Abstract
A C-to-T transition mutation in the neuraminidase gene from seasonal A/H1N1 causes a His-to-Tyr mutation at amino acid position 275 (H274Y, universal N2 numbering), conferring resistance against oseltamivir (Tamiflu). This mutation was first detected in clinical samples in Europe during the 2007-2008 influenza season. Viruses with this mutation reached a prevalence of ∼11% by the end of the season in North American isolates tested by the CDC. We developed a highly sensitive and specific quantitative real-time reverse transcriptase PCR assay to detect the H274Y mutation. This assay utilizes a 5′-methyl-isocytosine (isoC) residue and fluorescent reporters on genotype-specific primers. During PCR, a quencher coupled to isoguanine (isoG) is site-specifically incorporated complementary to the isoC/dye, resulting in loss of fluorescence. Optimization of primers and assay conditions produced a limit of detection of 100 gene copies per reaction for both wild-type and H274Y genotypes. In samples with mixed populations, it can reliably detect as little as a 1% wild-type or 0.1% H274Y component. This high sensitivity makes the assay usable on samples with viral loads too low for dideoxy or pyrosequencing analysis. Additionally, the assay distinguishes seasonal A/H1N1 from A/H3N2, influenza B, or 2009 pandemic A/H1N1, making it useful for influenza virus subtyping as well as for drug resistance detection. We probed seasonal A/H1N1 samples from the 2005-2006, 2006-2007, and 2007-2008 influenza seasons. Data from the new assay closely matched available drug resistance genotype data previously determined by dideoxy sequencing. The H274Y mutation was only found in samples from the 2007-2008 season.
Influenza viruses cause considerable annual worldwide morbidity and mortality. In the United States alone, greater than 200,000 persons are hospitalized each year due to influenza and approximately 36,000 die from influenza-related disease (16). Vaccination is considered the first and best defense against influenza. However, the efficacy of protection conferred by annual vaccination can be limited by the strength of the antigenic match of vaccine strains to the circulating strains. In addition, herd (community-level) immunity is limited by less than 100% vaccine coverage. These circumstances allow influenza to easily spread among susceptible persons and through populations. Antiviral drug treatment and prophylaxis are additional and necessary modes of defense against morbidity, mortality, and further spread of the virus.
Widespread resistance against the adamantane class of drugs among A/H3N2 viruses, beginning in the 2003-2004 season, prompted public health officials in the United States to recommend against the use of these drugs during the 2005-2006 season in favor of neuraminidase inhibitors (NAIs) (3). First approved for clinical use in the United States in 1999 (17), NAIs target the viral surface protein neuraminidase and are effective against both influenza A and B. There are currently two FDA-approved drugs in this class: oseltamivir (Tamiflu; Roche) and zanamivir (Relenza; GlaxoSmithKline). When given within the first 48 h of a patient becoming symptomatic, NAIs have been shown to reduce the duration and severity of influenza illness in both adults and children (11, 25).
In January 2008, nine European countries reported seasonal influenza A/H1N1 isolates showing resistance to oseltamivir (13, 25). In the following months, additional countries, including the United States, reported oseltamivir-resistant influenza viruses (24). These findings were alarming because drug resistance testing during the previous influenza season (2006-2007) had revealed no oseltamivir resistance in Europe (13). Fewer than 1% of North American seasonal A/H1N1 viruses from the same time period tested by the Centers for Disease Control and Prevention (CDC) showed resistance (4). In addition, during clinical trials with oseltamivir, shedding of drug-resistant virus was noted at a frequency of only 4% in children and in ≤1% in adults (28). Analysis revealed that all resistant viruses were seasonal A/H1N1, carrying the same C→T transition mutation in the neuraminidase gene, with a resulting histidine-to-tyrosine change at amino acid position 275 (“H274Y” in universal N2 numbering). By the end of the 2007-2008 season, the CDC reported this mutation in 111 of 1,020 tested seasonal A/H1N1 isolates; 4 were found in New York State (4). Oseltamivir-resistant seasonal A/H1N1 spread extensively, becoming the dominant variant in Oceania and Southeast Asia in May 2008 (10) and with virtually all seasonal A/H1N1 strains possessing the H274Y mutation during the 2008-2009 influenza season in the United States (5).
The need for continual monitoring for antiviral drug resistance among influenza viruses is highlighted by several factors. Use of antiviral medications as a treatment and prophylactic is an integral component of infection control during influenza outbreaks. With the advent of adamantane resistance, use of the neuraminidase inhibitors, most commonly oseltamivir, has risen dramatically. In addition, oseltamivir is a major component of influenza pandemic preparedness stockpiles in the United States. Continual improvement of methods for the survey and detection of oseltamivir resistance in all influenza isolates is imperative to ensure the lasting viability of this drug as an antiviral treatment.
Laboratory testing for NAI resistance can be performed via a phenotypic neuraminidase inhibition assay, giving a 50% inhibitory concentration (IC50) for a drug of interest. Laboratories can also use nucleic acid testing such as real-time quantitative reverse transcriptase (RT)-PCR as well as dideoxy sequencing and pyrosequencing methods. Sequencing methods provide detailed information about gene regions of interest and enable the identification of new mutations that may have an effect on drug resistance. Pyrosequencing, in particular, has been reported to be capable of detecting minor populations at concentrations as low as 10% within mixtures of neuraminidase gene targets (7, 8). However, both dideoxy sequencing and pyrosequencing require additional processing steps after an initial PCR, as well as specialized sequencing instruments, software, and training. Real-time quantitative RT-PCR provides a rapid, highly sensitive, and specific alternative to sequencing. This method is also capable of detecting targets in samples with viral loads too low for detection in sequencing assays. Furthermore, real-time platforms are available from multiple manufacturers and are more commonplace in clinical laboratories than sequencing equipment.
Here, we describe the development of a novel, highly sensitive real-time quantitative RT-PCR assay for the detection of H274Y in seasonal A/H1N1 influenza. The assay utilizes MultiCode technology, a system that employs an additional base pair set of 5′-methyl-isocytosine (isoC) and isoguanine (isoG) (12, 22). An isoC and isoG can only base pair to one another, and neither an isoC-G nor a C-isoG bond can form. In real-time RT-PCR applications, an isoC base is included at the 5′ end of a primer, directly linked to a fluorophore, such as 6-carboxyfluorescein (FAM) or hexachlorofluorescein (HEX) (Fig. 1). During PCR, a quencher (Dabcyl) directly linked to an isoG nucleotide is site specifically incorporated into the PCR product complementary to the isoC-fluorophore, resulting in a detectable decrease in fluorescence (23). Because multiple fluorophores can be used in the same reaction, multiplexed assays can be designed. In addition, because the fluorophores are not cleaved from the oligonucleotides, postamplification melt curve analysis is possible, facilitating analyses of assay specificity. This PCR technology has been successfully employed as part of a PCR panel targeting several respiratory viruses (15), in assays for other viruses including hepatitis C (20) and HIV (19, 26, 27), and in the detection of anthrax-related toxin genes (18).
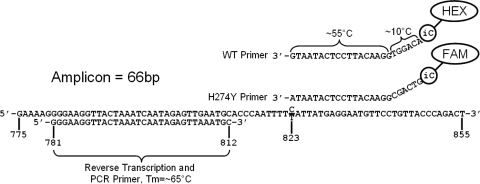
FIG. 1.
Primer alignment to target sequence. Primers were designed against a consensus sequence of N1 sequences from seasonal A/H1N1-positive clinical samples submitted for testing to the Wadsworth Center between 2004 and 2008. The genotype-specific primers are designed in two sections: one section anneals to the coding section of the N1 gene, and there is a noncoding tail which includes an isoC residue and a fluorophore. The WT-targeted primer is labeled in HEX, while the mutant-targeted primer is labeled in FAM. The nucleotide at the 3′ end of each of the genotype-specific primers targets the SNP responsible for coding for His or Tyr. A specific primer is used for reverse transcription. The same primer is paired against the genotype-specific primers during PCR. This primer system creates an amplicon of 66 bp.
This new assay was developed with analyses for both mutation detection and facilitation of resistance evolution in mind. To that end, we developed a novel multiplexed assay with high sensitivity and specificity and with a higher sensitivity for detecting a minor component mutant population compared to that detectable by pyrosequencing. The assay was applied to a collection of A/H1N1 influenza-positive samples collected between 2005 to 2008, in order to determine whether the H274Y mutation was present in clinical samples prior to its apparent emergence during the 2007-2008 season.
MATERIALS AND METHODS
Specimens and standard curve construction.
A total of 85 primary specimens and isolates used in this study were collected in New York State during the 2005-2006, 2006-2007, or 2007-2008 influenza season as part of normal surveillance. Only samples that were previously characterized as seasonal A/H1N1 influenza virus by the Wadsworth Center Virus Reference and Surveillance Laboratory using real-time reverse transcriptase PCR were included in the present study. For a subset of these samples, the neuraminidase gene had already been characterized by dideoxy sequencing (14).
Cultured influenza isolates, used in assay development and validation (see below), were maintained on rhesus monkey kidney cells (ViroMed Laboratories, Minnetonka, MN, or Diagnostic Hybrids, Athens, OH). Neuraminidase inhibitor-resistant influenza was maintained with oseltamivir carboxylate at a final concentration of 10 μg/ml (25.9 μM; kindly provided by Roche). Virus cultures were maintained until cytopathic effect was evident throughout the culture and then harvested.
Nucleic acids were extracted using the QIAcube (Qiagen USA, Valencia, CA) automated extractor with the Qiagen QIAamp viral RNA extraction kit according to manufacturer's instructions. Influenza A/H1N1 isolates to be used for controls and standard curve construction were first quantified in a separate quantitative real-time RT-PCR assay targeting the matrix gene segment. Standard curves were constructed by testing 10-fold serial dilutions of target, containing between 108 and 1 gene copy per reaction. Standard curve reactions were applied to the multiplexed assay in duplicate, and each test was performed at least three times.
Primer design.
Primers were designed against a database of compiled seasonal N1 gene sequences derived from New York State samples spanning from 2004 to 2008 and with the HYTHER online nucleic acid hybridization program, accessible at http://ozone3.chem.wayne.edu. The resulting primer designs used in the final assay are shown in Fig. 1. A specific primer was used for reverse transcription. The same primer was also used during PCR and was paired with genotype-specific primers. The primer targeted against wild-type (WT) N1 was labeled with HEX, while the H274Y-targeted primer was labeled with FAM. PCRs with these primers give a 66-bp amplicon.
Real-time reverse transcriptase PCR.
The real-time quantitative reverse transcriptase PCR was performed with the Plexor one-step RT-PCR system from Promega (Promega, Madison, WI). The reverse transcription step was performed in a 20-μl volume including Plexor system components, 5 μl target, and primer (Fig. 1) to a final concentration of 0.4 μM, at 45°C for 15 min, followed by 95°C for 2.5 min. After reverse transcription, FAM (0.4 μM final concentration)- and HEX (0.15 μM final concentration)-labeled primers were added, giving a final PCR volume of 25 μl. All PCRs were performed on the Bio-Rad iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA). The cycling conditions are as follows: 2.5 min at 95°C, 2 cycles of 5 s at 95°C and 35 s at 53°C, and 45 cycles of 5 s at 95°C and 35 s at 63°C. PCR was followed by melt curve analysis ranging from 55°C to 95°C, with 30 s per step with a 1°C increase per step. The iQ5 real-time PCR software is designed to interpret and display data to the user from reactions that show increases in fluorescence over time. Because the PCR system used in this study is based upon decreases in fluorescence over time, data from real-time PCR runs were imported into Promega Plexor software, version 1.1.4, for data analysis. When using MultiCode PCR chemistry, fluorescence is reduced with target amplification and is reported in relative fluorescence units (RFU). In the MultiCode PCR developed in this study for resistance mutation analysis, to define a sample as positive for the target, two criteria must be met: (i) the fluorescence trace on the amplification plot must cross the (user-defined) threshold line (giving the sample a defined threshold cycle [CT] value), and (ii) the corresponding trace on the melt curve graph must show a peak at 73 ± 1°C. As with all real-time PCR assays, CT values are inversely proportional to the target concentration.
Specificity.
Specificity was tested by application of the assay to a panel of nonseasonal A/H1N1 viruses, including the following: influenza B; influenza A/H3N2; 2009 pandemic A/H1N1; adenoviruses 3, 4, 5, and 8; coxsackieviruses A9, B3, B4, and B5; human metapneumovirus 1B and 2B; rhinovirus 1A; measles virus; mumps virus; severe acute respiratory syndrome (SARS) coronavirus; coronavirus 229E; lymphocytic choriomeningitis virus; enteroviruses 68 and 71; and echoviruses 6, 9, 11, and 30. Each virus in the panel was tested at a concentration of at least 106 genome copies per reaction, in duplicate. Specificity experiments were performed at least three times, except for coxsackieviruses A9 and B5, coronavirus 229E, human metapneumovirus 2B, and lymphocytic choriomeningitis virus, which were tested only twice. Seasonal A/H1N1 virus containing WT or H274Y neuraminidase 1 genes was used as the positive control.
RESULTS
We utilized MultiCode technology to detect and quantify seasonal A/H1N1 clinical samples containing populations of WT or H274Y mutant neuraminidase genes. Primers were designed against both the WT and mutant genotypes, such that the nucleotide at the 3′ end of each primer targets the single nucleotide polymorphism (SNP) coding for either His or Tyr. The primers are designed in two sections: an annealing segment that binds the coding region of the gene and a noncoding tail that includes the isoC nucleotide and fluorophore. As shown in Fig. 1, the neuraminidase-specific annealing region of each primer has an annealing temperature of approximately 55°C. The 6-base noncoding tails increase the overall annealing temperature by approximately 10°C, giving the primers a final melting temperature (Tm) of approximately 65°C. The WT- and H274Y-specific primers possess different tail sequences and fluorophores. This overall primer design allows a two-stage PCR cycling scheme (see Materials and Methods), which begins with a lower-specificity cycling Tm of 55°C, followed by additional cycles with higher specificity annealing/extension at 65°C.
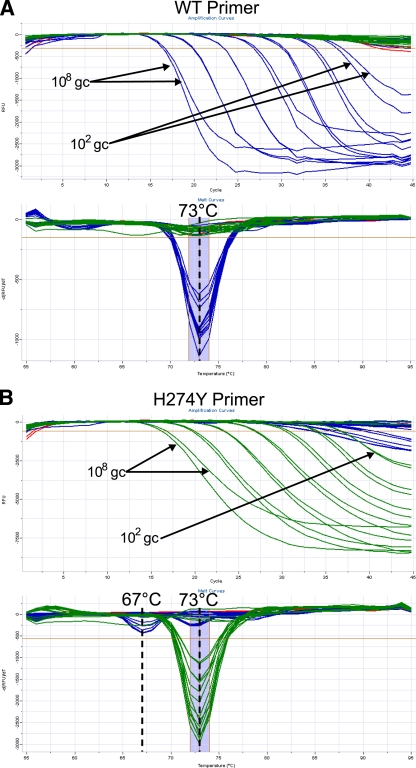
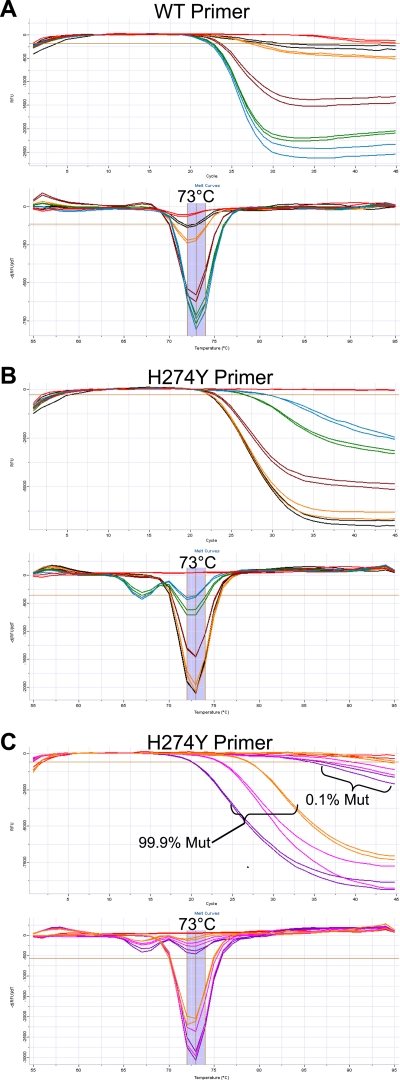
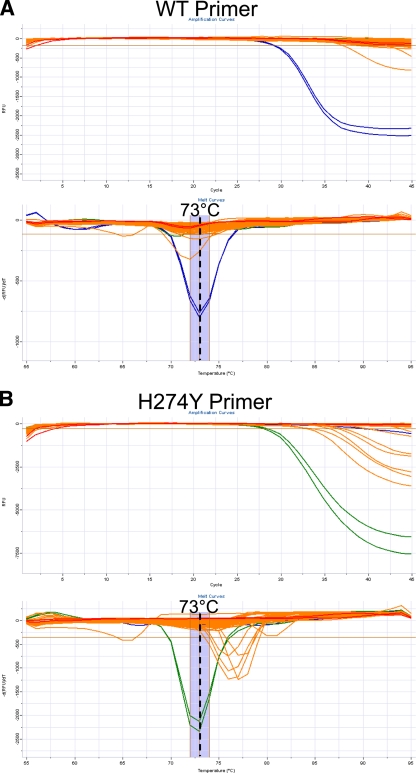
The assay employs a specific reverse transcription primer, targeted against the negative-stranded viral RNA. The same primer is also used during PCR, paired against both the WT- and H274Y-specific primers. These primers yield a 66-bp amplicon on a seasonal N1 target. Melt curve analysis of amplicons generated with either the WT- or H274Y-targeted primer gives a product with a Tm of 73°C, as shown in Fig. 2.
FIG. 2.
Limit of detection. The lower limit of detection was determined by application of the assay to standard curves from quantified nucleic acid extracts to give between 108 and 1 gene copy per reaction. Standard curve samples were derived from cultures containing seasonal A/H1N1 cultures with either pure WT or pure H274Y neuraminidase genes. The limit of detection for both the WT- and H274Y-targeted primers is 100 gene copies (gc), using the reaction conditions stated in Materials and Methods. In the amplification plots, data from a sample positive for the N1 target is shown by a loss of fluorescence (i.e., decreasing relative fluorescence units [RFU]) over increasing PCR cycles. In melt curve analysis, a sample positive for an N1 target gives a melt peak at 73°C. Cross-reactivity of H274Y primer against a WT target gives a secondary melt peak at 67°C. (A) WT primer; (B) H274Y primer. Blue, amplification on a WT target; green, amplification on a mutant target; red, no-template control.
Limit of detection.
Optimization of primer concentrations and cycling conditions under multiplexed conditions resulted in a lower limit of detection of 100 WT or H274Y gene copies per reaction. The limit of detection was determined using standard curves constructed from quantified influenza virus extracts from cultured isolates containing pure populations of either WT neuraminidase or neuraminidase with the H274Y mutation, as shown in Fig. 2. Some cross-reactivity of the H274Y primer with WT template was observed at concentrations of ≥105 WT gene copies. However, such cross-reactivity is distinguishable due to the presence of a secondary melt peak at 67°C, as shown in Fig. 2. When such cross-reactivity occurs on samples with a pure WT neuraminidase genotype, the melt peak at 67°C is stronger than the peak at 73°C.
Detection of mixed populations.
Due to multiplexing, the assay is capable of detecting mixed populations within the same reaction. To test assay sensitivity for mixed populations, quantified WT and H274Y viral nucleic acid extracts were combined at 1%, 5%, 50%, 95%, and 99% WT mixtures, with a total 106 gene copies per reaction. Additionally, 0.1% and 99.9% WT mixtures were created, with 105, 106, and 107 total gene copies. The duplexed assay is capable of detecting a WT component as low as 1% (104 gene copies) in an otherwise mutant background (9.9 × 105 gene copies), as shown in Fig. 3 A. As demonstrated in Fig. 3C, the assay is also capable of detecting a mutant population as low as 0.1% in an otherwise WT background containing 107, 106, or 105 total gene copies.
FIG. 3.
Mixed-sample analysis. For panels A and B, nucleic acid extracts from pure populations of WT and H274Y viruses were mixed in ratios of 1%, 5%, 50%, 95%, and 100% WT with 106 gene copies in total and applied to the multiplexed assay. Black, 1% WT; orange, 5% WT; brown, 50% WT; green, 95% WT; blue, 99% WT; red, no-template control. In panel C, nucleic acid extracts from pure populations of WT and H274Y viruses were mixed in a ratio of 99.9% major component to 0.1% minor component genotype with 107, 106, or 105 total gene copies (gc) applied to the multiplexed assay. Purple, 107 gc total; pink, 106 gc total; orange, 105 gc total; red, no-template control. Data from these plots are interpreted as described in the legend to Fig. 2.
Assay specificity.
To test assay specificity for seasonal A/H1N1, the assay was applied to a panel of nucleic acid extracts from influenza and noninfluenza viruses. Figure 4 shows negative signal results from a representative experiment. As can be seen in this figure, there is a limited amount of nonspecific amplification for nonseasonal A/H1N1 samples, including those with adenoviruses 4 and 8, coxsackievirus B3, enterovirus 68, and A/H3N2 influenza virus. However, these nonspecific amplifications are easily distinguished from specific PCR products generated on an N1 template, by melt curve analysis. The primers used in this assay do not strongly cross-react with the 2009 pandemic A/H1N1 target, which can be attributed to mismatches between the primers and the 2009 pandemic virus N1 target. Analysis of the primer sequences reveals 11 mismatches between the reverse transcription primer and the 2009 pandemic virus N1 sequence and 2 mismatches for each of the genotype-specific primers.
FIG. 4.
Specificity analysis. Nucleic acid extracts from a panel of viruses were tested against the assay. A representative experiment is pictured above. The following viruses are represented in these plots: WT and H274Y seasonal A/H1N1; seasonal A/H3N2; influenza B; 2009 pandemic A/H1N1; adenoviruses 3, 4, 5, and 8; coronavirus 229E; SARS coronavirus; coxsackieviruses A9, B3, B4, and B5; echoviruses 6, 9, 11, and 30; enteroviruses 68 and 71; human metapneumoviruses 1B and 2B; lymphocytic choriomeningitis virus; measles virus; mumps virus; and rhinovirus 1A. Blue, WT (positive) control (104 gene copies [gc]); green, H274Y (positive) control (104 gc); orange, specificity test sample; red, no-template control. Data from these plots is interpreted as described in the legend to Fig. 2.
Analysis of archived influenza samples.
The H274Y mutation was first detected during the 2007-2008 influenza season, but we were interested in investigating whether the mutation had gone undetected in seasonal A/H1N1 samples collected prior to the 2007-2008 season. A total of 85 archived respiratory samples positive for seasonal A/H1N1 were identified for testing with the new assay. These samples had been submitted to the Wadsworth Center for testing during the 2005-2006, 2006-2007, and 2007-2008 influenza seasons. Available dideoxy sequencing data on the N1 gene sequences from 2006-2007 (17 specimens) and 2007-2008 (44 specimens) seasonal A/H1N1-positive samples (14) were used as a basis for comparison to the data generated from the MultiCode-based assay.
Results from the new assay indicated that the 44 specimens from the 2007-2008 season contained either a 100% WT or 100% H274Y neuraminidase genotype. As shown in Table 1, the data generated by the new assay on samples from the 2007-2008 season closely matched available sequencing data. Importantly, the four samples that had previously tested positive for the H274Y oseltamivir resistance mutation by dideoxy sequencing also tested positive for the mutation by the new assay. The new assay was additionally able to identify as WT one sample that dideoxy sequencing had failed to characterize at all. A total of 41 additional seasonal A/H1N1-positive respiratory specimens from the 2005-2006 (4 specimens) and 2006-2007 (37 specimens) seasons were retrieved from sample archives at the Wadsworth Center. The new assay detected only WT neuraminidase in all samples from these seasons. No samples of mixed genotype were found in samples from any of the three seasons.
TABLE 1.
Comparative results between MultiCode-based testing and dideoxy sequencing of seasonal A/H1N1 samples from 2005 to 2008
| Period and test | Total no. of samples | No. (%) of samples |
||
|---|---|---|---|---|
| WT | H274Y | No result/failure | ||
| 2005-2006 | 4 | |||
| MultiCode | 4 | 4b (100) | 0 | 0 |
| Sequencing | NAc | |||
| 2006-2007 | 37 | |||
| MultiCode | 37 | 19b + 17 (97) | 0 | 1b (3) |
| Sequencinga | 17 | 17 (100) | 0 | 0 |
| 2007-2008 | 44 | |||
| MultiCode | 44 | 39 (89) | 4 (9) | 1 (2) |
| Sequencinga | 44 | 38 (86) | 4 (9) | 2 (5) |
Sequencing data from Laplante et al.(14).
Neuraminidase from these samples was not previously characterized by dideoxy sequencing.
NA, not applicable.
DISCUSSION
The adamantanes have been approved as antiinfluenza drugs since 1966 (1) and were commonly prescribed prior to recommendations against their use during the 2005-2006 season. Amino acid changes in the M2 protein provide resistance to these drugs and have been reported to emerge within as little as 6 days after the initiation of rimantadine prophylaxis (2, 9, 21). Resistance to the NAI oseltamivir was first detected in the 2007-2008 influenza season among seasonal A/H1N1 viruses, all with the H274Y neuraminidase mutation. Viruses possessing the H274Y mutation spread rapidly, even in the absence of drug pressure, and oseltamivir-resistant seasonal A/H1N1 became the dominant virus of this subtype during the 2008-2009 season. In addition, the H274Y mutation in seasonal A/H1N1 is the equivalent of the H275Y mutation detected in some 2009 pandemic A/H1N1 viruses.
Common molecular methodologies for identifying influenza virus and detecting drug-resistant strains include dideoxy sequencing, pyrosequencing, and quantitative real-time PCR. Used as part of quantitative reverse transcriptase PCR, the methodology employed here has several advantages that would make it attractive to hospital and public health laboratories. MultiCode technology permits multiplexing as well as melt curve analysis in real-time PCR, enabling testing of smaller sample volumes, and a high level of discrimination between specific and nonspecific amplification products. These advantages make isoC/isoG-based technology highly effective at detecting specified targets and at quantifying mutant subpopulations based on discrimination of a single nucleotide polymorphism. As with all real-time PCR systems, this methodology is best suited to laboratory facilities possessing real-time equipment and having staff sufficiently trained and skilled in molecular biology techniques. As such, this test system is not appropriate for use in physician's office laboratories.
For the detection of viral populations containing the H274Y mutation and for discrimination of them from WT populations, the assay has advantages over sequencing methods. Primers and cycling conditions were optimized to achieve a limit of detection of 100 gene copies per reaction, making the assay usable on samples with viral loads too low to yield results by either dideoxy sequencing or pyrosequencing. In samples containing both WT and mutant neuraminidase gene targets, the assay has the ability to detect as low as a 1% WT component or a 0.1% H274Y component. This ability to reliably detect mixed populations at low percentages is important in the monitoring of patients for emergent drug resistance during treatment. Due to its higher sensitivity compared to sequencing methods, the assay presented here would be a preferable method for detecting emergent drug resistance, as current pyrosequencing methods for the H274Y mutation are reported to only reliably detect minor populations down to approximately a 10% component (7, 8). Specificity testing demonstrated the ability to distinguish seasonal A/H1N1 from seasonal A/H3N2, influenza B, and pandemic 2009 A/H1N1 viruses. While this assay was designed to detect the H274Y oseltamivir-resistance mutation, its level of specificity also makes the new assay useful as an influenza subtyping test. Additionally, since the assay requires no additional sample processing or manipulation after the PCR step, results are obtained faster than dideoxy or pyrosequencing. This assay is also more deployable than either sequencing method, due to the ready availability of real-time PCR instrumentation in clinical laboratory settings.
Testing of New York State influenza A/H1N1-positive primary samples from the 2007-2008 season confirmed the presence of the H274Y neuraminidase mutation in four samples and only the WT genotype in all other samples from this season. These real-time RT-PCR results matched all available dideoxy sequencing data, lending validity to the data generated by the new assay. The ability of the new assay to characterize one additional sample as WT demonstrates its increased sensitivity compared to that of dideoxy sequencing. The original characterization of this specimen in 2008 indicated the specimen had an extremely low viral load. Previous attempts to amplify this sample by conventional PCR for use in dideoxy sequencing failed, producing no visible band in gel analysis. Samples tested by the new assay from the 2005-2006 and 2006-2007 seasons (including 24 samples not previously characterized by dideoxy sequencing) did not reveal the presence of H274Y, in agreement with the documented emergence of this mutation in the 2007-2008 influenza season. These data are consistent with several possibilities: (i) only seasonal A/H1N1 viruses with WT neuraminidase were circulating in New York State prior to the 2007-2008 season, (ii) the H274Y mutation was present in specimens at a level lower than the 0.1% detectable by the new assay, or (iii) the H274Y mutation was present in seasonal A/H1N1 influenza prior to the 2007-2008 season but not at a frequency that would be detected without a larger sample set than that available for this study.
Intensive and effective public health surveillance for drug resistance in influenza is integral to the maintenance of neuraminidase inhibitors as useful treatment and prophylactic agents. While oseltamivir is not currently effective against seasonal A/H1N1, it remains effective against A/H3N2, influenza B, and 2009 pandemic A/H1N1. The WHO has identified 285 cases of oseltamivir-resistant 2009 pandemic A/H1N1, with 64 cases identified by the CDC (6, 29). Viruses from these oseltamivir-resistant cases contain the H275Y neuraminidase mutation, and approximately 8% of the global cases have had no drug exposure or are suspected to have contracted resistant virus by person-to-person transmission. Further intensified surveillance efforts and improved detection methods for drug resistance will be needed to ensure that resistance can be identified in a timely manner and rapidly contained.
Acknowledgments
We thank Amy Dean, Sarah DuVall, Meghan Fuschino, Sara Griesemer, Daryl Lamson, Jennifer Laplante, and Kim Rush-Wilson at the Wadsworth Center and Scott Johnson at EraGen Biosciences for technical assistance and helpful discussions, as well as Gino Battaglioli at the Wadsworth Center for critical readings of the manuscript.
This work was supported in part by NIH grant T32 AI05542903A1, CDC Cooperative Agreement no. U90/CCU216988, and CDC Cooperative Agreement no. U50/CCU223671.
This work is solely the responsibility of the authors and does not necessarily represent the official views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Belshe, R. B., M. H. Smith, C. B. Hall, R. Betts, and A. J. Hay. 1988. Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J. Virol. 62:1508-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bright, R. A., D. K. Shay, B. Shu, N. J. Cox, and A. I. Klimov. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 295:891-894. [DOI] [PubMed] [Google Scholar]
- 3.Bright, R. A., D. Shay, J. Bresee, A. Klimov, N. Cox, and J. Ortiz. 2006. High levels of adamantane resistance among influenza A (H3N2) viruses and interim guidelines for use of antiviral agents—United States, 2005-2006 influenza season. MMWR Morb. Mortal. Wkly. Rep. 55:44-46. [PubMed] [Google Scholar]
- 4.CDC. 2008. 2007-08 U.S. influenza season summary. Centers for Disease Control and Prevention, Atlanta, GA.
- 5.CDC. 2009. 2008-2009 influenza season week 39 ending October 3, 2009. Centers for Disease Control and Prevention, Atlanta, GA.
- 6.CDC. 2010. 2009-2010 influenza season week 14 ending April 10, 2010. Centers for Disease Control and Prevention, Atlanta, GA.
- 7.Deyde, V. M., and L. V. Gubareva. 2009. Influenza genome analysis using pyrosequencing method: current applications for a moving target. Expert Rev. Mol. Diagn. 9:493-509. [DOI] [PubMed] [Google Scholar]
- 8.Deyde, V. M., M. Okomo-Adhiambo, T. G. Sheu, T. R. Wallis, A. Fry, N. Dharan, A. I. Klimov, and L. V. Gubareva. 2009. Pyrosequencing as a tool to detect molecular markers of resistance to neuraminidase inhibitors in seasonal influenza A viruses. Antiviral Res. 81:16-24. [DOI] [PubMed] [Google Scholar]
- 9.Drinka, P. J., and T. Haupt. 2007. Emergence of rimantadine-resistant virus within 6 days of starting rimantadine prophylaxis with oseltamivir treatment of symptomatic cases. J. Am. Geriatr. Soc. 55:923-926. [DOI] [PubMed] [Google Scholar]
- 10.Hurt, A. C., J. Ernest, Y. M. Deng, P. Iannello, T. G. Besselaar, C. Birch, P. Buchy, M. Chittanganpitch, S. C. Chiu, D. Dwyer, A. Guigon, B. Harrower, I. P. Kei, T. Kok, C. Lin, K. McPhie, A. Mohd, R. Olveda, T. Panayotou, W. Rawlinson, L. Scott, D. Smith, H. D'Souza, N. Komadina, R. Shaw, A. Kelso, and I. G. Barr. 2009. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 83:90-93. [DOI] [PubMed] [Google Scholar]
- 11.Jefferson, T., M. Jones, P. Doshi, and C. Del Mar. 2009. Neuraminidase inhibitors for preventing and treating influenza in health adults: systematic review and meta-analysis. BMJ 339:b5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, S. C., C. B. Sherrill, D. J. Marshall, M. J. Moser, and J. R. Prudent. 2004. A third base pair for the polymerase chain reaction: inserting isoC and isoG. Nucleic Acids Res. 32:1937-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lackenby, A., O. Hungnes, S. G. Dudman, A. Meijer, W. J. Paget, A. J. Hay, and M. C. Zambon. 2008. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill. 13:8026. [DOI] [PubMed] [Google Scholar]
- 14.Laplante, J. M., S. A. Marshall, M. Shudt, T. T. Van, E. S. Reisdorf, L. A. Mingle, P. A. Shult, and K. St. George. 2009. Influenza antiviral resistance testing in New York and Wisconsin, 2006-2008: methodology and surveillance data. J. Clin. Microbiol. 47:1372-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall, D. J., E. Reisdorf, G. Harms, E. Beaty, M. J. Moser, W. M. Lee, J. E. Gern, F. S. Nolte, P. Shult, and J. R. Prudent. 2007. Evaluation of a multiplexed PCR assay for detection of respiratory viral pathogens in a public health laboratory setting. J. Clin. Microbiol. 45:3875-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monto, A. S. 2009. The risk of seasonal and pandemic influenza: prospects for control. Clin. Infect. Dis. 1:S20-S25. [DOI] [PubMed] [Google Scholar]
- 17.Monto, A. S., J. L. M. McKimm-Breschkin, C. Macken, A. W. Hampson, A. Hay, A. Klimov, M. Tashiro, R. G. Webster, M. Aymard, F. G. Hayden, and M. Zambon. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 50:2395-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moser, M. J., D. R. Christensen, D. Norwood, and J. R. Prudent. 2006. Multiplexed detection of anthrax-related toxin genes. J. Mol. Diagn. 8:89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moser, M. J., M. Ruckstuhl, C. A. Larsen, A. J. Swearingen, M. Kozlowski, L. Bassit, P. L. Sharma, R. F. Schinazi, and J. R. Prudent. 2005. quantifying mixed populations of drug-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 49:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulligan, E. K., J. J. Germer, M. Q. Arens, K. L. D'Amore, A. Di Bisceglie, N. A. Ledeboer, M. J. Moser, A. C. Newman, A. K. O'Guin, P. D. Olivo, D. S. Podzorski, K. A. Vaughan, J. D. Yao, S. A. Elagin, and S. C. Johnson. 2009. Detection and quantification of hepatitis C (HCV) by MultiCode-RTx real-time PCR targeting the HCV 3′ untranslated region. J. Clin. Microbiol. 47:2635-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson, M. I., L. Simonsen, C. Viboud, M. A. Miller, and E. C. Holmes. 2009. The origin and global emergence of adamantane resistant A/H3N2 influenza viruses. Virology 388:270-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prudent, J. R. 2006. Using expanded genetic alphabets to simplify high-throughput genetic testing. Expert Rev. Mol. Diagn. 6:245-252. [DOI] [PubMed] [Google Scholar]
- 23.Sherrill, C. B., D. J. Marshall, M. J. Moser, C. A. Larsen, L. Daude-Snow, and J. R. Prudent. 2004. Nucleic acid analysis using an expanded genetic alphabet to quench fluorescence. J. Am. Chem. Soc. 126:4550-4556. [DOI] [PubMed] [Google Scholar]
- 24.Sheu, T. G., V. M. Deyde, M. Okomo-Adhiambo, R. J. Garten, X. Xu, R. A. Bright, E. N. Butler, T. R. Wallis, A. I. Klimov, and L. V. Gubareva. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004-2008. Antimicrob. Agents Chemother. 52:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shun-Shin, M., M. Thompson, C. Heneghan, R. Perera, A. Harnden, and D. Mant. 2009. Neuraminidase inhibitors for treatment and prophylaxis in children: systematic review and meta-analysis of randomised controlled trials. BMJ 339:b3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svarovskaia, E. S., J. Y. Feng, N. A. Margot, F. Myrick, D. Goodman, J. K. Ly, K. L. White, N. Kutty, R. Wang, K. Borroto-Esoda, and M. D. Miller. 2008. The A62V and S68G mutations in HIV-1 reverse transcriptase partially restore the replication defect associated with the K65R mutation. J. Acquir. Immune Defic. Syndr. 48:428-436. [DOI] [PubMed] [Google Scholar]
- 27.Svarovskaia, E. S., M. J. Moser, A. S. Bae, J. R. Prudent, M. D. Miller, and K. Borroto-Esoda. 2006. MultiCode-RTx real-time PCR system for detection of subpopulations of K65R human immunodeficiency virus type 1 reverse transcriptase mutant viruses in clinical samples. J. Clin. Microbiol. 44:4237-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward, P., I. Small, J. Smith, P. Suter, and R. Dutkowski. 2005. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J. Antimicrob. Chemother. 55:i5-i21. [DOI] [PubMed] [Google Scholar]
- 29.WHO. 2010. Weekly update on oseltamivir resistance to pandemic influenza A (H1N1) 2009 viruses—April 14, 2010. World Health Organization, Geneva, Switzerland.