Abstract
The reference standard methods for drug susceptibility testing of Mycobacterium tuberculosis, such as culture on Lowenstein-Jensen or Middlebrook 7H10/11 medium, are very slow to give results; and due to the emergence of multidrug-resistant M. tuberculosis and extensively drug-resistant M. tuberculosis, there is an urgent demand for new, rapid, and accurate drug susceptibility testing methods. PCR-single-strand conformational polymorphism (PCR-SSCP) analysis has been proposed as a rapid method for the detection of resistance to rifampin, but its accuracy has not been systematically evaluated. We performed a systematic review and meta-analysis to evaluate the accuracy of PCR-SSCP analysis for the detection of rifampin-resistant tuberculosis. We searched the Medline, Embase, Web of Science, BIOSIS, and LILACS databases and contacted authors if additional information was required. Ten studies met our inclusion criteria for rifampin resistance detection. We applied the summary receiver operating characteristic (SROC) curve to perform the meta-analysis and to summarize diagnostic accuracy. The sensitivity of PCR-SSCP analysis for the rapid detection of rifampin-resistant tuberculosis was 0.79 (95% confidence interval [CI], 0.75 to 0.82), the specificity was 0.96 (95% CI, 0.94 to 0.98), the positive likelihood ratio was 16.10 (95% CI, 5.87 to 44.13), the negative likelihood ratio was 0.20 (95% CI, 0.10 to 0.40), and the diagnostic odds ratio was 100.93 (95% CI, 31.95 to 318.83). PCR-SSCP analysis is a sensitive and specific test for the rapid detection of rifampin-resistant M. tuberculosis. Additional studies in countries with a high prevalence of multidrug-resistant M. tuberculosis and also cost-effectiveness analysis are required in order to obtain a complete picture on the utility of this method for rapid drug resistance detection in M. tuberculosis.
Tuberculosis (TB) is a major global health problem. Early detection of drug resistance in patients with tuberculosis allows the use of appropriate treatment regimens for the patient, which has an important impact for the better control of the disease. The development of rapid methods for drug susceptibility testing is very important due to the increasing rates of multidrug-resistant tuberculosis (MDR-TB) worldwide and the recently described extensively drug-resistant tuberculosis (XDR-TB). The World Health Organization (WHO) urgently called for expanded access to culture and drug susceptibility testing in response to the spread of MDR-TB and XDR-TB, declared serious emerging threats to public health (28). This poses significant challenges for TB laboratory capacity and the need for faster drug susceptibility testing methods.
Conventional culture methods using egg- or agar-based media are still the most commonly used approaches in many countries. To test for drug resistance, the standard methods using Lowenstein-Jensen (LJ) medium include the proportion method, the absolute concentration method, and the resistant ratio method, which are well standardized with clinical isolates, at least for the major antituberculosis drugs. The proportion method was developed in the 1960s and is still the “gold standard” method used in many laboratories, especially in developing countries, because it is an inexpensive method easily accessible in these settings (1, 2). In recent years, due to the long turnaround times of conventional drug susceptibility testing methods, several new approaches have been proposed for the faster detection of MDR Mycobacterium tuberculosis, including both genotypic and phenotypic methods (23, 24). Among the genotypic methods proposed, PCR-single-strand conformational polymorphism (PCR-SSCP) analysis has been an important tool for detecting point mutations underlying genetic diseases since it was established in 1989 (9, 21). Under nondenaturing conditions, single-stranded DNA (ssDNA) has a folded structure that is determined by intramolecular interactions and, therefore, by its sequence. Even a single point mutation can alter the conformation of ssDNA, so that the altered conformation affects the migration of ssDNA, which can be detected as abnormal bands on a nondenaturing gel. Thus, SSCP analysis has a high resolving power to distinguish most conformational changes caused by subtle sequence differences even in a several-hundred-base fragment (9). The PCR-SSCP assay is relatively simple and was initially promising. Recently, the PCR-SSCP assay has been used in some studies for the rapid detection of rifampin-resistant M. tuberculosis (3, 11, 12, 17). However, individually, these trials found that the sensitivity and specificity were statistically inconsistent. The purpose of the present study was to perform a systematic review and meta-analysis to synthesize all available literature on the PCR-SSCP assay for the rapid detection of rifampin-resistant M. tuberculosis and to evaluate the overall accuracy of this method for the detection of rifampin resistance in isolates and in sputum samples.
MATERIALS AND METHODS
Search strategy and study selection.
We searched the following electronic databases: Medline (from 1966 to 2010), Embase (from 1980 to 2010), Web of Science (from 1990 to 2010), BIOSIS (from 1994 to 2010), and LILACS (from 1980 to 2010). All searches were up to date as of March 2010. The search terms used included PCR, polymerase chain reaction, PCR-SSCP, PCR-single-strand conformation polymorphism, tuberculosis, Mycobacterium tuberculosis, drug susceptibility, drug resistance, diagnosis, rifampin, and rifampicin. All titles and abstracts retrieved were scrutinized for relevant studies on detection of drug resistance in M. tuberculosis using the PCR-SSCP assay.
The search through the electronic databases returned studies that used the PCR-SSCP assay for rapid drug susceptibility testing of M. tuberculosis. We identified results from all primary studies evaluating the accuracy (sensitivity and specificity) of the PCR-SSCP assay for the rapid detection of rifampin resistance in M. tuberculosis isolates or in sputum samples. We included studies that met the following predetermined criteria: studies that compared the PCR-SSCP assay with a reference standard method (including the proportion method, absolute concentration method, or radiometric Bactec 460-TB method); studies that reported on the detection of rifampin resistance; and studies that reported data on false-positive, true-positive, false-negative, and true-negative results. Our initial search had no language restrictions, but studies not available in the English language were excluded from the data extraction process.
Data extraction.
Two independent reviewers (H.-B.X. and R.-H.J.) examined the titles and abstracts of all studies identified to confirm that they had fulfilled the inclusion criteria defined above. Titles and abstracts were first read independently by the two reviewers, and then all papers considered possibly eligible were reviewed independently by the authors, who assessed whether the paper was concerned with the PCR-SSCP assay for drug susceptibility testing. The bibliographies of selected articles were screened for potentially suitable references, which were then retrieved. Those studies that did not match with our requirements were taken out. Data from each article were extracted by one reviewer, and a sample of these was assessed by a second reviewer to check the accuracy of data extraction. The articles were examined in detail, and any disagreement was resolved by consensus with a third author. We classified data according to the following parameters included in Table 1 : the reference standard method used, type of sample (isolates or sputum), the sample size, and the outcome data (sensitivity and specificity, determined by comparison with the reference standard). For each study included, data were also extracted to generate a two-by-two table to estimate the sensitivity and the specificity of the PCR-SSCP assay. All extracted data were double-checked by a second author. Sensitivity (the true-positive rate [TPR]) was defined as the proportion of isolates determined to be rifampin resistant by the reference method correctly identified to be rifampin resistant by the PCR-SSCP method. Specificity (true-negative rate or false-positive rate [FPR]) was defined as the proportion of isolates determined to be rifampin susceptible by the reference method correctly identified to be rifampin susceptible by the PCR-SSCP method.
TABLE 1.
Summary of studies included
| Author, yr (reference) | Country | No. of samples | Specimen type | Reference test | No. of samples with the indicated test resulta |
Sensitivity (%) | Specificity (%) | QUADAS quality score | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FP | FN | TN | ||||||||
| Cheng et al., 2007 (3) | China | 116 | Isolates | Bactec 460 | 54 | 4 | 12 | 46 | 82 | 92 | 10 |
| Isfahani et al., 2006 (11) | Iran | 37 | Isolates | Proportion method | 7 | 1 | 14 | 15 | 33 | 94 | 11 |
| Kim et al., 2004 (12) | South Korea | 127 | Isolates | Proportion method | 14 | 0 | 3 | 110 | 82 | 100 | 12 |
| Mani et al., 2003 (17) | India | 201 | Isolates | Proportion method | 77 | 0 | 24 | 100 | 76 | 100 | 11 |
| Kim et al., 2001 (13) | South Korea | 56 | Sputum | Proportion method | 20 | 8 | 3 | 25 | 87 | 76 | 11 |
| Miriam et al., 2001 (18) | Mexico | 46 | Sputum | Bactec 460 | 11 | 0 | 24 | 11 | 31 | 100 | 10 |
| Wu et al., 1999 (37) | China | 85 | Isolates | Absolute concn method | 42 | 0 | 3 | 40 | 93 | 100 | 10 |
| Lee et al., 1998 (16) | South Korea | 77 | Isolates | Absolute concn method | 50 | 0 | 17 | 10 | 75 | 100 | 11 |
| Telenti et al., 1997 (31) | Spain | 95 | Isolates | Proportion method | 72 | 0 | 3 | 20 | 96 | 100 | 11 |
| Kim et al., 1997 (14) | South Korea | 58 | Isolates | Absolute concn method | 32 | 2 | 0 | 24 | 100 | 92 | 11 |
TP, true positive; FP, false positive; FN, false negative; TN, true negative.
Assessment of study quality.
We assessed the quality of the individual studies using criteria based on the Quality Assessment of Studies of Diagnostic Accuracy Included in Systematic Reviews (QUADAS) tools for the assessment of quality of diagnostic studies (36).
Statistical analysis.
We used standard methods recommended for meta-analysis of diagnostic test evaluations (5). Measures of test accuracy for each study, sensitivity, specificity, positive likelihood ratio (PLR), negative-likelihood ratio (NLR), and diagnostic odds ratio (DOR), were computed using two statistical software programs (Stata, version 10 [Stata Corporation, College Station, TX], and Meta-Disc, version 1.4 [XI Cochrane Colloquium, Barcelona, Spain]). The analysis was based on a summary receiver operating characteristic (SROC) curve (5, 19). The sensitivity and specificity for the single test threshold identified for each study were used to plot an SROC curve (15, 19). A random-effects model was used to calculate the average sensitivity and specificity and the average values of the other measures across studies (10, 34). The term heterogeneity, when it is used in relation to meta-analysis, refers to the degree of variability. Heterogeneity was detected using chi-square and I-square tests. To assess the effects of the QUADAS scores on the diagnostic values of the PCR-SSCP assay, we included them as covariates in univariate meta-regression analysis (inverse variance weighted). We also analyzed the effects of other covariates on DOR (i.e., cross-sectional design, consecutive or random sampling of patients, single or double interpretation of determination and reference standard results, and prospective data collection). The relative DOR (RDOR) was calculated to analyze the change in diagnostic precision in the study per unit increase in the covariate (30, 35). Because publication bias is of concern for meta-analysis of diagnostic studies, we tested the potential presence of this bias using the Egger test (6).
RESULTS
Sixteen publications dealing with PCR-SSCP analysis for the detection of rifampin-resistant M. tuberculosis were considered to be eligible for inclusion in the analysis (3, 11, 12-14, 16, 17, 18, 20, 26, 27, 29, 31, 32, 33, 37). Of those, three studies were excluded because only the PCR-SSCP assay was performed with rifampin-resistant M. tuberculosis isolates (27, 29, 32) and three did not calculate the sensitivity or specificity (20, 26, 32). Subsequently, 10 studies (3, 11, 12-14, 16, 17, 18, 31, 37) including 482 specimens with rifampin resistance and 416 rifampin-susceptible specimens were available for analysis, and the characteristics of these studies, along with the QUADAS scores, are outlined in Table 1.
Quality of reporting and study characteristics.
The average sample size of the studies included was 90 (range, 37 to 201). Eight studies performed the PCR-SSCP assay on culture isolates, and two performed it on sputum samples. Eight studies used a standard phenol-chloroform method to extract chromosomal DNA for PCR amplification (3, 12-14, 16, 18, 31, 37), two studies used the cetyltrimethylammonium bromide (CTAB)-NaCl method (11, 17) to extract DNA. Seven studies used the same PCR amplification primers (12-14, 16, 17, 18, 31), and three studies used other primers (3, 11, 37). All studies used the same SSCP analysis method. In addition, most laboratories used the proportion method for conventional drug susceptibility testing.
Our initial data were affected by the poor quality of reporting in the primary studies. To overcome this problem, we contacted all authors of the 10 studies included in the analysis by air mail as well as e-mail, when e-mail addresses were available. Three authors who could provide additional data responded (3, 11, 17). As shown in Table 2, 9 of 10 studies (90%) were cross-sectional in design, the samples were collected from consecutive patients in 2 studies (22%), 1 study (10%) was blind, and all (100%) were prospective.
TABLE 2.
Characteristics of studies included
| Author, yr (reference) | Country | Sample size (no. resistant/no. susceptible) | Cross-sectional design | Consecutive or random | Blinded design | Prospective |
|---|---|---|---|---|---|---|
| Cheng et al., 2007 (3) | China | 66/50 | Yes | No | No | Yes |
| Isfahani et al., 2006 (11) | Iran | 21/16 | Yes | Yes | No | Yes |
| Kim et al., 2004 (12) | South Korea | 17/110 | No | Unknown | No | Yes |
| Mani et al., 2003 (17) | India | 101/100 | Yes | Unknown | No | Yes |
| Kim et al., 2001 (13) | South Korea | 23/33 | Yes | Unknown | No | Yes |
| Miriam et al., 2001 (18) | Mexico | 35/11 | Yes | Unknown | No | Yes |
| Wu et al., 1999 (37) | China | 45/40 | Yes | Unknown | No | Yes |
| Lee et al., 1998 (16) | South Korea | 67/10 | Yes | Unknown | No | Yes |
| Telenti et al., 1997 (31) | Spain | 75/20 | Yes | Yes | Yes | Yes |
| Kim et al., 1997 (14) | South Korea | 32/26 | Yes | Unknown | No | Yes |
Diagnostic accuracy.
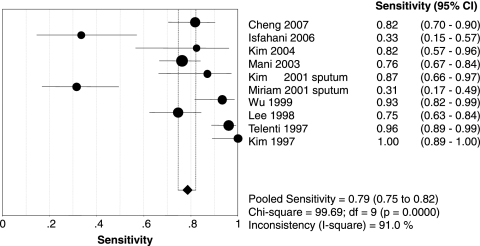
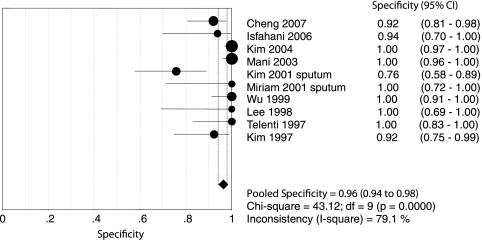
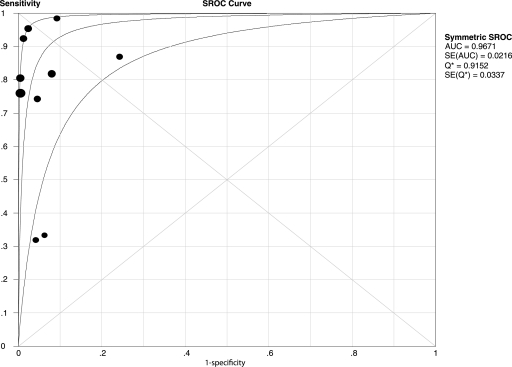
Figures 1 and 2 show forest plots of the sensitivities and specificities of the 10 PCR-SSCP assays for the detection of rifampin resistance in Mycobacterium tuberculosis. The sensitivity and specificity ranged from 0.31 to 1.00 (mean, 0.79; 95% confidence interval [CI], 0.75 to 0.82) and from 0.76 to 1.00 (mean, 0.96; 95% CI, 0.94 to 0.98), respectively. PLR was 16.10 (95% CI, 5.87 to 44.13), NLR was 0.20 (95% CI, 0.10 to 0.40), and DOR was 100.93 (95% CI, 31.95 to 318.83). The chi-square and I-square tests for heterogeneity in the summary results suggested significant heterogeneity for sensitivity, specificity, PLR, NLR, and DOR across studies. Unlike a traditional ROC plot that explores the effects of various thresholds on sensitivity and specificity in a single study, each data point in the SROC plot represents a separate study. The SROC curve presents a global summary of test performance and shows the trade-off between sensitivity and specificity. A graph of the SROC curve for the PCR-SSCP assay determination showing true-positive rates versus false-positive rates from individual studies is shown in Fig. 3. As a global measure of test efficacy, we used the Q value, the intersection point of the SROC curve with a diagonal line from the left upper corner to the right lower corner of the ROC space, which corresponds to the highest common value of sensitivity and specificity for the test. This point does not indicate the only or even the best combination of sensitivity and specificity for a particular clinical setting but represents an overall measure of the discriminatory power of a test. Our data showed that the SROC curve was positioned near the upper left corner of the SROC curve and the Q value was 0.92, while the area under the curve (AUC) was 0.97, indicating a high level of overall accuracy.
FIG. 1.
Forest plot of estimate of sensitivity for PCR-SSCP assays for the rapid detection of rifampin-resistant tuberculosis. •, point estimates of sensitivity from each study; error bars, 95% CIs. The pooled estimate of sensitivity for the PCR-SSCP assays was 0.79 (95% CI, 0.75 to 0.82). df, degrees of freedom.
FIG. 2.
Forest plot of estimate of specificity for PCR-SSCP assays for the rapid detection of rifampin-resistant tuberculosis. •, point estimates of specificity from each study; error bars, 95% CIs. The pooled estimate of specificity for PCR-SSCP assays was 0.96 (95% CI, 0.94 to 0.98). df, degrees of freedom.
FIG. 3.
SROC curves for PCR-SSCP assays. •, each study in the meta-analysis (the size of each study is indicated by the size of the solid circle). The maximum joint sensitivity and specificity (i.e., the Q value) was 0.9152; the AUC was 0.9671. SROC curves summarize the overall diagnostic accuracy.
Multiple-regression analysis and publication bias.
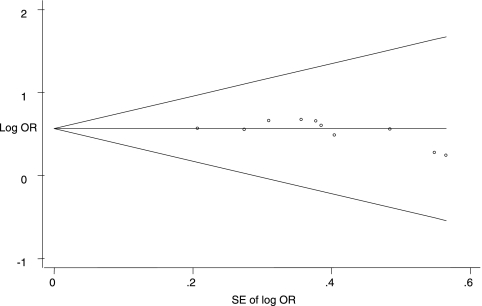
The quality for every study was scored using QUADAS criteria, where a 1 or a 0 was given if all criteria were fulfilled or not achieved, respectively (Table 1; see also Table S1 in the supplemental material) (37). These scores were used in the meta-regression analysis to assess the effect of study quality on the RDOR of the PCR-SSCP assay for the detection of rifampin resistance in Mycobacterium tuberculosis. As shown in Table 3, there was no statistical significance in RDOR values between studies with higher and lower qualities. There was no significant difference between studies with or without cross-sectional, consecutive/random, and blinded designs. The evaluation of publication bias showed that the Egger test was not significant (P = 0.152). The funnel plots for publication bias (Fig. 4) also did not reveal the presence of asymmetry.
TABLE 3.
Weighted meta-regression of the effects of methodological quality and study design on diagnostic precision of PCR-SSCP in 10 assays
| Covariate | No. of studies | Coefficient | RDOR (95% CI) | P value |
|---|---|---|---|---|
| QUADAS score ≥ 10 | 10 | 0.883 | 2.42 (0.16-36.79) | 0.4681 |
| Cross-sectional design | 9 | −1.420 | 0.24 (0.01-4.45) | 0.2869 |
| Consecutive or random | 2 | −0.007 | 1.01 (0.05-18.98) | 0.9958 |
| Blinded | 1 | 1.137 | 3.12 (0.19-50.25) | 0.3654 |
FIG. 4.
Funnel graph for the assessment of potential publication bias in PCR-SSCP assays. The funnel graph plots the log of the odds ratio (OR) against the standard error (SE) of the log of the odds ratio (an indicator of sample size). •, each study in the meta-analysis. The result of the Egger test for publication bias was not significant (P = 0.152).
DISCUSSION
The purpose of drug susceptibility testing is the early detection of drug resistance. This allows the better management and treatment of patients. The early identification of drug-resistant TB cases would decrease the risk of disease and possible amplification of drug resistance. The methods currently available for drug susceptibility testing of M. tuberculosis are cheap but slow in most areas with a high incidence of TB. There is obviously a great need for fast, reliable, and inexpensive methods for drug susceptibility testing of M. tuberculosis. However, any new rapid drug susceptibility testing method must be carefully calibrated with representative isolates of M. tuberculosis in order to determine in vitro the cutoff for resistant and susceptible isolates with acceptable reproducibility.
The present meta-analysis showed that the mean values of the sensitivity and specificity of the PCR-SSCP assay were 0.79 and 0.96, respectively, and that the maximum joint sensitivity and specificity (Q value) was 0.92 and the AUC was 0.97, indicating a high level of overall accuracy. We also noted that two studies (11, 18) showed relatively low sensitivities (<0.70) and one study (13) demonstrated a low specificity (<0.90) when they used PCR-SSCP assays for the rapid detection of rifampin resistance in Mycobacterium tuberculosis.
The DOR is a single indicator of test accuracy that combines the data from sensitivity and specificity into a single number and the ratio of the odds of a positive test result for a patient with disease or without disease (7). The value of a DOR ranges from 0 to infinity, with higher values indicating better discriminatory test performance (i.e., higher accuracy). In the present meta-analysis, we found that the mean DOR was 100.93, also indicating a high level of overall accuracy, and also presented both PLR and NLR as diagnostically accurate, since the SROC curve and the DOR are not easy to interpret and use in clinical practice and ratios are considered to be more clinically meaningful (4, 8). The PLR value was 16.10, indicating that the isolates in specimens with M. tuberculosis have an approximately 16-fold higher chance of being rifampin resistant than the isolates in specimens with susceptible M. tuberculosis. On the other hand, if the PCR-SSCP assay result was negative, the probability that the isolates in the sample are resistant is approximately 20%, which is not low enough to rule out resistance. These data suggest that a negative PCR-SSCP assay result should not be used alone as a justification to deny rifampin resistance in Mycobacterium tuberculosis.
An exploration of the reasons for heterogeneity rather than the computation of a single summary measure is an important goal of meta-analysis (25). In our meta-analysis, QUADAS scores were used to assess the effect of study quality on RDOR. We found significant heterogeneity for sensitivity, specificity, PLR, NLR, and DOR among the studies analyzed, although the exact mechanism responsible for the significance was unable to be explained. There were no differences between studies with or without cross-sectional, consecutive/random, and blinded designs, which may contribute to the quality of the test performance. However, PCR-SSCP assay-related technical issues in the individual studies, such as the preanalytical steps and primers used and the method of DNA quantification, may be responsible for the heterogeneity.
The meta-analysis had several strengths. First, a standard protocol was used for carrying out the systematic review (22), including a comprehensive search strategy. Moreover, two reviewers independently carried out various stages of the systematic review process, including article selection and data extraction. Lastly, rigorous methods were used for data analysis.
The meta-analysis was limited by the relatively small number of available studies and the types of outcomes reported in the studies. Some studies presented only data on sensitivity and specificity. An obvious limitation is the lack of data on whether or not the PCR-SSCP assay has a clinical impact on patient management and treatment outcomes and how much value the assay contributes beyond that offered by conventional tests. Data on cost-effectiveness and feasibility in routine program settings are also lacking. Furthermore, there is little current evidence on how the PCR-SSCP assay may fit into existing diagnostic and treatment algorithms. Despite using publication bias analysis, the considerable heterogeneity in the results remained unexplained; further work is necessary to determine why values vary across settings. Geographic and genetic variations in the distribution of drug-resistant strains of M. tuberculosis might partially explain the present findings. Finally, the present authors excluded two studies, published in Chinese and Japanese. The exclusion of non-English-language studies, combined with potential publication bias, may have resulted in an overly optimistic estimate of the accuracy of the PCR-SSCP assay.
A notable advantage of molecular tests is their rapid turnaround time, which may have implications for patient management and transmission of drug-resistant M. tuberculosis. The turnaround time for the PCR-SSCP assay is less than 48 h, making it substantially faster than conventional drug susceptibility testing methods. An important issue that remains, however, is the affordability of molecular assays and the associated laboratory infrastructure needs in resource-constrained settings. Molecular tests, whose prices are typically higher than those of conventional tests, may be popular in resource-rich settings. However, the most resource-constrained countries tend to have the highest burden of MDR-TB cases and are the least likely to benefit from expensive technologies because of high costs and a lack of appropriate laboratory capacity.
In conclusion, the PCR-SSCP assay demonstrates a high level of overall accuracy for the detection of rifampin resistance, which is a proxy for MDR-TB. This suggests that it has good utility as a rapid screening tool, especially in settings with high rates of MDR-TB. Use of the PCR-SSCP assay is currently limited to culture isolates and direct testing of smear-positive sputum specimens, and it is not commercially available. The PCR-SSCP assay is not recommended as a replacement for conventional culture and drug susceptibility testing for the detection of rifampin resistance in M. tuberculosis.
Supplementary Material
Footnotes
Published ahead of print on 28 July 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Canetti, G., W. Fox, A. Khomenko, H. T. Mahler, N. K. Menon, D. A. Mitchison, N. Rist, and N. A. Šmelev. 1969. Advances in techniques of testing mycobacterial drug sensitivity and the use of sensitivity tests in tuberculosis control programs. Bull. World Health Organ. 41:21-43. [PMC free article] [PubMed] [Google Scholar]
- 2.Canetti, G., S. Froman, J. Grosset, P. Hauduroy, M. Langerová, H. T. Mahler, G. Meissner, D. A. Mitchison, and L. Šula. 1963. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull. World Health Organ. 29:565-578. [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, X. D., J. F. Zhang, L. Yang, X. L. Xu, J. Y. Liu, W. B. Yu, M. Q. Su, and X. K. Hao. 2007. A new multi-PCR-SSCP assay for simultaneous detection of isoniazid and rifampin resistance in Mycobacterium tuberculosis. J. Microbiol. Methods 70:301-305. [DOI] [PubMed] [Google Scholar]
- 4.Deeks, J. J. 2001. Systematic reviews in health care: systematic reviews of evaluations of diagnostic and screening tests. BMJ 323:157-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deville, W. L., F. Buntinx, L. M. Bouter, V. M. Montori, H. C. W. Vet, D. A. Windt, and P. D. Bezemer. 2002. Conducting systematic reviews of diagnostic studies: didactic guidelines. BMC Med. Res. Methodol. 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egger, M., S. G. Davey, M. Schneider, and C. Minder. 1997. Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glas, A. S., J. G. Lijmer, M. H. Prins, G. J. Bonsel, and P. M. Bossuyt. 2003. The diagnostic odds ratio: a single indicator of test performance. J. Clin. Epidemiol. 56:1129-1135. [DOI] [PubMed] [Google Scholar]
- 8.Graves, R. S. 2002. Users' guides to the medical literature: a manual for evidence-based clinical practice. J. Med. Libr. Assoc. 90:482-483. [Google Scholar]
- 9.Hayashi, K. 1991. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1:34-38. [DOI] [PubMed] [Google Scholar]
- 10.Irwig, L., A. N. Tosteson, C. Gatsonis, J. Lau, G. Colditz, T. C. Chalmers, and F. Mosteller. 1999. Guidelines for meta-analyses evaluating diagnostic tests. Ann. Intern. Med. 120:667-676. [DOI] [PubMed] [Google Scholar]
- 11.Isfahani, B. N., A. Tavakoli, M. Salehi, and M. Tazhibi. 2006. Detection of rifampin resistance patterns in Mycobacterium tuberculosis strains isolated in Iran by polymerase chain reaction-single-strand conformation polymorphism and direct sequencing methods. Mem. Inst. Oswaldo Cruz 101:597-602. [DOI] [PubMed] [Google Scholar]
- 12.Kim, B. J., K. H. Lee, Y. J. Yun, E. M. Park, Y. G. Park, G. H. Bai, C. Y. Cha, and Y. H. Kook. 2004. Simultaneous identification of rifampin-resistant Mycobacterium tuberculosis and nontuberculous mycobacteria by polymerase chain reaction-single strand conformation polymorphism and sequence analysis of the RNA polymerase gene (rpoB). J. Microbiol. Methods 58:111-118. [DOI] [PubMed] [Google Scholar]
- 13.Kim, B. J., K. H. Lee, B. N. Perk, S. J. Kim, E. M. Park, Y. G. Park, G. H. Bai, S. J. Kim, and Y. H. Kook. 2001. Detection of rifampin-resistant Mycobacterium tuberculosis in sputa by nested PCR-linked single-strand conformation polymorphism and DNA sequencing. J. Clin. Microbiol. 39:2610-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, B. J., S. Y. Kim, B. H. Park, M. A. Lyu, I. K. Park, G. H. Bai, S. J. Kim, C. Y. Cha, and Y. H. Kook. 1997. Mutations in the rpoB gene of Mycobacterium tuberculosis that interfere with PCR-single-strand conformation polymorphism analysis for rifampin susceptibility testing. J. Clin. Microbiol. 35:492-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau, J., J. P. Ioannidis, E. M. Balk, C. Milch, N. Terrin, P. Chew, and D. Salem. 2001. Diagnosing acute cardiac ischemia in the emergency department: a systematic review of the accuracy and clinical effect of current technologies. Ann. Emerg. Med. 37:453-460. [DOI] [PubMed] [Google Scholar]
- 16.Lee, H., S. N. Cho, H. E. Bang, J. H. Lee, G. H. Bae, S. J. Kim, and J. D. Kim. 1998. Molecular analysis of rifampin-resistant Mycobacterium tuberculosis isolated from Korea by polymerase chain reaction-single strand conformation polymorphism sequence analysis. Int. J. Tuberc. Lung Dis. 2:585-589. [PubMed] [Google Scholar]
- 17.Mani, C., N. Selvakumar, V. Kumar, S. Narayanan, and P. R. Narayanan. 2003. Comparison of DNA sequencing, PCR-SSCP and PhaB assays with indirect sensitivity testing for detection of rifampicin resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 7:652-659. [PubMed] [Google Scholar]
- 18.Miriam, B. V., P. L. Alfredo, A. H. Catalina, V. A. Gilberto, K. M. Midori, M. S. Peter, C. Patricia, M. R. P. Guillermo, and S. O. Jose. 2001. RpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis identified by polymerase chain reaction single-stranded conformational polymorphism. Emerg. Infect. Dis. 7:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moses, L. E., D. Shapiro, and B. Littenberg. 1993. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat. Med. 12:1293-1316. [DOI] [PubMed] [Google Scholar]
- 20.Noman, S., M. Shamim, N. K. Jain, A. Rattan, A. Amin, V. M. Katoch, S. K. Sharma, and S. E. Hasnain. 1998. Molecular genetic analysis of multi-drug resistance in Indian isolates of Mycobacterium tuberculosis. Mem. Inst. Oswaldo Cruz 93:589-594. [DOI] [PubMed] [Google Scholar]
- 21.Orita, M., H. Iwahana, H. Kanazawa, K. Hayashi, and T. Sekiya. 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single strand conformation polymorphisms. Proc. Natl. Acad. Sci. U. S. A. 86:2766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pai, M., M. McCulloch, W. Enanoria, and J. M. Colford. 2004. Systematic reviews of diagnostic test evaluations: what's behind the scenes? ACP J. Club 141:A11-A13. [PubMed] [Google Scholar]
- 23.Palomino, J. C. 2005. Nonconventional and new methods in the diagnosis of tuberculosis: feasibility and applicability in the field. Eur. Respir. J. 26:339-350. [DOI] [PubMed] [Google Scholar]
- 24.Palomino, J. C. 2006. Newer diagnostics for tuberculosis and multi-drug resistant tuberculosis. Curr. Opin. Pulm. Med. 12:172-178. [DOI] [PubMed] [Google Scholar]
- 25.Petitti, D. B. 2001. Approaches to heterogeneity in meta-analysis. Stat. Med. 20:3625-3633. [DOI] [PubMed] [Google Scholar]
- 26.Ramasoota, P., P. Pitaksajjakul, W. Phatihattakorn, V. Pransujarit, and J. Boonyasopun. 2006. Mutations in the rpoB gene of rifampicin-resistant Mycobacterium tuberculosis strains from Thailand and its evolutionary implication. Southeast Asian J. Trop. Med. Public Health 37:136-147. [PubMed] [Google Scholar]
- 27.Raphael, C. Y. C., H. Mamie, W. C. C. Edward, T. K. Au, M. L. Chin, K. Y. Chun, K. W. A. Carrie, Y. L. Y. Christina, K. M. Kam, C. W. Y. Peter, and F. B. C. Augustine. 2007. Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J. Antimicrob. Chemother. 59:866-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah, N. S., A. Wright, G. H. Bai, L. Barrera, F. Boulahbal, N. Martín-Casabona, F. Drobniewski, C. Gilpin, M. Havelková, R. Lepe, R. Lumb, B. Metchock, F. Portaels, M. F. Rodrigues, S. Rüsch-Gerdes, A. V. Deun, V. Vincent, K. Laserson, C. Wells, and J. P. Cegielski. 2007. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg. Infect. Dis. 13:380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheng, J., J. Li, G. Sheng, H. Yu, H. Huang, H. Cao, Y. Lu, and X. Deng. 2008. Characterization of rpoB mutations associated with rifampin resistance in Mycobacterium tuberculosis from eastern China. J. Appl. Microbiol. 105:904-911. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki, S., T. Morooka, and N. K. Choudhry. 2004. The conditional relative odds ratio provided less biased results for comparing diagnostic test accuracy in meta-analyses. J. Clin. Epidemiol. 57:461-469. [DOI] [PubMed] [Google Scholar]
- 31.Telenti, A., N. Honore, C. Bernasconi, J. March, A. Ortega, B. Heym, H. E. Takifff, and S. T. Cole. 1997. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J. Clin. Microbiol. 35:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telenti, A., P. Imboden, F. Marchesi, T. Schmidheini, and T. Bodmer. 1993. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob. Agents Chemother. 37:2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres, M. J., A. Criado, N. Gónzalez, J. C. Palomares, and J. Aznar. 2002. Rifampin and isoniazid resistance associated mutations in Mycobacterium tuberculosis clinical isolates in Seville, Spain. Int. J. Tuberc. Lung Dis. 6:160-163. [PubMed] [Google Scholar]
- 34.Vamvakas, E. C. 2004. Meta-analyses of studies of the diagnostic accuracy of laboratory tests: a review of the concepts and methods. Arch. Pathol. Lab. Med. 122:675-686. [PubMed] [Google Scholar]
- 35.Westwood, M. E., P. F. Whiting, and J. Kleijnen. 2005. How does study quality affect the results of a diagnostic meta-analysis? BMC Med. Res. Methodol. 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whiting, P., A. W. Rutjes, J. B. Reitsma, P. M. Bossuyt, and J. Kleijnen. 2003. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med. Res. Methodol. 3:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, X. Q., J. X. Zhang, Y. H. Zhuang, X. G. Zhang, G. L. Li, and X. Y. He. 1999. Molecular mechanisms of drug resistance in Mycobacterium tuberculosis clinical isolates. Chin. Med. J. (Engl.) 112:524-528. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.