant tuberculosis in central Asia. Emerg. Infect. Dis. 10:865-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, H. S., K. Sibilia, S. Feuerriegel, S. Kalon, J. Polonsky, A. K. Khamraev, S. Rusch-Gerdes, C. Mills, and S. Niemann. 2008. Emergence of extensive drug resistance during treatment for multidrug-resistant tuberculosis. N. Engl. J. Med. 359:2398-2400. [DOI] [PubMed] [Google Scholar]
- 12.Drobniewski, F., Y. Balabanova, V. Nikolayevsky, M. Ruddy, S. Kuznetzov, S. Zakharova, A. Melentyev, and I. Fedorin. 2005. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA 293:2726-2731. [DOI] [PubMed] [Google Scholar]
- 13.Gagneux, S., C. D. Long, P. M. Small, T. Van, G. K. Schoolnik, and B. J. Bohannan. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944-1946. [DOI] [PubMed] [Google Scholar]
- 14.Gegia, M., N. Mdivani, R. E. Mendes, H. Li, M. Akhalaia, J. Han, G. Khechinashvili, and Y. W. Tang. 2008. Prevalence of and molecular basis for tuberculosis drug resistance in the Republic of Georgia: validation of a QIAplex system for detection of drug resistance-related mutations. Antimicrob. Agents Chemother. 52:725-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homolka, S., E. Post, B. Oberhauser, A. G. George, L. Westman, F. Dafae, S. Rusch-Gerdes, and S. Niemann. 2008. High genetic diversity among Mycobacterium tuberculosis complex strains from Sierra Leone. BMC Microbiol. 8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ignatova, A., S. Dubiley, V. Stepanshina, and I. Shemyakin. 2006. Predominance of multidrug-resistant LAM and Beijing family strains among Mycobacterium tuberculosis isolates recovered from prison inmates in Tula Region, Russia. J. Med. Microbiol. 55:1413-1418. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, R., R. M. Warren, G. D. van der Spuy, N. C. Gey van Pittius, D. Theron, E. M. Streicher, M. Bosman, G. J. Coetzee, P. D. van Helden, and T. C. Victor. 2010. Drug-resistant tuberculosis epidemic in the Western Cape driven by a virulent Beijing genotype strain. Int. J. Tuberc. Lung Dis. 14:119-121. [PubMed] [Google Scholar]
- 18a.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovalev, S. Y., E. Y. Kamaev, M. A. Kravchenko, N. E. Kurepina, and S. N. Skorniakov. 2005. Genetic analysis of mycobacterium tuberculosis strains isolated in Ural region, Russian Federation, by MIRU-VNTR genotyping. Int. J. Tuberc. Lung Dis. 9:746-752. [PubMed] [Google Scholar]
- 20.Lomtadze, N., R. Aspindzelashvili, M. Janjgava, V. Mirtskhulava, A. Wright, H. M. Blumberg, and A. Salakaia. 2009. Prevalence and risk factors for multidrug-resistant tuberculosis in the Republic of Georgia: a population-based study. Int. J. Tuberc. Lung Dis. 13:68-73. [PMC free article] [PubMed] [Google Scholar]
- 21.Mokrousov, I. 2008. Genetic geography of Mycobacterium tuberculosis Beijing genotype: a multifacet mirror of human history? Infect. Genet. Evol. 8:777-785. [DOI] [PubMed] [Google Scholar]
- 22.Mokrousov, I., O. Narvskaya, A. Vyazovaya, J. Millet, T. Otten, B. Vishnevsky, and N. Rastogi. 2008. Mycobacterium tuberculosis Beijing genotype in Russia: in search of informative variable-number tandem-repeat loci. J. Clin. Microbiol. 46:3576-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mokrousov, I., T. Otten, B. Vyshnevskiy, and O. Narvskaya. 2002. Detection of embB306 mutations in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis from Northwestern Russia: implications for genotypic resistance testing. J. Clin. Microbiol. 40:3810-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mokrousov, I., T. Otten, T. Zozio, E. Turkin, V. Nazemtseva, A. Sheremet, B. Vishnevsky, O. Narvskaya, and N. Rastogi. 2009. At Baltic crossroads: a molecular snapshot of Mycobacterium tuberculosis population diversity in Kaliningrad, Russia. FEMS Immunol. Med. Microbiol. 55:13-22. [DOI] [PubMed] [Google Scholar]
- 25.Mokrousov, I., V. Valcheva, N. Sovhozova, A. Aldashev, N. Rastogi, and J. Isakova. 2009. Penitentiary population of Mycobacterium tuberculosis in Kyrgyzstan: exceptionally high prevalence of the Beijing genotype and its Russia-specific subtype. Infect. Genet. Evol. 9:1400-1405. [DOI] [PubMed] [Google Scholar]
- 26.Oelemann, M. C., R. Diel, V. Vatin, W. Haas, S. Rusch-Gerdes, C. Locht, S. Niemann, and P. Supply. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J. Clin. Microbiol. 45:691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardini, M., E. Iona, F. Varaine, H. Karakozian, H. Arzumanian, L. Brunori, G. Orefici, and L. Fattorini. 2005. Mycobacterium tuberculosis drug resistance, Abkhazia. Emerg. Infect. Dis. 11:501-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardini, M., S. Niemann, F. Varaine, E. Iona, F. Meacci, G. Orru, H. Yesilkaya, T. Jarosz, P. Andrew, M. Barer, F. Checchi, H. Rinder, G. Orefici, S. Rusch-Gerdes, L. Fattorini, M. R. Oggioni, and M. Bonnet. 2009. Characteristics of drug-resistant tuberculosis in Abkhazia (Georgia), a high-prevalence area in Eastern Europe. Tuberculosis 89:317-324. [DOI] [PubMed] [Google Scholar]
- 29.Parwati, I., B. Alisjahbana, L. Apriani, R. D. Soetikno, T. H. Ottenhoff, A. G. van der Zanden, J. van der Meer, D. van Soolingen, and R. van Crevel. 2010. Mycobacterium tuberculosis Beijing genotype is an independent risk factor for tuberculosis treatment failure in Indonesia. J. Infect. Dis. 201:553-557. [DOI] [PubMed] [Google Scholar]
- 30.Shah, N. S., A. Wright, G. H. Bai, L. Barrera, F. Boulahbal, N. Martin-Casabona, F. Drobniewski, C. Gilpin, M. Havelkova, R. Lepe, R. Lumb, B. Metchock, F. Portaels, M. F. Rodrigues, S. Rusch-Gerdes, A. Van Deun, V. Vincent, K. Laserson, C. Wells, and J. P. Cegielski. 2007. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg. Infect. Dis. 13:380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toungoussova, O. S., G. Bjune, and D. A. Caugant. 2006. Epidemic of tuberculosis in the former Soviet Union: social and biological reasons. Tuberculosis 86:1-10. [DOI] [PubMed] [Google Scholar]
- 33.Toungoussova, O. S., A. Mariandyshev, G. Bjune, P. Sandven, and D. A. Caugant. 2003. Molecular epidemiology and drug resistance of Mycobacterium tuberculosis isolates in the Archangel prison in Russia: predominance of the W-Beijing clone family. Clin. Infect. Dis. 37:665-672. [DOI] [PubMed] [Google Scholar]
- 34.van Soolingen, D., K. Kremer, and E. Vynycky. 2003. New perspectives in the molecular epidemiology of tuberculosis, p. 17-45. In S. Kaufmann and H. Hahn (ed.), Mycobacteria and TB. Karger, Berlin, Germany.
- 35.Victor, T. C., E. M. Streicher, C. Kewley, A. M. Jordaan, G. D. van der Spuy, M. Bosman, H. Louw, M. Murray, D. Young, P. D. van Helden, and R. M. Warren. 2007. Spread of an emerging Mycobacterium tuberculosis drug-resistant strain in the western Cape of South Africa. Int. J. Tuberc. Lung Dis. 11:195-201. [PubMed] [Google Scholar]
- 36.World Health Organization. 2008. Anti-tuberculosis drug resistance in the world. Report 4. World Health Organization, Geneva, Switzerland.
- 37.Wright, A., M. Zignol, D. A. Van, D. Falzon, S. R. Gerdes, K. Feldman, S. Hoffner, F. Drobniewski, L. Barrera, S. D. van, F. Boulabhal, C. N. Paramasivan, K. M. Kam, S. Mitarai, P. Nunn, and M. Raviglione. 2009. Epidemiology of antituberculosis drug resistance 2002-07: an updated analysis of the Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Lancet 373:1861-1873. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Y., and W. R., Jr. Jacobs. 2008. Mechanisms of drug action, drug resistance, and drug tolerance in Mycobacterium tuberculosis: expected phenotypes from evolutionary pressure from a highly successful pathogen, p. 323-378. In S. H. Kaufmann and E. Rubin (ed.), Handbook of tuberculosis. Wiley-VCH Verlag, Weinheim, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.