Abstract
We report five new IS711 chromosomal locations that are specific for marine mammal Brucella groups of strains and useful for their identification and classification. Our data support their current classification into two species, Brucella ceti and B. pinnipedialis, with subgroups in each, but also the possibility of additional species.
Brucellae are Gram-negative, facultative, intracellular bacteria that can infect many species of animals and humans. Six species were initially recognized within the genus Brucella: Brucella abortus, B. melitensis, B. suis, B. ovis, B. canis, and B. neotomae (13, 20, 27). This classification is based mainly on differences in pathogenicity, host preference, and phenotypic characteristics. Four additional species have been included in the genus Brucella since 2007. B. ceti and B. pinnipedialis have been isolated from marine mammals, with cetaceans (dolphin, porpoise, and whale species) and pinnipeds (various seal species), respectively, as the preferred hosts (11, 12). B. microti, first described in 2008, was isolated initially from the common vole but later also from the red fox and from soil (21-23). The latest species is B. inopinata, isolated from a human breast implant infection, representing the Brucella species most distant from the others at the phenotypic and molecular levels (9, 24). The animal or environmental reservoir of B. inopinata is not known.
Molecular and phenotypic typing of marine mammal Brucella strains has led to their classification into two species, i.e., B. ceti and B. pinnipedialis (12). However, several subgroups within each species have been identified by molecular typing methods, such as multilocus sequence analysis (MLSA), multilocus variable-number tandem-repeat (VNTR) analysis (MLVA), and omp2a and omp2b porin gene diversity analysis (1, 3, 5, 7, 8, 14, 17, 26, 27). Among these subgroups, one within B. ceti, which is composed exclusively of strains isolated from various dolphin species, has been proposed to constitute a separate species with the name B. delphini (1, 14, 27). The isolates from cetaceans from the Pacific may also constitute a separate species (17). Three human cases with naturally acquired infection by Brucella strains presumably of marine origin have been reported, one case of spinal osteomyelitis from a patient in New Zealand (19) and two neurobrucellosis cases from Peruvian patients (25). Interestingly, these human isolates presented the same genotype as strains from cetaceans from the Pacific (28).
The presence of the mobile genetic element IS711 (GenBank accession no. M94960) has been a useful target for molecular characterization of classical terrestrial mammal Brucella species and biovars based on the number and distribution of IS711 copies within the bacterial genomes. Among classical Brucella species, IS711-based fingerprints are stable, species specific (except B. canis), and to some extent biovar specific (2). Marine mammal Brucella strains have been shown to carry a higher number of the IS711 element in their genomes than terrestrial mammal Brucella species and biovars, with the exception of B. ovis (2, 3, 8, 18). Consequently, infrequent restriction site PCR (IRS-PCR) methods were applied, taking into account this higher number of IS711 elements, to study the genomic diversity of marine mammal strains (5, 16). These studies confirmed the classification into two marine mammal Brucella species, with subgroups in each. In addition, three specific IS711-containing DNA fragments were detected, allowing the molecular identification of B. ceti and its subgroup composed exclusively of dolphin isolates (5, 8, 16).
In this study, to further analyze the distribution of IS711 elements in the genomes of Brucella strains of marine origin, we applied ligation-mediated PCR (LM-PCR) specifically targeting IS711. This method aimed to determine other specific chromosomal locations of IS711 that would allow molecular identification of the different B. ceti and B. pinnipedialis subgroups or presumed new species.
The Brucella strains of marine mammal origin used in this study are listed in Table 1. They were from diverse marine mammal species (seal, otter, dolphin, porpoise, and whale species) and geographic origins (France, Norway, and Scotland). The human isolate from New Zealand presumably of marine origin was also included in this study. Bacterial culture and DNA extraction were performed as described previously (6). LM-PCR was performed on representative groups of Brucella strains, including terrestrial mammal Brucella control strains, classified previously by IS711 Southern blot analysis (Table 1) (18). DNA was digested with a restriction enzyme cutting outside the IS711 element (EcoRI, BglII, or BamHI), and an adapter specific for each enzyme was used with a primer specific for the IS711 element. Thus, only DNA fragments containing part of the IS711 element were amplified, and sequencing of these fragments enabled us to locate IS711 copies in the genomes of marine mammal Brucella isolates. Primers and adapters used are listed in Table 2. Ligation of adapters and PCR amplifications were performed as described previously (10). The DNA fragments generated were run on agarose gels, and five fragments of interest that seemed to be specific to marine mammal Brucella strains were sequenced at Genome Express (Meylan, France) (data not shown). Specific PCRs were then performed by using a forward primer in the sequence identified upstream of IS711 and a reverse primer designed for the IS711 element (Fig. 1). These primers are listed in Table 2.
TABLE 1.
Brucella strains of marine mammal origin used in this study
| Species | Strain | Host or source | Latin name of host or source | Geographic origin of strain | MLVA-16 cluster (panel 1 genotype)a | MLSA STb | omp2b-omp2a RFLP patternc | IS711 RFLP patternd (clustere) | IS711 copy no.f | Presence or absence, by fragment-specific PCR, of fragmentg: |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | ||||||||||
| B. pinnipedialis | 9a-1 | Hooded seal | Cystophora cristata | Norway | C3 (76) | P-I | C | 19 | + | − | − | − | − | |
| 17a-1 | Hooded seal | Cystophora cristata | Norway | C3 (76) | P-I | C | 19 | + | − | − | − | − | ||
| 22a-2 | Hooded seal | Cystophora cristata | Norway | C3 (76) | P-I | C | 19 | + | − | − | − | − | ||
| 23a-1 | Hooded seal | Cystophora cristata | Norway | C3 (75) | P-I | C | 19 | + | − | − | − | − | ||
| 24a-2 | Hooded seal | Cystophora cristata | Norway | C3 (75) | P-I | C | 19 | + | − | − | − | − | ||
| 25a-1 | Hooded seal | Cystophora cristata | Norway | C3 (76) | P-I | C | 19 | + | − | − | − | − | ||
| 30a-1 | Hooded seal | Cystophora cristata | Norway | C3 (74) | P-I | C | 19 | + | − | − | − | − | ||
| 37a-1 | Hooded seal | Cystophora cristata | Norway | C3 (76) | P-I | C | 19 | + | − | − | − | − | ||
| 38g-1 | Hooded seal | Cystophora cristata | Norway | C3 (76) | P-I | C | 19 | + | − | − | − | − | ||
| 39a-1 | Hooded seal | Cystophora cristata | Norway | C3 (76) | P-I | C | 19 | + | − | − | − | − | ||
| 53c-1 | Hooded seal | Cystophora cristata | Norway | C3 (76) | P-I | C | 19 | + | − | − | − | − | ||
| M163/99/10 | Hooded seal | Cystophora cristata | Scotland | C3 (76) | ST25 | P-I | C (4) | 19 | + | − | − | − | − | |
| M603/99/7 | Hooded seal | Cystophora cristata | Scotland | C3 (76) | ST25 | P-I | C (4) | 19 | + | − | − | − | − | |
| M2006/94/6 | Hooded seal | Cystophora cristata | Scotland | C3 (76) | ST25 | P-I | C (4) | 19 | + | − | − | − | − | |
| B2/94 (NCTC 12890; BCCN 94-73h) | Common seal | Phoca vitulina | Scotland | C2 (25) | L-I | D (1) | 23 | + | + | − | − | − | ||
| M13/01/1 | Common seal | Phoca vitulina | Scotland | C2 (25) | ST25 | L-I | D (1) | 23 | + | + | − | − | − | |
| M336/94/1 | Common seal | Phoca vitulina | Scotland | C2 (25) | L-I | D (1) | 23 | + | + | − | − | − | ||
| M339/94/1 | Common seal | Phoca vitulina | Scotland | C2 (25) | L-I | D (1) | 23 | + | + | − | − | − | ||
| M445/99/2 | Common seal | Phoca vitulina | Scotland | C2 (25) | ST25 | L-I | D (1) | 23 | + | + | − | − | − | |
| M514/96/4 | Common seal | Phoca vitulina | Scotland | C2 (25) | ST25 | L-I | D (1) | 23 | + | + | − | − | − | |
| M972/94/1 | Common seal | Phoca vitulina | Scotland | C2 (25) | L-I | D (1) | 23 | + | + | − | − | − | ||
| M621/99/2 | Gray seal | Halichoerus grypus | Scotland | C2 (25) | ST25 | L-I | D (1) | 23 | + | + | − | − | − | |
| M1771/94/1 | Otter | Lutra lutra | Scotland | C2 (25) | ST25 | L-I | D (1) | 23 | + | + | − | − | − | |
| M292/94/1 | Common seal | Phoca vitulina | Scotland | C1 (71) | ST24 | O-I | E (1) | 24 | + | + | − | − | − | |
| M449/02/2 | Common seal | Phoca vitulina | Scotland | C1 (71) | O-I | E (1) | 24 | + | + | − | − | − | ||
| M2466/93/4 | Common seal | Phoca vitulina | Scotland | C2 (72) | L-I | E (1) | 24 | + | + | − | − | − | ||
| M2533/93/1 | Common seal | Phoca vitulina | Scotland | C1 (71) | ST24 | O-I | E (1) | 24 | + | + | − | − | − | |
| M2375/94/3 | Gray seal | Halichoerus grypus | Scotland | C2 (25) | ST25 | L-I | E (1) | 24 | + | + | − | − | − | |
| M194/00/1 | Gray seal | Halichoerus grypus | Scotland | C2 (25) | L-I | E (1) | 24 | + | + | − | − | − | ||
| M192/00/1 | Minke whale | Balaenoptera acutorostrata | Scotland | C1 (71) | ST24 | O-I | E (1) | 24 | + | + | − | − | − | |
| B. ceti | M490/95/1 | Common seal | Phoca vitulina | Scotland | B (23) | ST23 | M-J | F (2) | 30 | + | − | + | + | − |
| 05-0684-1144 | Porpoise | Phocoena phocoena | France | B (23) | M-J | F | 30 | + | − | + | + | − | ||
| M23/03/4 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M39/94/1 | Porpoise | Phocoena phocoena | Scotland | B (23) | ST23 | M-J | F (2) | 30 | + | − | + | + | − | |
| M51/04/2 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M58/05/1 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F | 30 | + | − | + | + | − | ||
| M78/05/2 | Porpoise | Phocoena phocoena | Scotland | B (23) | ST23 | M-J | F | 30 | + | − | + | + | − | |
| M93/04/3 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M117/01/1 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M165/03/6 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M195/03/10 | Porpoise | Phocoena phocoena | Scotland | B (70) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M234/05/2 | Porpoise | Phocoena phocoena | Scotland | B (23) | ST23 | M-J | F | 30 | + | − | + | + | − | |
| M291/03/2 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M499/99/10 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M515/96/2 | Porpoise | Phocoena phocoena | Scotland | B (23) | ST23 | M-J | F | 30 | + | − | + | + | − | |
| M615/99/1 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M854/98/8 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M1570/94/1 | Porpoise | Phocoena phocoena | Scotland | B (23) | ST23 | M-J | F | 30 | + | − | + | + | − | |
| 7763/2 | Bottlenose dolphin | Tursiops truncatus | France | B (23) | ST23 | M-J | F | 30 | + | − | + | + | − | |
| M452/97/2 | Common dolphin | Delphinus delphis | Scotland | B (23) | ST23 | M-J | F (2) | 30 | + | − | + | + | − | |
| M2/00/1 | White-sided dolphin | Lagenorhynchus acutus | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M52/01/1 | White-sided dolphin | Lagenorhynchus acutus | Scotland | B (23) | M-J | F (2) | 30 | + | − | + | + | − | ||
| M181/97/1 | White-sided dolphin | Lagenorhynchus acutus | Scotland | B (23) | ST23 | M-J | F | 30 | + | − | + | + | − | |
| M187/00/1 | White-sided dolphin | Lagenorhynchus acutus | Scotland | B (23) | ST23 | M-J | F (2) | 30 | + | − | + | + | − | |
| M2438/95/1 | White-sided dolphin | Lagenorhynchus acutus | Scotland | B (23) | M-J | F | 30 | + | − | + | + | − | ||
| B202R | Minke whale | Balaenoptera acutorostrata | Norway | B (23) | ST23 | M-J | F (2) | 30 | + | − | + | + | − | |
| B1/94 (NCTC 12891; BCCN 94-74h) | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | G (2) | 29 | + | − | + | + | − | ||
| M12/00/3 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | G (2) | 29 | + | − | + | + | − | ||
| M38/04/3 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | G (2) | 29 | + | − | + | + | − | ||
| M103/99/1 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | G (2) | 29 | + | − | + | + | − | ||
| M199/04/2 | Porpoise | Phocoena phocoena | Scotland | B (23) | ST23 | M-J | G | 29 | + | − | + | + | − | |
| M1747/98/3 | Porpoise | Phocoena phocoena | Scotland | B (23) | M-J | G (2) | 29 | + | − | + | + | − | ||
| M870/97/1 | White-beaked dolphin | Lagenorhynchus albirostris | Scotland | B (23) | M-J | G | 29 | + | − | + | + | − | ||
| M997/94/2 | White-sided dolphin | Lagenorhynchus acutus | Scotland | B (69) | M-J | G (2) | 29 | + | − | + | + | − | ||
| M2788/97/1 | White-sided dolphin | Lagenorhynchus acutus | Scotland | B (23) | M-J | G (2) | 29 | + | − | + | + | − | ||
| B14/94 | Common dolphin | Delphinus delphis | Scotland | A1 (24) | ST26 | N-K | H (3) | 26 | + | − | − | − | + | |
| M13/05/1 | Striped dolphin | Stenella coeruleoalba | Scotland | A1 (24) | ST26 | N-K | H | 26 | + | − | − | − | + | |
| M40/95/1 | Striped dolphin | Stenella coeruleoalba | Scotland | A1 (24) | ST26 | N-K | H (3) | 26 | + | − | − | − | + | |
| M642/99/2 | Striped dolphin | Stenella coeruleoalba | Scotland | A1 (24) | ST26 | N-K | H (3) | 26 | + | − | − | − | + | |
| M654/99/1 | Striped dolphin | Stenella coeruleoalba | Scotland | A2 (77) | ST26 | N-K | H (3) | 26 | + | − | − | − | + | |
| M656/99/1 | Striped dolphin | Stenella coeruleoalba | Scotland | A1 (24) | ST26 | N-K | H (3) | 26 | + | − | − | − | + | |
| M2194/94/1 | Striped dolphin | Stenella coeruleoalba | Scotland | A1 (24) | ST26 | N-K | H (3) | 26 | + | − | − | − | + | |
| M18/96/1 | White-sided dolphin | Lagenorhynchus acutus | Scotland | A1 (24) | N-K | H | 26 | + | − | − | − | + | ||
| Brucella spp. | 02/611 | Human | Homo sapiens | New Zealand | (78) | ST27 | Q-I | I | 23 | + | − | − | − | − |
According to Maquart et al. (17).
According to Groussaud et al. (14). ST, sequence type.
According to Maquart et al. (18) and results of this study.
According to Dawson et al. (8) for the strains indicated.
According to Maquart et al. (18) and results of this study.
Fragments as numbered in Fig. 1.
Reference strains.
TABLE 2.
Primers and adapters used in this study
| Procedure and adapter/primer name | Adapter/primer locationc | Fragment no. | Fragment size (bp) | Nucleotide sequence (5′-3′) |
|---|---|---|---|---|
| Ligation-mediated PCR | ||||
| Eco1 | NAd | NA | NA | GAGTCGTGTTACTCCCATCG |
| Eco2 | NA | NA | NA | AATTCGATGGGAGTAACACG |
| ABg1a | NA | NA | NA | GAGTCGTGTTACTCCCATCG |
| ABg2a | NA | NA | NA | GATCGATGGGAGTAACACGACTC |
| IS1b | IS711 | NA | NA | CGGTTCCATTGCTCATCTGT |
| Fragment-specific PCR | ||||
| I1 | BR1657 | I | 253 | CTACATAGGCATCGCCAGAG |
| II1 | BRA0655 | II | 750 | TGCCATAGGGCACATTGTTC |
| III1 | BR1612 | III | 523 | TTATATGATGCGCCCGGAGG |
| IV1 | BR0613 | IV | 670 | GAAGGTAGAGCCGGAATATC |
| V1 | BR0625 | V | 195 | GCTTTCGCATTGGAGCTTAC |
| IS2 | IS711 | I through V | NA | GCTCACGGCTGTTCTCCTTT |
The same adapters and primers were used for LM-PCR with restriction enzymes BglII and BamHI.
All LM-PCRs were performed with primer IS1.
According to the genome sequence of B. suis 1330 chromosome I or II.
NA, not applicable.
FIG. 1.
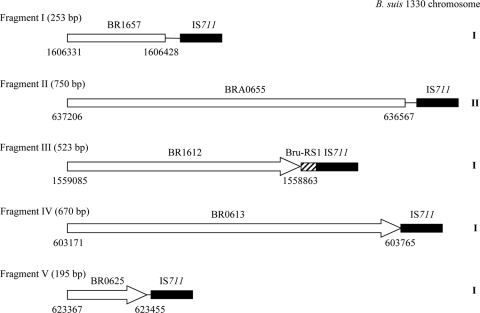
Schematic representation of IS711-containing DNA fragments I to V identified by LM-PCR. The nucleotide positions relative to the genome sequences of B. suis 1330 chromosomes I and II (GenBank accession numbers NC_004310 and NC_004311, respectively) are indicated. Open reading frame BR1657 (in fragment I) codes for a putative two-component response regulator. BRA0655, or the ugpB gene (in fragment II), codes for a putative glycerol-3-phosphate ABC transporter. BR1612 (in fragment III) codes for a putative spermidine/putrescine ABC transporter. BR0613 (in fragment IV) codes for a putative sensor histidine kinase. BR0625 (in fragment V) codes for a hypothetical protein. The hatched region upstream from IS711 in fragment III represents a Bru-RS1-like element.
The IS711-specific locations identified relative to the B. suis 1330 genome sequences of chromosomes I and II (GenBank accession numbers NC_004310 and NC_004311, respectively) are shown in Fig. 1. Specific PCRs performed on the strains listed in Table 1 and the reference strains of terrestrial mammal Brucella species and biovars showed that all of these locations are specific to marine mammal Brucella strains. Among the novel IS711 locations identified in this study, there was one near a Bru-RS1-like element (86% nucleotide identity) (in fragment III [Fig. 1]), previously described as being a hot spot for IS711 insertion (15). The novel specific IS711 locations were distributed as follows according to the PCR-amplified DNA fragments. (Interestingly, they correlated well with other molecular classification methods and, in particular, as expected with their IS711 Southern blot restriction fragment length polymorphism [RFLP] profiles [Table 1].) Fragment I was detected in all marine mammal strains studied (n = 74) and also in the human isolate from New Zealand. Fragment II was found to be specific to B. pinnipedialis strains but was not detected in the hooded seal isolates constituting a particular group of pinniped strains with some distinct molecular characteristics (1, 16, 17, 18). These strains were previously shown to carry fewer IS711 elements in their genomes, and this could explain why we were not able to detect specific IS711-containing DNA fragments in this group of strains (Table 1). They could be ancestral to the other B. pinnipedialis strains, as suggested previously (16). Fragments III, IV, and V were found to be specific to B. ceti strains, with III and IV on the one hand and V on the other hand corresponding to the two major groups of cetacean strains classified by other molecular methods (Table 1). As indicated above, the latter group of strains corresponds to that composed exclusively of dolphin isolates. Unfortunately, under our conditions, no specific IS711-containing DNA fragment could be detected in the human isolate from New Zealand studied, although it presented a distinct IS711 RFLP pattern, likely with other specific IS711 locations (18). Nevertheless, this finding as well as the distinct IS711 locations in the dolphin group of strains and other molecular data supports the possibility of additional Brucella species in marine mammal Brucella isolates besides B. ceti and B. pinnipedialis (27). The specific IS711 locations found in this study, together with others previously reported (4, 5, 29), may be of further usefulness for the molecular identification of Brucella isolates of marine origin.
Nucleotide sequence accession numbers.
The nucleotide sequences of the five DNA fragments from this study have been deposited in GenBank under accession numbers HM352552 (fragment I), HM352553 (fragment II), HM352554 (fragment III), HM352555 (fragment IV), and HM352556 (fragment V).
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Bourg, G., D. O'Callaghan, and M. L. Boschiroli. 2007. The genomic structure of Brucella strains isolated from marine mammals gives clues to evolutionary history within the genus. Vet. Microbiol. 125:375-380. [DOI] [PubMed] [Google Scholar]
- 2.Bricker, B. J., D. R. Ewalt, A. P. MacMillan, G. Foster, and S. Brew. 2000. Molecular characterization of Brucella strains isolated from marine mammals. J. Clin. Microbiol. 38:1258-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clavareau, C., V. Wellemans, K. Walravens, M. Tryland, J. M. Verger, M. Grayon, A. Cloeckaert, J. J. Letesson, and J. Godfroid. 1998. Phenotypic and molecular characterization of a Brucella strain isolated from a minke whale (Balaenoptera acutorostrata). Microbiology 144:3267-3273. [DOI] [PubMed] [Google Scholar]
- 4.Cloeckaert, A., M. Grayon, and O. Grépinet. 2000. An IS711 element downstream of the bp26 gene is a specific marker of Brucella spp. isolated from marine mammals. Clin. Diagn. Lab. Immunol. 7:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloeckaert, A., M. Grayon, O. Grépinet, and K. Sidi Boumedine. 2003. Classification of Brucella strains isolated from marine mammals by infrequent restriction site PCR and development of specific PCR identification tests. Microbes Infect. 5:593-602. [DOI] [PubMed] [Google Scholar]
- 6.Cloeckaert, A., J. M. Verger, M. Grayon, and O. Grépinet. 1995. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology 141:2111-2121. [DOI] [PubMed] [Google Scholar]
- 7.Cloeckaert, A., J. M. Verger, M. Grayon, J. Y. Paquet, B. Garin-Bastuji, G. Foster, and J. Godfroid. 2001. Classification of Brucella spp. isolated from marine mammals by DNA polymorphism at the omp2 locus. Microbes Infect. 3:729-738. [DOI] [PubMed] [Google Scholar]
- 8.Dawson, C. E., E. J. Stubberfield, L. L. Perrett, A. C. King, A. M. Whatmore, J. B. Bashiruddin, J. A. Stack, and A. P. MacMillan. 2008. Phenotypic and molecular characterisation of Brucella isolates from marine mammals. BMC Microbiol. 8:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De, B. K., L. Stauffer, M. S. Koylass, S. E. Sharp, J. E. Gee, L. O. Helsel, A. G. Steigerwalt, R. Vega, T. A. Clark, M. I. Daneshvar, P. P. Wilkins, and A. M. Whatmore. 2008. Novel Brucella strain (BO1) associated with a prosthetic breast implant infection. J. Clin. Microbiol. 46:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doublet, B., G. R. Golding, M. R. Mulvey, and A. Cloeckaert. 2008. Secondary chromosomal attachment site and tandem integration of the mobilizable Salmonella genomic island 1. PLoS One 3:e2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, G., A. P. MacMillan, J. Godfroid, F. Howie, H. M. Ross, A. Cloeckaert, R. J. Reid, S. Brew, and I. A. P. Patterson. 2002. A review of Brucella sp. infection of sea mammals with particular emphasis on isolates from Scotland. Vet. Microbiol. 90:563-580. [DOI] [PubMed] [Google Scholar]
- 12.Foster, G., B. S. Osterman, J. Godfroid, I. Jacques, and A. Cloeckaert. 2007. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int. J. Syst. Evol. Microbiol. 57:2688-2693. [DOI] [PubMed] [Google Scholar]
- 13.Godfroid, J., A. Cloeckaert, J. P. Liautard, S. Kohler, D. Fretin, K. Walravens, B. Garin-Bastuji, and J. J. Letesson. 2005. From the discovery of the Malta fever's agent to the discovery of a marine mammal reservoir, brucellosis has continuously been a re-emerging zoonosis. Vet. Res. 36:313-326. [DOI] [PubMed] [Google Scholar]
- 14.Groussaud, P., S. J. Shankster, M. S. Koylass, and A. M. Whatmore. 2007. Molecular typing divides marine mammal strains of Brucella into at least three groups with distinct host preferences. J. Med. Microbiol. 56:1512-1518. [DOI] [PubMed] [Google Scholar]
- 15.Halling, S. M., and B. J. Bricker. 1994. Characterization and occurrence of two repeated palindromic DNA elements of Brucella spp.: Bru-RS1 and Bru-RS2. Mol. Microbiol. 14:681-689. [DOI] [PubMed] [Google Scholar]
- 16.Maquart, M., Y. Fardini, M. S. Zygmunt, and A. Cloeckaert. 2008. Identification of novel DNA fragments and partial sequence of a genomic island specific of Brucella pinnipedialis. Vet. Microbiol. 132:181-189. [DOI] [PubMed] [Google Scholar]
- 17.Maquart, M., P. Le Flèche, G. Foster, M. Tryland, F. Ramisse, B. Djonne, S. Al Dahouk, I. Jacques, H. Neubauer, K. Walravens, J. Godfroid, A. Cloeckaert, and G. Vergnaud. 2009. MLVA-16 typing of 295 marine mammal Brucella isolates from different animal and geographic origins identifies 7 major groups within Brucella ceti and Brucella pinnipedialis. BMC Microbiol. 9:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maquart, M., M. S. Zygmunt, and A. Cloeckaert. 2009. Marine mammal Brucella isolates with different genomic characteristics display a differential response when infecting human macrophages in culture. Microbes Infect. 11:361-366. [DOI] [PubMed] [Google Scholar]
- 19.McDonald, W. L., R. Jamaludin, G. Mackereth, M. Hansen, S. Humphrey, P. Short, T. Taylor, J. Swingler, C. E. Dawson, A. M. Whatmore, E. Stubberfield, L. L. Perrett, and G. Simmons. 2006. Characterization of a Brucella strain as a marine-mammal type despite isolation from a patient with spinal osteomyelitis in New Zealand. J. Clin. Microbiol. 44:4363-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno, E., A. Cloeckaert, and I. Moriyon. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90:209-227. [DOI] [PubMed] [Google Scholar]
- 21.Scholz, H. C., E. Hofer, G. Vergnaud, P. Le Fleche, A. M. Whatmore, S. Al Dahouk, M. Pfeffer, M. Krüger, A. Cloeckaert, and H. Tomaso. 2009. Isolation of Brucella microti from mandibular lymph nodes of red foxes, Vulpes vulpes, in Lower Austria. Vector Borne Zoonotic Dis. 9:153-156. [DOI] [PubMed] [Google Scholar]
- 22.Scholz, H. C., Z. Hubalek, J. Nesvadbova, H. Tomaso, G. Vergnaud, P. Le Flèche, A. M. Whatmore, S. Al Dahouk, M. Krüger, C. Lodri, and M. Pfeffer. 2008. Isolation of Brucella microti from soil. Emerg. Infect. Dis. 14:1316-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholz, H. C., Z. Hubalek, I. Sedlacek, G. Vergnaud, H. Tomaso, S. Al Dahouk, F. Melzer, P. Kampfer, H. Neubauer, A. Cloeckaert, M. Maquart, M. S. Zygmunt, A. Whatmore, E. Falsen, P. Bahn, C. Gollner, M. Pfeffer, B. Huber, H. J. Busse, and K. Nockler. 2008. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int. J. Syst. Evol. Microbiol. 58:375-382. [DOI] [PubMed] [Google Scholar]
- 24.Scholz, H. C., K. Nöckler, C. Göllner, P. Bahn, G. Vergnaud, H. Tomaso, S. Al Dahouk, P. Kämpfer, A. Cloeckaert, M. Maquart, M. S. Zygmunt, A. M. Whatmore, M. Pfeffer, B. Huber, H. J. Busse, and B. K. De. 2010. Brucella inopinata sp. nov., isolated from a breast implant infection. Int. J. Syst. Evol. Microbiol. 60:801-808. [DOI] [PubMed] [Google Scholar]
- 25.Sohn, A. H., W. S. Probert, C. A. Glaser, N. Gupta, A. W. Bollen, J. D. Wong, E. M. Grace, and W. C. McDonald. 2003. Human neurobrucellosis with intracerebral granuloma caused by a marine mammal Brucella spp. Emerg. Infect. Dis. 9:485-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vizcaino, N., P. Caro-Hernandez, A. Cloeckaert, and L. Fernandez-Lago. 2004. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6:821-834. [DOI] [PubMed] [Google Scholar]
- 27.Whatmore, A. M. 2009. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 9:1168-1184. [DOI] [PubMed] [Google Scholar]
- 28.Whatmore, A. M., C. E. Dawson, P. Groussaud, M. S. Koylass, A. C. King, S. J. Shankster, A. H. Sohn, W. S. Probert, and W. L. McDonald. 2008. Marine mammal Brucella genotype associated with zoonotic infection. Emerg. Infect. Dis. 14:517-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zygmunt, M. S., J. M. Blasco, J. J. Letesson, A. Cloeckaert, and I. Moriyon. 2009. DNA polymorphism analysis of Brucella lipopolysaccharide genes reveals marked differences in O-polysaccharide biosynthetic genes between smooth and rough Brucella species and novel species-specific markers. BMC Microbiol. 9:92. [DOI] [PMC free article] [PubMed] [Google Scholar]